Abstract
Background and Objectives: With the growing incidence and disability associated with myocardial infarction (MI), there is an increasing focus on cardiac rehabilitation post-MI. Kuanxiongzhuyu decoction (KXZY), a traditional Chinese herbal formula, has been used in the rehabilitation of patients after MI. However, the chemical composition, protective effects, and underlying mechanism of KXZY remain unclear. Materials and Methods: In this study, the compounds in KXZY were identified using a high-performance liquid chromatography-mass spectrometry (HPLC-MS) analytical method. Based on the compounds identified in the KXZY, we predictively selected the potential targets of MI and then constructed a protein–protein interaction (PPI) network to identify the key targets. Furthermore, the DAVID database was used for the GO and KEGG analyses, and molecular docking was used to verify the key targets. Finally, the cardioprotective effects and mechanism of KXZY were investigated in post-MI mice. Results: A total of 193 chemical compounds of KXZY were identified by HPLC-MS. In total, 228 potential targets were obtained by the prediction analysis. The functional enrichment studies and PPI network showed that the targets were largely associated with AKT-pathway-related apoptosis. The molecular docking verified that isoguanosine and adenosine exhibited excellent binding to the AKT. In vivo, KXZY significantly alleviated cardiac dysfunction and suppressed AKT phosphorylation. Furthermore, KXZY significantly increased the expression of the antiapoptotic proteins Bcl-2 and Bcl-xl and decreased the expression of the proapoptotic protein BAD. Conclusions: In conclusion, the network pharmacological and experimental evidence suggests that KXZY manifests anti-cardiac dysfunction behavior by alleviating cardiomyocyte apoptosis via the AKT pathway in MI and, thus, holds promising therapeutic potential.
1. Background
Myocardial infarction (MI) is currently a major cause of morbidity and mortality worldwide, posing a significant public health challenge with enormous medical and societal consequences [1]. While thrombolysis, percutaneous coronary intervention, and coronary-artery-bypass grafts remain the most commonly used and effective treatments for MI, they are unable to prevent the irreversible damage to myocardial cells and the series of repair and wound-healing responses following MI. Excessive and extended remodeling of the left ventricle can often result in further deterioration of cardiac function and ultimately lead to heart failure, with a 5-year mortality rate approaching 50% [2]. Consequently, new and validated treatment agents or targets are urgently needed to alleviate the cardiomyocyte injury, prevent the deterioration of irreversible myocardial function, and improve prognoses for patients post-MI.
The use of natural and herbal remedies to treat human problems has been well known for a significant length of time. Because of its holistic and systematic nature, traditional Chinese medicine (TCM) has recently attracted global attention [3]. Thus, herbal remedies are valuable in the search for novel agents to treat myocardial damage post-MI. In the clinic, post-MI patients are mainly associated with the Qi-deficiency-and-blood-stasis syndrome in TCM. This Qi-deficiency-and-blood-stasis syndrome is one of the most common syndromes in TCM, characterized by chest distress, shortness of breath, heart palpitations, tiredness, rapid pulse, etc. Its pathogenic process resembles that of restricted blood flow and cardiac dysfunction, as revealed by modern science [4]. Thus, Qi-supplementation and blood-stimulation therapies are widely used in TCM. In clinical settings, Kuanxiongzhuyu decoction (KXZY), a classical Chinese herbal formula derived from Siwu decoction, showed effectiveness in treating MI patients. As one of the characteristic treatments for the syndrome of Qi deficiency and blood stasis, KXZY can enhance blood regeneration, promote blood circulation, and eliminate blood stasis. Modern medicine has confirmed that several of these herb-extract mixtures have lipid-reduction, anti-inflammatory, immunomodulatory, and antiatherogenic activities [5,6]. In particular, Conioselinum Chuanxiong and Angelica sinensis (Oliv.) Diels improve myocardial energy metabolism, promote hematopoiesis, and reduce blood deposition [7,8]. Nevertheless, investigations elucidating the protective effect and mechanism of KXZY remain insufficient due to its complex components and the absence of efficient study approaches. Consequently, there is an urgent need to establish an applicable and effective strategy to elucidate complex herbal formulas.
The approaches of network biology and multitarget drug development have made tremendous strides over the past several years. Thus, some new research concepts, such as network pharmacology, have been created, providing an applicable and efficient research strategy for modern drug research and novel therapy discovery [9]. The primary concept of network pharmacology is quite similar to the holistic approach of TCM. It enables researchers to use a holistic view to completely understand the effectiveness and mechanisms of complex herbal formulas. Therefore, network pharmacology is widely utilized in contemporary TCM research. In our study, we integrated HPLC-MS analytical techniques, network pharmacology, and in vitro experiments to determine the chemical composition, potential protective effects, and mechanism of KXZY. We utilized the research strategy of network pharmacology to predict the potential targets based on the results of our HPLC-MS analysis. An MI mouse model was established to evaluate the protective effects of KXZY and to validate the underlying mechanism (Figure 1).
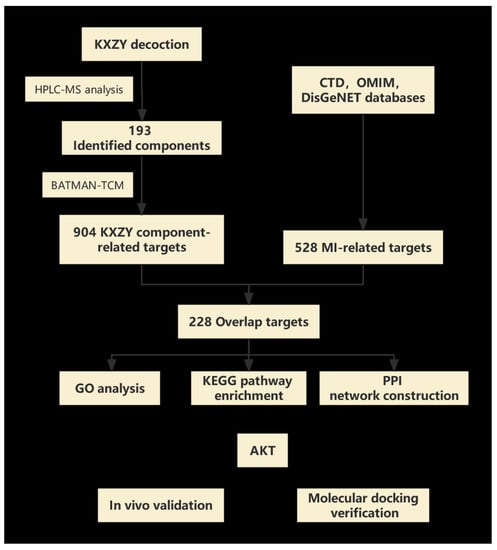
Figure 1.
Study flowchart. Flowchart showing the process of this study.
2. Methods
2.1. Preparation for KXZY
KXZY is composed of sixteen Chinese herbs (Table 1), namely, Conioselinum Chuanxiong (Chuanxiong), Paeonia lactiflora Pall. (Chishao), Rehmannia glutinosa (Gaertn.) DC. (Dihuang), Angelica sinensis (Oliv.) Diels (Danggui), Prunus persica (L.) Batsch (Taoren), Carthamus tinctorius L. (Honghua), Citrus aurantium L. (Zhike), Bupleurum chinense DC. (Chaihu), Achyranthes bidentata Blume (niuxi), Allium macrostemon Bunge (Xiebai), Trichosanthes kirilowii Maxim. (Gualou), Ziziphus jujuba Mill. (Hongzao), Hirudo nipponica Whitman. (Shuizhi), Alpinia katsumadai Hayata (Doukou), Platycodon grandiflorus A.DC. (Jiegeng), and Glycyrrhiza uralensis Fisch. ex DC. (Gancao). All of these ingredients were provided by the Hangzhou First People’s Hospital (Hangzhou, China). First, the herbal mixture was soaked in water (1:10, w/v) for 30 mins and then extracted twice at reflux for 2 times (1 h per cycle). The extracted herbal mixtures were then blended, concentrated, and freeze-dried into a powder for storage.

Table 1.
The composition of the KXZY decoction.
2.2. HPLC-MS Characterization of Primary Chemical Constituents in KXZY
HPLC-MS analysis was conducted on a Q Exactive plus Orbitrap LC-MS/MS (Thermo Fisher, Waltham, MA, USA) equipped with a Waters ACQUITY UPLC HSS T3 column (100 mm 2.1 mm i.d., 1.8 m); the column temperature was 35 °C. The parameter conditions were as follows: injection volume: 10 L; flow rate: 0.3 mL/min; mobile phase: A (deionized water, 0.1% v/v formic acid) and B (acetonitrile, 0.1% v/v formic acid). Using a gradient program, the following profile was created: 0 min (100% A, 0% B), 10 min (70% A, 30% B), 25 min (60% A, 40% B), 30 min (50% A, 50% B) 0% B, 40 min (30% A, 70% B), 45 min (0% A, 100% B), 60 min (0% A, 100% B), and 70 min (100% A, 0% B). Q Exactive Orbitrap high-resolution mass spectrometry was used for mass spectrometry (MS) data acquisition, and the detection mode was Full MS-ddMS2 with separate scanning in positive and negative ion modes. The parameter conditions were as follows: mass range: 100–1200 m/z; MS1 resolution: 70,000; MS2 resolution: 17,500; ion source voltage: 3.2 kV; capillary temp: 320 °C; aux gas heater temp: 350 °C; sheath gas flow rate: 40 L/min; aux gas flow rate: 15 L/min.
Compound Discover 3.2 software was utilized to extract the characteristic peaks from the raw mass spectrometry data. The mass deviation for elemental matching, molecular formula prediction, and isotopic distribution matching was set to within 5 ppm. To identify the characteristic peaks, two natural product databases, mzcloud and mzVault, were employed. To consider a result positive, it had to meet the following criteria: a mass deviation of less than 5 ppm, isotope distribution matching, and a database match score greater than 70 from the mzVault best match database. All positive results were verified manually, and duplicate results were eliminated.
2.3. Prediction and Screening of Targets
Based on the aforementioned prototype components, the possible targets were predicted by a bioinformatics research tool called TCM (BATMAN-TCM) [10]. The predicted targets were rated based on the interactions between possible targets and their likeness to existing targets; those with scores >20 were chosen.
MI-associated targets were compiled by merging the Comparative Toxicogenomics (CTD), Online Mendelian Inheritance in Man (OMIM), and DisGeNET databases [11,12], and “Homo sapiens” was the only species considered.
Potential KXZY targets for treating MI were the targets shared by KXZY component-related targets and MI-related targets.
2.4. Protein–Protein Interaction (PPI) Network Construction
To examine the probable interactions between target-related genes, we imported these genes into the public STRING database [13]. STRING is often applied to collect comprehensive information on protein interactions between genes, and targets with interaction scores of 0.400 were chosen. Afterward, we loaded all of the above results into Cytoscape to display the connections and screened the key genes using plugins. The Cytoscape plugin CytoNCA was used to filter hub genes by combining several centrality measure calculations, analyses, and assessments. “Betweenness centrality” (BC), “closeness centrality” (CC), and “degree centrality” (DC) are the topological parameters.
2.5. Gene and KEGG Analysis
We used the DAVID V6.8 database to perform GO and KEGG pathway enrichment analyses to show the roles of targets for the active components of TCM in terms of gene function and particular signaling pathways [14]. GO analysis was then utilized to annotate gene function via the use of three modules: biological process (BP), molecular function (MF), and cellular component (CC). The cutoff threshold for statistical significance in the DAVID analysis was a p value of less than 0.05.
2.6. Molecular Docking
AutoDockTools 1.5.6 was used to accomplish molecular docking. The critical target’s atomic coordinates were collected from the Protein Data Bank (PDB) and generated in AutoDockTools-1.5.6 by eliminating water molecules, adding charge, and parameterizing. Downloaded from the TCMSP Database, the 3D structures of active components were built in AutoDockTools by calculating atomic partial charges and parameterizing. The docking site was positioned in the middle of the original ligand inside a cuboid box, and a grid map of each atom type within the box was calculated. Molecular docking of possible targets and components was simulated using the AutoDockTools-1.5.6 program. Each compound’s highest-scoring conformer was examined and displayed using AutoDockTools-1.5.6 and PyMOL.
2.7. Animal Model and Groups
Zhejiang Chinese Medical University (Animal Care and Use Committee) approved the procedures for this investigation (Approval No. IACUC-20211101-07). Shanghai SLAC Laboratory Animal Co. provided male C57/BL6 mice weighing between 22 and 24 g at the outset (Shanghai, China). Prior to surgical modeling, experimental mice were acclimated to the laboratory setting for one week. Following surgery, mice were randomly divided into four groups (five to eight mice per group): the sham group (sham), the MI model group (MI), the KXZY group (KXZY, 3.7 g/kg/d), and the captopril group (Cap, 30 mg/kg/d). The medication dosage was determined using the body surface area. As mentioned earlier, surgery was performed in the Model and KXZY groups by ligating the left anterior descending (LAD) coronary artery. Without ligating the LAD coronary artery, a suture was passed across the LAD of the sham group. All groups received intragastric delivery of water or medication for 28 days. Four weeks after an acute myocardial infarction, the heart tissues of sacrificed mice were swiftly removed. A part of the heart was preserved in 4% paraformaldehyde, and another part was kept at −80 °C for future research.
2.8. Echocardiography
The supine posture was maintained after intraperitoneal injections of 1% pentobarbital sodium rendered the experimental mice unconscious. The left ventricular function in mice was evaluated by transthoracic echocardiography (Vevo TM 2100, VisualSonics, Toronto, ON, Canada). Under the parasternal short-axis (SAX) ultrasound section, we captured the LVIDd and LVIDs. After this, the EF and FS were separately assessed.
2.9. NT-proBNP in Serum
The abdominal aorta of mice was used to collect whole blood, which was then processed to isolate serum and stored at −80 °C for further research. The levels of NT-proBNP in serum were measured using a commercial ELISA kit (Elabscience Biotechnology Co., Ltd., Wuhan, China).
2.10. Histopathological
Cardiac tissues treated with paraformaldehyde were embedded in paraffin and cut into 4 μm thick slices. Subsequently, the sections were processed by hematoxylin-eosin (H&E) and Sirius red staining to evaluate the extent of fibrosis and pathological changes. The reagents were purchased from Abcam (Cambridge, MA, USA).
2.11. Western Blotting
Total protein was extracted from cardiac tissues using radioimmunoprecipitation assay (RIPA) buffer, including a protease inhibitor cocktail. A 12% SDS polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate the proteins. Then, the proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Burlington, MA, USA), which were incubated overnight at 4 °C. Primary antibodies against BAD, AKT, and p-AKT were obtained from Cell Signaling Technology (Danvers, MA, USA), while Bcl-2, Bcl-xl, and GAPDH were purchased from Abcam (Cambridge, MA, USA). Then, secondary antibodies (Abcam, Cambridge, MA, USA) were incubated at 25 °C for 1 h. Bands were visualized using enhanced chemiluminescence (ECL) plus Western blotting detection reagents (Bio-Rad, Hercules, CA, USA) and subsequently captured by the ChemiDoc Touch Imaging System and Image Lab software (Bio-Rad, Hercules, CA, USA).
2.12. Statistical Analysis
All animal experiment results are reported as the mean ± SD. SPSS 17.0 statistical software was used for the statistical analysis (IBM, Armonk, NY, USA). For numerous comparisons across groups, one-way analysis of variance (ANOVA) was applied in line with tests for normality and homogeneity of variance. p values less than 0.05 were considered statistically significant.
3. Results
3.1. Characterization of the Chemical Constituents in KXZY
Figure 2 depicts the representative chromatogram acquired by HPLC-MS analysis. Based on the HPLC-MS results, we matched the previous research and databases, and a total of 193 compounds (Table 2) were identified [15,16,17,18,19]. The abundance of the compounds was inferred from the relative peak areas, and the compounds with high relative contents included amygdalin, naringin, paeoniflorin, neohesperidin, diammonium glycyrrhizinate, betaine, sucrose, stachydrine, safflomin A, 18 β-glycyrrhetintic acid, 2-pyrrolidinecarboxylic acid, citric acid, and naringenin. In addition, we searched the herb sources of the compounds and summed the abundance of compounds attributed to each herb to calculate the relative content percentages. Among them, the highest content of compounds was found for Gancao, followed by Jiegeng and Chishao, as shown in Figure 3.
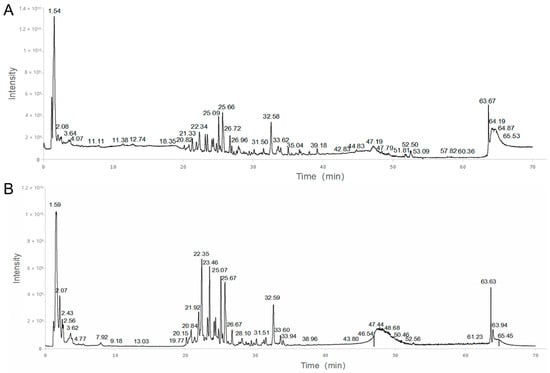
Figure 2.
Total ion chromatogram (TIC) in positive mode (A) and negative mode (B) of KXZY.

Table 2.
Characterization of chemical constituents in KXZY.

Figure 3.
Relative content percentages of each herb. Different colors indicate different herbs, and font size indicates the content.
3.2. Screening of the Potential Targets of KXZY in Treating MI
Then, the above compounds were input into BATMAN to predict the related targets, and targets with scores > 20 were considered meaningful. After eliminating duplicated genes, a total of 90 vital components with 904 targets were retrieved.
In total, 757 genes were identified from the CTD, OMIM, and DisGeNET databases that met the requirements for a connection with MI. After eliminating duplicated genes, 528 MI-related genes were retrieved. As shown in Figure 4, the screened drug and disease targets were intersected to obtain 228 common targets between KXZY and MI, They were used as the potential targets of KXZY in treating MI in subsequent network construction and pathway enrichment analysis.
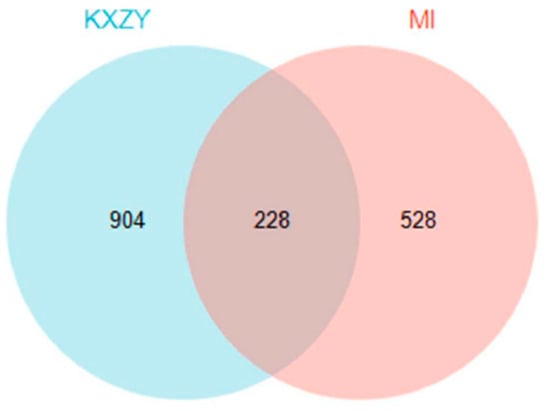
Figure 4.
The common targets between KXZY and MI. The blue circles indicate KXZY targets, and the pink circles indicate MI targets.
3.3. GO and KEGG Analysis
As Figure 5 shows, GO analysis revealed that the following terms were significantly enriched: For BP, the top 3 terms were response to decreased oxygen levels, response to oxygen levels, and response to hypoxia. For CC, the top 3 terms were membrane raft, membrane microdomain, and apical part of cell. For MF, the top 3 terms were receptor ligand activity, signaling receptor activator activity, and protein heterodimerization activity. KEGG enrichment results indicated that these targets were mostly enriched in lipid and atherosclerosis, the PI3K-Akt signaling pathway, the cAMP signaling pathway, and the HIF-1 signaling pathway.
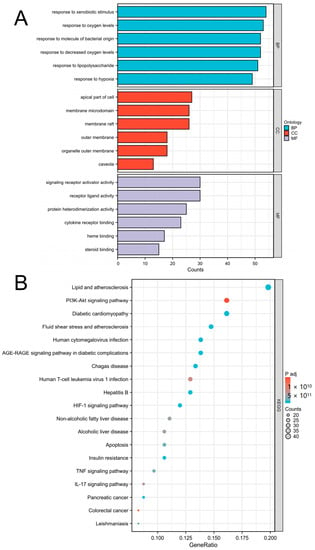
Figure 5.
Major enrichment analyses of GO terms and KEGG pathways. (A) GO term analysis included BP (blue), CC (red), and MF (gray). (B) KEGG pathways. The area of the dot indicates the number of enriched genes; the larger the dot is, the greater the number of genes. The color of the dot represents the significance of the p value; the pathway with an intense red color indicates a significant p value.
3.4. PPI Network Analysis
To analyze potential protein interactions, the predicted targets of KXZY were uploaded into the SPRING database. As shown in Figure 6, a PPI network was constructed involving 228 nodes and 1327 edges. CytoNCA, a plug-in for Cytoscape, was applied to conduct topological analysis. According to the outcomes of the CytoNCA analysis, there were 62 key nodes whose betweenness centrality (BC) was greater than the mean (betweenness centrality = 379.2568782); this represented 28.44% of the total number of nodes. The top 10 hub genes were as follows: AKT1, ALB, SRC, TP53, EGFR, CTNNB1, JUN, PPARA, TNF, and F2. The complete results are illustrated in Table 3. Our findings revealed that these genes play an important role as network hubs.
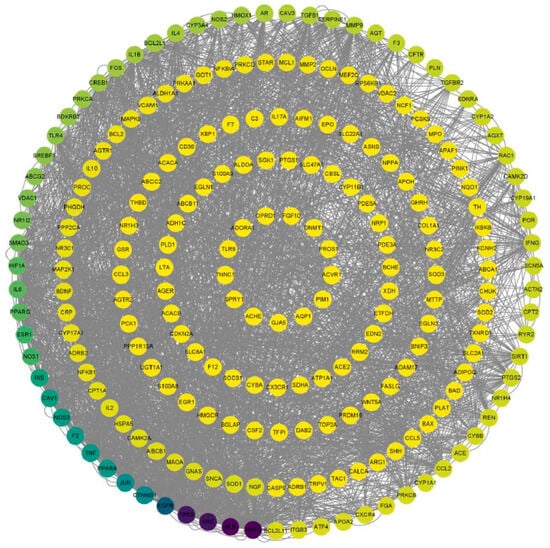
Figure 6.
Topological analysis of the PPI network. Nodes represent target proteins (the color of the dot represents the significance of BC, and the dot with an intense black color indicates a significant BC). Edge represents protein–protein association.

Table 3.
The top 10 targets of the PPI network.
KEGG pathway enrichment results indicated that there was significant enrichment in the PI3K/AKT signaling pathway. AKT1 was the top hub target in the structural network. Therefore, we inferred that the PI3K/AKT signaling pathway plays a critical role after MI. Thus, we further mapped the top-ranked hub targets in the PI3K/AKT pathway. As shown in Figure 7, a significant proportion of hub targets were associated with the PI3K/AKT pathway. Notably, apoptosis regulation-related genes, including BAD, Bcl-2, and Bcl-xl, were significantly labeled. Based on the above results, we inferred that KXZY might exert anti-apoptotic effects by regulating the AKT protein. The top hub gene was AKT1.
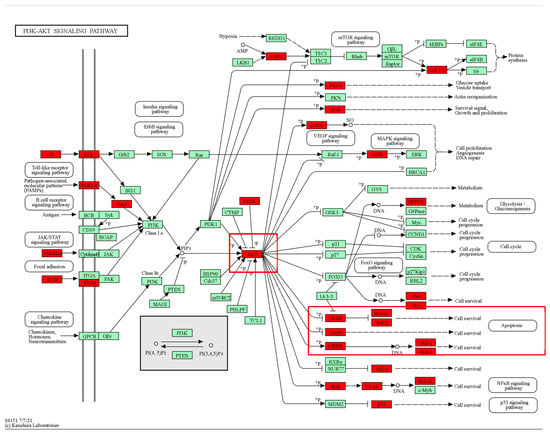
Figure 7.
Mapping of the top targets in the PI3K-AKT signaling pathway. Red markers indicate predicted targets. The red box indicates that hub targets are enriched in the AKT/Bcl-2/Bax-associated apoptosis pathway.
3.5. Molecular Docking Verification
Finally, we screened the key active compounds that precisely hit AKT1, which is the key target protein, based on the above target prediction results (isoguanosine, naringenin, liquiritigenin, isosakuranetin, hesperetin, dehydrodiisoeugenol, glabridin, and adenosine). As shown in Table 4, isoguanosine and adenosine were the most effective combinations with AKT1. However, adenosine was relatively low in the HPLC analysis. The PyMOL results are visualized in Figure 8.

Table 4.
The docking affinity and interactions of compounds binding to key targets.
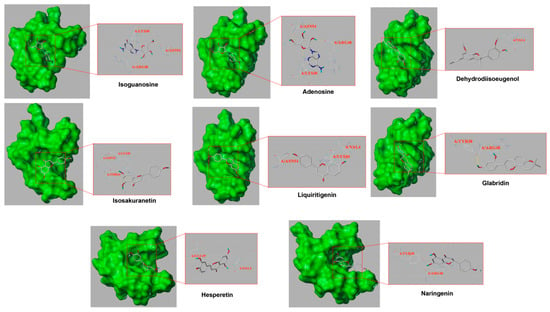
Figure 8.
Part of the molecular docking results. The left half represents the atom binding diagram of the molecule and protein, and the right half represents the enlarged picture. The AKT1 structure is presented as a green sphere, the skinny stick represents the residue, and the compound structure is presented as a thick stick (gray represents carbon atoms, red oxygen atoms, blue nitrogen atoms, and light blue hydrogen atoms, and the dotted yellow line represents the interactions).
3.6. KXZY Ameliorated Cardiac Dysfunction in Post-MI Mice
To confirm the cardioprotective benefits of KXZY after MI, KXZY was intragastrically administered to post-MI mice for 4 weeks. Subsequently, we evaluated the function and histopathology of the LV. As shown in Figure 9, echocardiographic data revealed that the EF and FS values in the MI group were significantly lower than those in the sham group (p < 0.05). Conversely, in mice treated with KXZY or captopril, the EF and FS levels were markedly increased compared with those in the MI group (p < 0.05). The results demonstrated that KXZY could improve cardiac function after MI. In addition, our study indicated that the serum levels of NT-proBNP, which is a cardiac dysfunction marker, were markedly elevated in the model group (p < 0.05). In contrast, KXZY appreciably attenuated the elevation of NT-proBNP levels (p < 0.05).
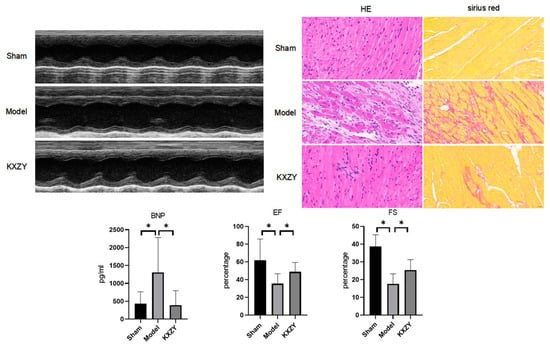
Figure 9.
The effects of KXZY on cardiac function and histology in post-MI mice. n = 3; ×40, scale bars, 50 μM; * p < 0.05.
Moreover, the histopathological results of cardiac tissue revealed disorganized cardiac muscle fiber arrangements, disruption of cardiac structure, and significant interstitial edema in the MI group. The total cardiac fibrosis area was markedly enlarged in the MI group versus the control group. In the KXZY group, these histopathologic abnormalities were restored. These results provide vital evidence that KXZY can attenuate the extent of myocardial injury in post-MI mice.
3.7. KXZY Activates the AKT Protein and Attenuates Myocardial Apoptosis
As shown in Figure 10, compared to the sham group, the levels of Bcl-2 and Bcl-xl proteins significantly decreased, while the expression of the BAD protein was significantly increased. Conversely, KXZY increased the expression of Bcl-2 and Bcl-xl proteins and decreased the expression of the BAD protein. Furthermore, KXZY increased the phosphorylation of the AKT protein. When AKT protein becomes activated after phosphorylation, it can inhibit the activity of the proapoptotic protein BAD, thereby leading to an increase in the expression of antiapoptotic proteins such as bcl-2 and bcl-xl. Thus, the results of our study show that FKZF inhibits cardiomyocyte apoptosis by regulating AKT phosphorylation levels and apoptosis-related protein expression.
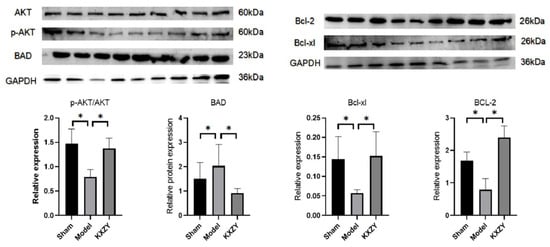
Figure 10.
The effects of KXZY on AKT and apoptosis proteins in post-MI mice. n = 3; * p < 0.05.
4. Discussion
TCM’s “holistic perspective” and “syndrome distinction” concepts provide unique advantages in the treatment of diseases. As one of the classic prescriptions for promoting blood circulation and removing blood stasis, KXZY has certain effects on the prevention and treatment of cardiovascular diseases. It is usually added according to clinical syndrome differentiation and disease, such as in post-MI patients with chest distress, shortness of breath, heart palpitations, and tiredness. Wang et al.’s research shows that supplementing qi and activating blood therapy were effective and safe in the treatment of coronary heart disease [20]. In addition, during the last decade, tremendous progress has been made in the treatment and study of a variety of illnesses employing a single active component derived from herbs [21,22]. Due to their complicated composition, however, current research into TCM formulas continues to encounter significant obstacles and problems. Due to the rapid growth of bioinformatics and polypharmacy, network pharmacology has been creatively applied in research for TCM formula distinction. In our research, HPLC-MS was used to identify the structural composition of KXZY extracts. Furthermore, we conducted a detailed network pharmacology analysis for target prediction based on the above results. Finally, molecular docking and related experiments were used for validation.
We first identified 193 substances using the HPLC-MS technique. Among these, quercetin, liquiritigenin, and naringin have been reported to have antiapoptotic, antihypertensive, anti-inflammatory, and anti-cardiovascular injury properties [23,24,25]. Then, we constructed the pharmacological network of KXZY based on previous HPLC-MS results. A total of 228 targets were predicted in combination with the MI gene database. GO and KEGG pathway enrichment results indicated that KXZY ameliorated MI damage through multiple biological processes, including response to oxygen levels, response to hypoxia, lipid and atherosclerosis, and regulation of apoptosis. The differential targets were then found to be predominantly enriched in pathways of neurodegeneration—multiple diseases, the PI3K-Akt signaling pathway, the cAMP signaling pathway, and the HIF-1 signaling pathway. Moreover, we screened hub genes using the cytoNCA plug-in and observed that AKT1 was the top target, and the majority of targets were associated with the PI3K-Akt signaling pathway. Specifically, the targets of functional genes were primarily associated with the regulation of apoptosis by the AKT signaling pathway. These findings indicated that AKT-mediated anti-apoptotic alterations may be the crucial target of KXZY.
Cardiomyocyte apoptosis plays an important role during chronic cardiac remodeling and heart failure. Especially in the acute and subacute phases of MI, apoptosis is a prevalent pathogenic characteristic [26]. Studies have shown that the detrimental effect of apoptosis may be more apparent after MI [27]. Moe et al. discovered that the apoptosis of cardiomyocytes occurs over time and correlates strongly with the severity of cardiac dysfunction in heart failure [28]. Baldi et al. found that there was a sustained apoptotic reaction after MI [29]. Wang et al. noted an increase in the number of apoptotic cells 28 days after AMI [30]. Nevertheless, suppression of apoptosis might dramatically increase heart survival and decrease chronic cardiac remodeling and dysfunction [31].
The AKT protein is one of the serine/threonine protein kinase families that regulates several biological activities, including cell growth, metabolism, and survival [32]. The AKT family consists of three isoforms: AKT1, AKT2, and AKT3. AKT1 is the most extensively studied isoform and is widely expressed in a variety of tissues. AKT2 is mainly expressed in skeletal muscle, liver, and adipose tissue. AKT3 is expressed in brain tissue and is involved in regulating neuronal development and function, as well as cell survival and proliferation [33]. The AKT isoform that is mainly expressed in myocardial tissue is AKT1. AKT1 plays a critical role in the regulation of cardiac function, including the control of cardiac hypertrophy, apoptosis (programmed cell death), and contractility. Among them, AKT1 promotes cell survival by inhibiting multiple targets, such as BAD and Bcl-2, in the apoptosis signaling cascade [34]. A growing number of studies have shown that AKT activation prevents cardiomyocyte death, while AKT inhibition exacerbates cardiomyocyte apoptosis and cardiac dysfunction [35]. AKT inactivation was responsible for MI-induced myocardial damage, whereas AKT phosphorylation increased cardiomyocyte survival and prevented MI-induced cardiac dysfunction, according to previous findings.
Therefore, based on the above results, further investigation was conducted to explore the potential anti-apoptotic mechanism of KXZY. To assess the potential cardioprotective and anti-apoptotic effects of KXZY, a post-MI mouse model was utilized in this study. Our study shows that post-MI mice exhibit better EF and FS values with the treatment of KXZY. Our research suggested that KXZY could improve left ventricular systolic function in post-MI mice. Furthermore, we assessed the AKT-related anti-apoptotic effects of KXZY after MI damage. AKT phosphorylation was significantly inhibited, and BAD expression was markedly reduced. There was an increase in the expression of Bcl-2 and Bcl-xl. We therefore infer that KXZY may exert its anti-apoptotic effects by regulating the phosphorylation and activation of AKT1, which in turn inhibits the pro-apoptotic protein BAD, allowing Bcl-2 and bcl-xl to inhibit apoptosis and maintain cell survival. These findings indicated that AKT-related antiapoptotic mechanisms might be the main mechanism of KXZY in post-MI damage.
Finally, molecular docking was used to verify the mode of interaction between compounds of KXZY and AKT1 protein molecules. Eight compounds of KXZY were predicted to bind to AKT1: isoguanosine, naringenin, liquiritigenin, isosakuranetin, hesperetin, dehydrodiisoeugenol, glabridin, and adenosine. Our study indicated that isoguanosine exhibits high binding energy through molecular docking. Thus, we inferred that isoguanosine might improve cardiac dysfunction after MI by inhibiting AKT-related apoptosis.
5. Conclusions
This study identified 193 chemical compounds of KXZY by HPLC-MS. In addition, 228 potential targets were predicted, and a PPI network was constructed for further network pharmacology research. The findings of functional enrichment analysis suggest that the apoptotic process is strongly associated with AKT1. Additionally, we used a post-MI mouse model to verify the anti-apoptotic and anti-cardiac dysfunction behaviors of KXZY, and isoguanosine and adenosine were found to be two important pharmacodynamic compounds. Consequently, the herbal method provides a wide repository for the development of novel therapeutic agents for the treatment of post-MI cardiac damage, and KXZY might serve as an important supplementary substance.
Author Contributions
Conceived/designed this study, Y.Z. and Y.X. Acquired/analysed/interpreted data, Y.W. and M.L. Drafted manuscript, Y.W. and M.L. Provided essential research tools/samples, L.C. and L.X. Reviewed manuscript, Y.Z., Y.X., L.C. and L.X. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Construction Fund of Key Medical Disciplines of Hangzhou (OO20200121) and the Traditional Chinese Medicine Science and Technology Plan Project of Zhejiang Province (2022ZZ028).
Institutional Review Board Statement
The animal study protocol was approved by the Laboratory animal management and ethics committee of Zhejiang Chinese Medical University (IACUC-20211101-07, 1 November 2021).
Informed Consent Statement
Not applicable.
Data Availability Statement
The original data contributing to the findings presented in this study are included in the article. Further inquiries can be addressed to the corresponding authors.
Acknowledgments
We are grateful to all those who contributed to this research. We thank Yue Zhu for the excellent technical support in HPLC analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Duran, E.K.; Aday, A.W.; Cook, N.R.; Buring, J.E.; Ridker, P.M.; Pradhan, A.D. Triglyceride-Rich Lipoprotein Cholesterol, Small Dense LDL Cholesterol, and Incident Cardiovascular Disease. J. Am. Coll. Cardiol. 2020, 75, 2122–2135. [Google Scholar] [CrossRef] [PubMed]
- Danielson, K.M.; Shah, R.; Yeri, A.; Liu, X.; Camacho Garcia, F.; Silverman, M.; Tanriverdi, K.; Das, A.; Xiao, C.; Jerosch-Herold, M.; et al. Plasma Circulating Extracellular RNAs in Left Ventricular Remodeling Post-Myocardial Infarction. EBioMedicine 2018, 32, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Zhang, P.; Zhang, Z.; Youn, J.Y.; Wang, C.; Zhang, H.; Cai, H. Traditional Chinese Medicine (TCM) in the treatment of COVID-19 and other viral infections: Efficacies and mechanisms. Pharmacol. Ther. 2021, 225, 107843. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Tian, S.; Zhao, J.; Zhang, W. Exploring the mechanism of TCM formulae in the treatment of different types of coronary heart disease by network pharmacology and machining learning. Pharmacol. Res. 2020, 159, 105034. [Google Scholar] [CrossRef]
- Hua, Y.Q.; Zeng, Y.; Xu, J.; Xu, X.L. Naringenin alleviates nonalcoholic steatohepatitis in middle-aged Apoe(−/−)mice: Role of SIRT1. Phytomedicine 2021, 81, 153412. [Google Scholar] [CrossRef] [PubMed]
- Zhongyin, Z.; Wei, W.; Juan, X.; Guohua, F. Isoliquiritin apioside relieves intestinal ischemia/reperfusion-induced acute lung injury by blocking Hif-1alpha-mediated ferroptosis. Int. Immunopharmacol. 2022, 108, 108852. [Google Scholar] [CrossRef]
- Gu, P.; Wusiman, A.; Wang, S.; Zhang, Y.; Liu, Z.; Hu, Y.; Liu, J.; Wang, D. Polyethylenimine-coated PLGA nanoparticles-encapsulated Angelica sinensis polysaccharide as an adjuvant to enhance immune responses. Carbohydr. Polym. 2019, 223, 115128. [Google Scholar] [CrossRef]
- Li, J.; Gong, X. Tetramethylpyrazine: An Active Ingredient of Chinese Herbal Medicine With Therapeutic Potential in Acute Kidney Injury and Renal Fibrosis. Front. Pharmacol. 2022, 13, 820071. [Google Scholar] [CrossRef]
- Zhong, Y.; Li, M.; Zhang, X.; Chen, L.; Wang, Y.; Xu, Y. Dissecting Chemical Composition and Cardioprotective Effects of Fuzhengkangfu Decoction against Doxorubicin-Induced Cardiotoxicity by LC-MS and Bioinformatics Approaches. ACS Omega 2020, 5, 14051–14060. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, F.; Wang, Y.; Li, C.; Zhang, X.; Li, H.; Diao, L.; Gu, J.; Wang, W.; Li, D.; et al. BATMAN-TCM: A Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine. Sci. Rep. 2016, 6, 21146. [Google Scholar] [CrossRef]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015, 43, D789–D798. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2021. Nucleic Acids Res. 2021, 49, D1138–D1143. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Gong, M.; Wu, S.; Qiu, F.; Ma, L. Identification and quantification of the quality markers and anti-migraine active components in Chuanxiong Rhizoma and Cyperi Rhizoma herbal pair based on chemometric analysis between chemical constituents and pharmacological effects. J. Ethnopharmacol. 2020, 246, 112228. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Han, X.; Lai, Y.; Fu, Y.; Li, K.; Zhang, W.; Wang, Q.; Jiang, X.; Zhou, Y.; Liang, H.; et al. Systems pharmacological study based on UHPLC-Q-Orbitrap-HRMS, network pharmacology and experimental validation to explore the potential mechanisms of Danggui-Shaoyao-San against atherosclerosis. J. Ethnopharmacol. 2021, 278, 114278. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.J.; Sun, Y.; Song, Y.; Ma, J.N.; Wang, Z.Q.; Ding, X.Q.; Chen, H.Y.; Zhang, X.B.; Song, M.M.; Hu, X.M. Mechanism and Molecular Targets of Ejiao Siwu Decoction for Treating Primary Immune Thrombocytopenia Based on High-Performance Liquid Chromatograph, Network Pharmacology, Molecular Docking and Cytokines Validation. Front. Med. 2022, 9, 891230. [Google Scholar] [CrossRef]
- Chen, Y.; Dai, Y.; Xia, J.; Liu, J.; Zhou, G.; Chen, C.; Jiang, B.; Yin, L.; Li, G. Serum Pharmacochemistry Combining Network Pharmacology to Discover the Active Constituents and Effect of Xijiao Dihuang Tang Prescription for Treatment of Blood-Heat and Blood-Stasis Syndrome-Related Disease. Oxid. Med. Cell Longev. 2022, 2022, 6934812. [Google Scholar] [CrossRef]
- Zhu, Y.L.; Wang, H.J.; Xue, H.; Zhang, Y.; Cheng, Q.S.; Chen, L.Y.; Qiu, X.J. Simultaneous Determination of Five Components of Chaihu-Shugan-San in Beagle Plasma by HPLC-MS/MS and Its Application to a Pharmacokinetic Study after a Single Dose of Chaihu-Shugan-San. J. Anal. Methods Chem. 2020, 2020, 8831938. [Google Scholar] [CrossRef]
- Wang, X.; Cui, H. Application of Entropy Method to Quantify Future Ecological Flow in the Yellow River Basin. Entropy 2021, 24, 72. [Google Scholar] [CrossRef]
- Shen, P.; Lin, W.; Deng, X.; Ba, X.; Han, L.; Chen, Z.; Qin, K.; Huang, Y.; Tu, S. Potential Implications of Quercetin in Autoimmune Diseases. Front. Immunol. 2021, 12, 689044. [Google Scholar] [CrossRef]
- Ding, B.; Lin, C.; Liu, Q.; He, Y.; Ruganzu, J.B.; Jin, H.; Peng, X.; Ji, S.; Ma, Y.; Yang, W. Tanshinone IIA attenuates neuroinflammation via inhibiting RAGE/NF-kappaB signaling pathway in vivo and in vitro. J. Neuroinflamm. 2020, 17, 302. [Google Scholar] [CrossRef]
- Dhanya, R. Quercetin for managing type 2 diabetes and its complications, an insight into multitarget therapy. Biomed. Pharmacother. 2022, 146, 112560. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.H.; Park, K.I.; Kim, K.Y.; Lee, J.H.; Jang, E.J.; Ku, S.K.; Kim, S.C.; Suk, H.Y.; Park, J.Y.; Baek, S.Y.; et al. Liquiritigenin inhibits hepatic fibrogenesis and TGF-beta1/Smad with Hippo/YAP signal. Phytomedicine 2019, 62, 152780. [Google Scholar] [CrossRef]
- Aihaiti, Y.; Song Cai, Y.; Tuerhong, X.; Ni Yang, Y.; Ma, Y.; Shi Zheng, H.; Xu, K.; Xu, P. Therapeutic Effects of Naringin in Rheumatoid Arthritis: Network Pharmacology and Experimental Validation. Front. Pharmacol. 2021, 12, 672054. [Google Scholar] [CrossRef]
- Vasamsetti, S.B.; Coppin, E.; Zhang, X.; Florentin, J.; Koul, S.; Gotberg, M.; Clugston, A.S.; Thoma, F.; Sembrat, J.; Bullock, G.C.; et al. Apoptosis of hematopoietic progenitor-derived adipose tissue-resident macrophages contributes to insulin resistance after myocardial infarction. Sci. Transl. Med. 2020, 12, eaaw0638. [Google Scholar] [CrossRef] [PubMed]
- Mohammadinejad, R.; Moosavi, M.A.; Tavakol, S.; Vardar, D.O.; Hosseini, A.; Rahmati, M.; Dini, L.; Hussain, S.; Mandegary, A.; Klionsky, D.J. Necrotic, apoptotic and autophagic cell fates triggered by nanoparticles. Autophagy 2019, 15, 4–33. [Google Scholar] [CrossRef]
- Wang, Z.; Qiu, Z.; Hua, S.; Yang, W.; Chen, Y.; Huang, F.; Fan, Y.; Tong, L.; Xu, T.; Tong, X.; et al. Nuclear Tkt promotes ischemic heart failure via the cleaved Parp1/Aif axis. Basic. Res. Cardiol. 2022, 117, 18. [Google Scholar] [CrossRef] [PubMed]
- Baldi, A.; Abbate, A.; Bussani, R.; Patti, G.; Melfi, R.; Angelini, A.; Dobrina, A.; Rossiello, R.; Silvestri, F.; Baldi, F.; et al. Apoptosis and post-infarction left ventricular remodeling. J. Mol. Cell Cardiol. 2002, 34, 165–174. [Google Scholar] [CrossRef]
- Wang, Y.; Li, C.; Ouyang, Y.; Shi, T.; Yang, X.; Yu, J.; Qiu, Q.; Han, J.; Wu, Y.; Tang, B.; et al. QSYQ Attenuates Oxidative Stress and Apoptosis Induced Heart Remodeling Rats through Different Subtypes of NADPH-Oxidase. Evid. Based Complement. Alternat Med. 2013, 2013, 824960. [Google Scholar] [CrossRef]
- Wang, K.; Li, Z.; Sun, Y.; Liu, X.; Ma, W.; Ding, Y.; Hong, J.; Qian, L.; Xu, D. Dapagliflozin Improves Cardiac Function, Remodeling, Myocardial Apoptosis, and Inflammatory Cytokines in Mice with Myocardial Infarction. J. Cardiovasc. Transl. Res. 2022, 15, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, N.; Gavin, M.; Loro, E.; Sostre-Colon, J.; Roberson, P.A.; Uehara, K.; Rivera-Fuentes, N.; Neinast, M.; Arany, Z.; Kimball, S.R.; et al. AKT controls protein synthesis and oxidative metabolism via combined mTORC1 and FOXO1 signalling to govern muscle physiology. J. Cachexia Sarcopenia Muscle 2022, 13, 495–514. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, Y.; Wu, J.; Guo, Y.; Xiao, H.; Zhang, Y.; Ma, K. Resveratrol Targets AKT1 to Inhibit Inflammasome Activation in Cardiomyocytes Under Acute Sympathetic Stress. Front. Pharmacol. 2022, 13, 818127. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Lu, H.; Xu, H. Galangin Reverses Hepatic Fibrosis by Inducing HSCs Apoptosis via the PI3K/Akt, Bax/Bcl-2, and Wnt/beta-Catenin Pathway in LX-2 Cells. Biol. Pharm. Bull. 2020, 43, 1634–1642. [Google Scholar] [CrossRef] [PubMed]
- Jing, Z.T.; Liu, W.; Xue, C.R.; Wu, S.X.; Chen, W.N.; Lin, X.J.; Lin, X. AKT activator SC79 protects hepatocytes from TNF-alpha-mediated apoptosis and alleviates d-Gal/LPS-induced liver injury. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 316, G387–G396. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).