A Phylogeographical Analysis of the Beetle Pest Species Callosobruchus chinensis (Linnaeus, 1758) in China
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection
2.2. DNA Extraction, Amplification, Sequencing and Sequence Editing
2.3. Ecological Niche Modelling
2.4. Genetic Polymorphism Analysis and Isolation by Distance
2.5. Phylogeographical Analysis
2.6. Reconstructions of Divergence Time and Historical Demography
2.7. Visualization of Dispersal Corridors
3. Results
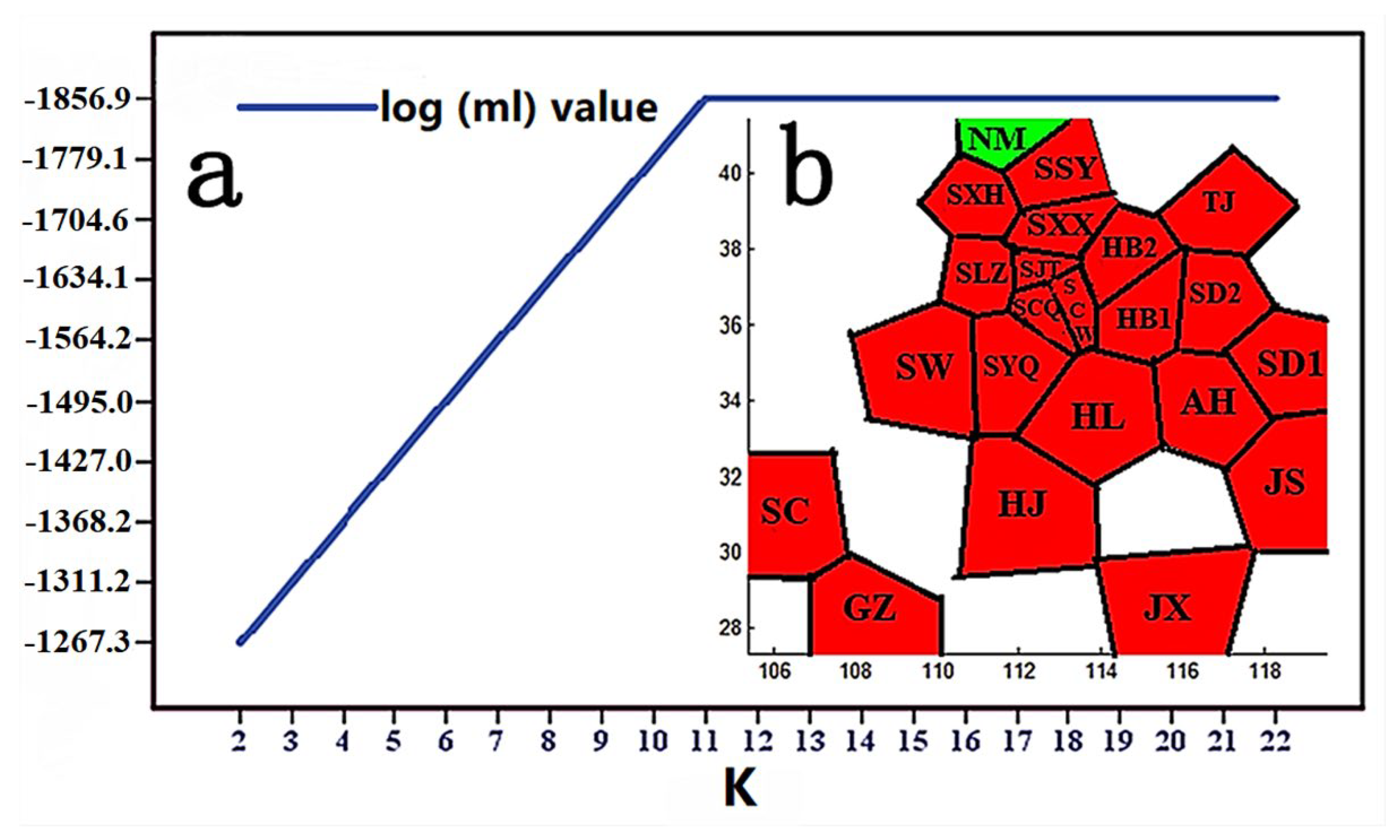
3.1. MaxEnt Model Evaluation
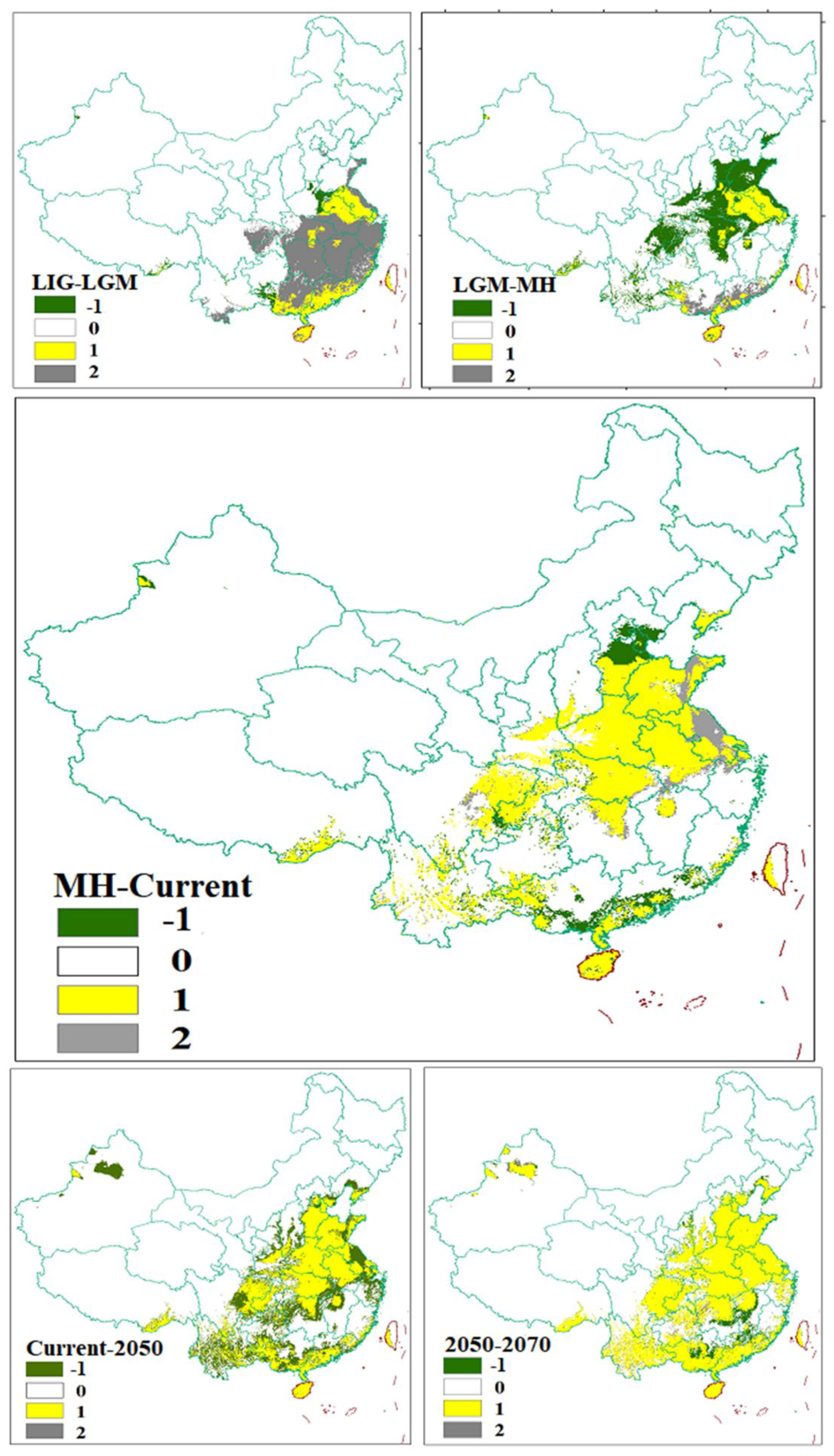
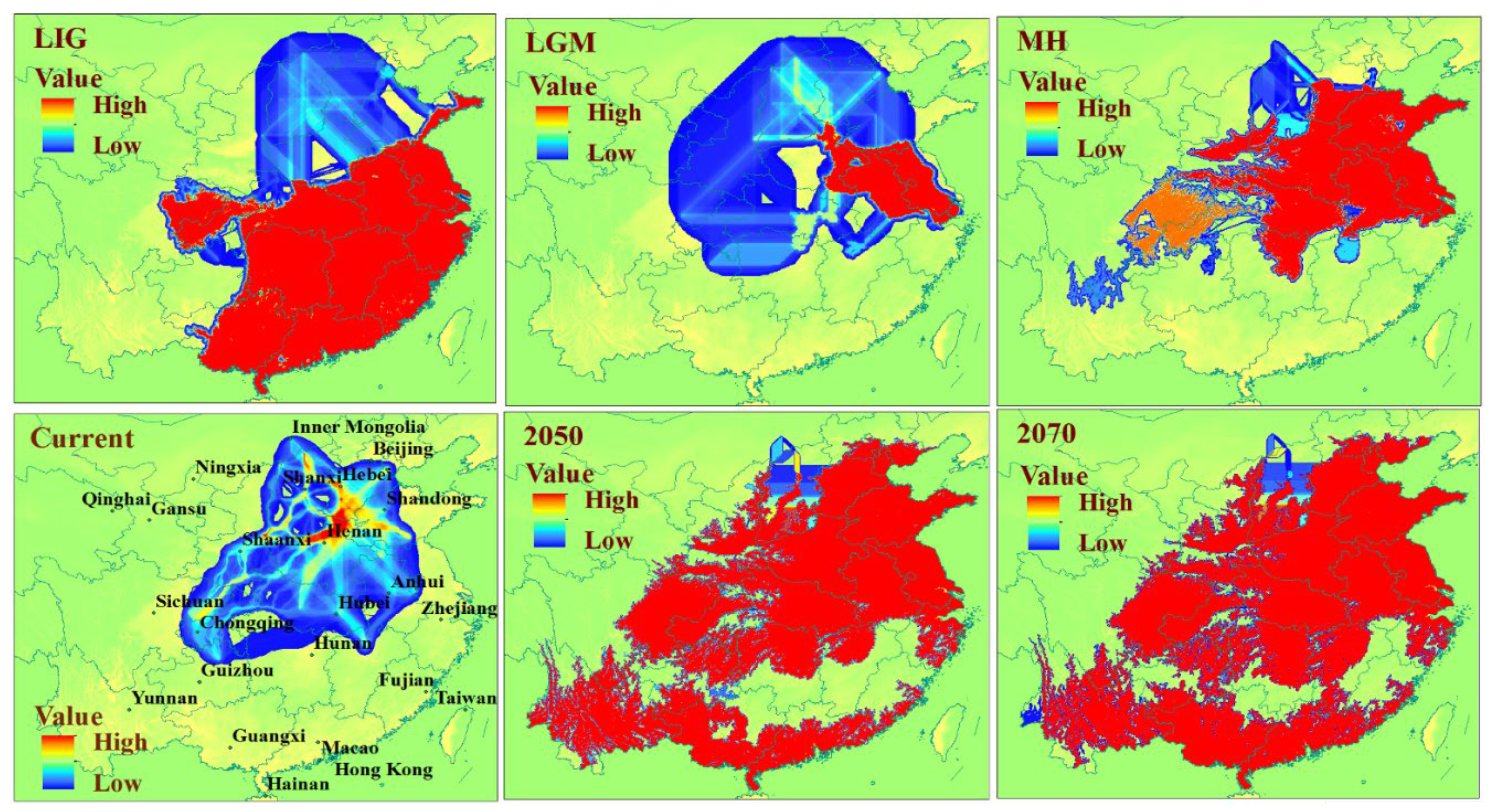
3.2. Prediction of the Range and Change in Habitat Suitability of C. chinensis
3.3. Genetic Polymorphism Analysis
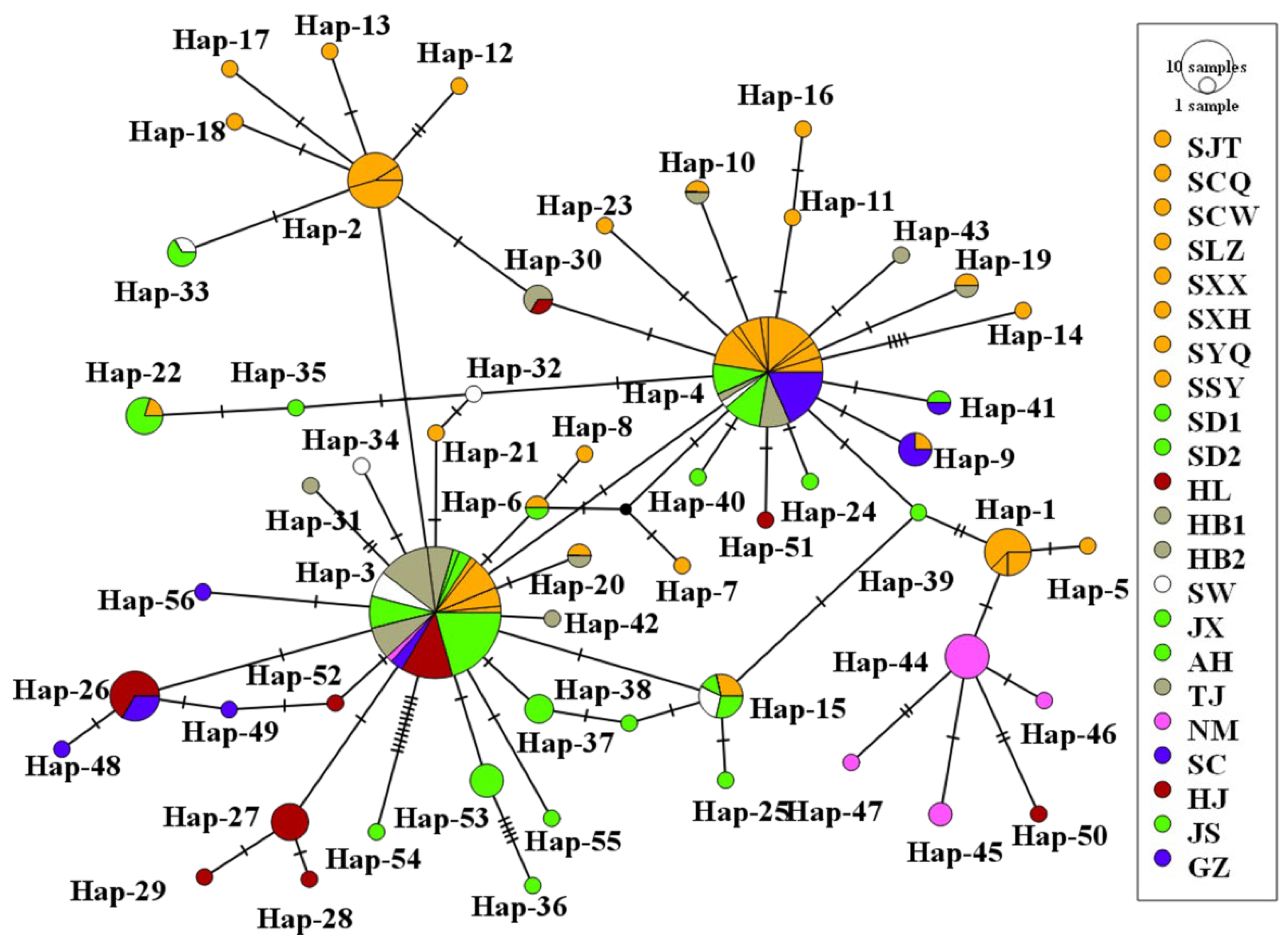
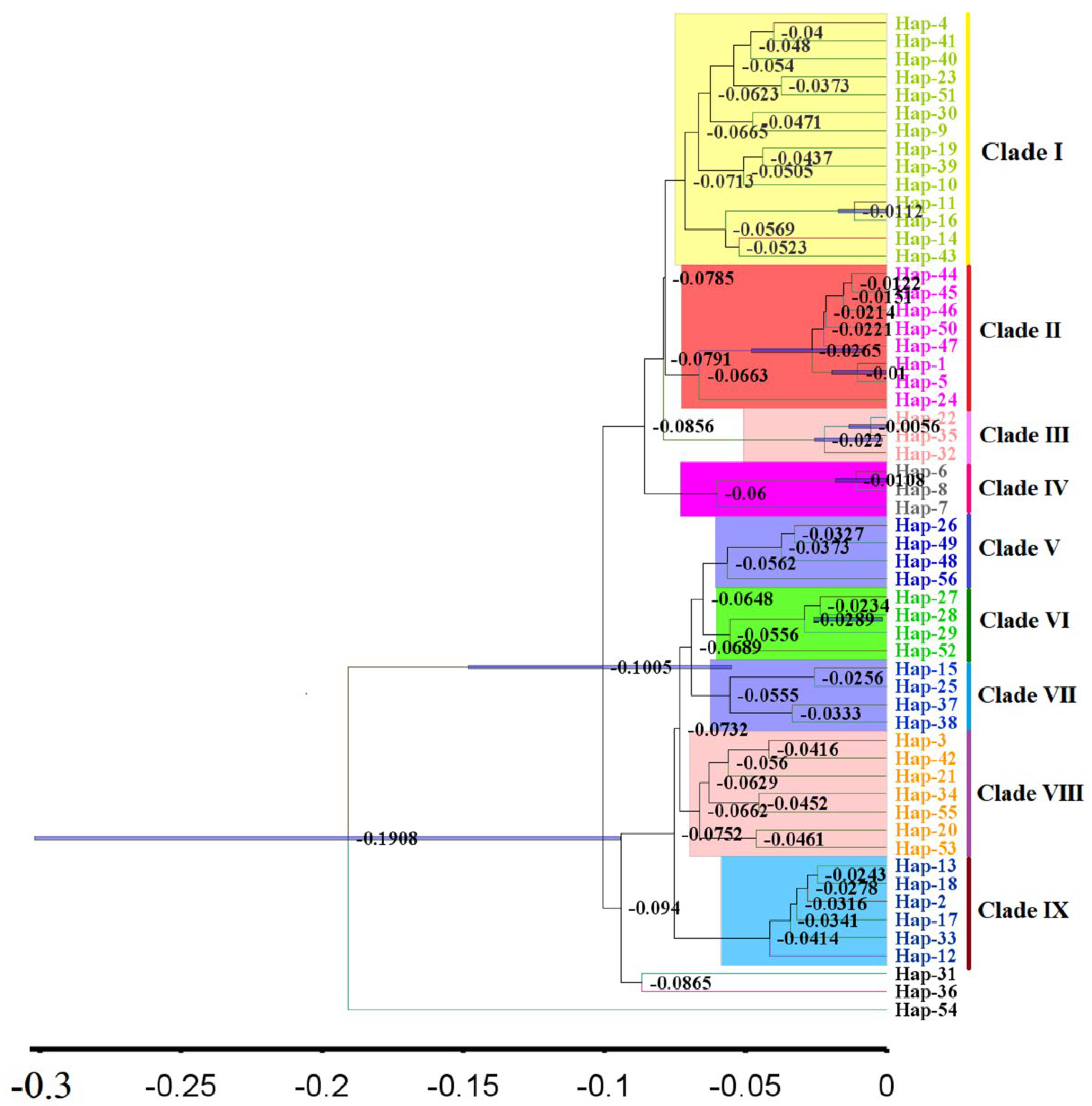
3.4. Phylogenetic and Phylogeographical Analyses
3.5. Isolation by Distance
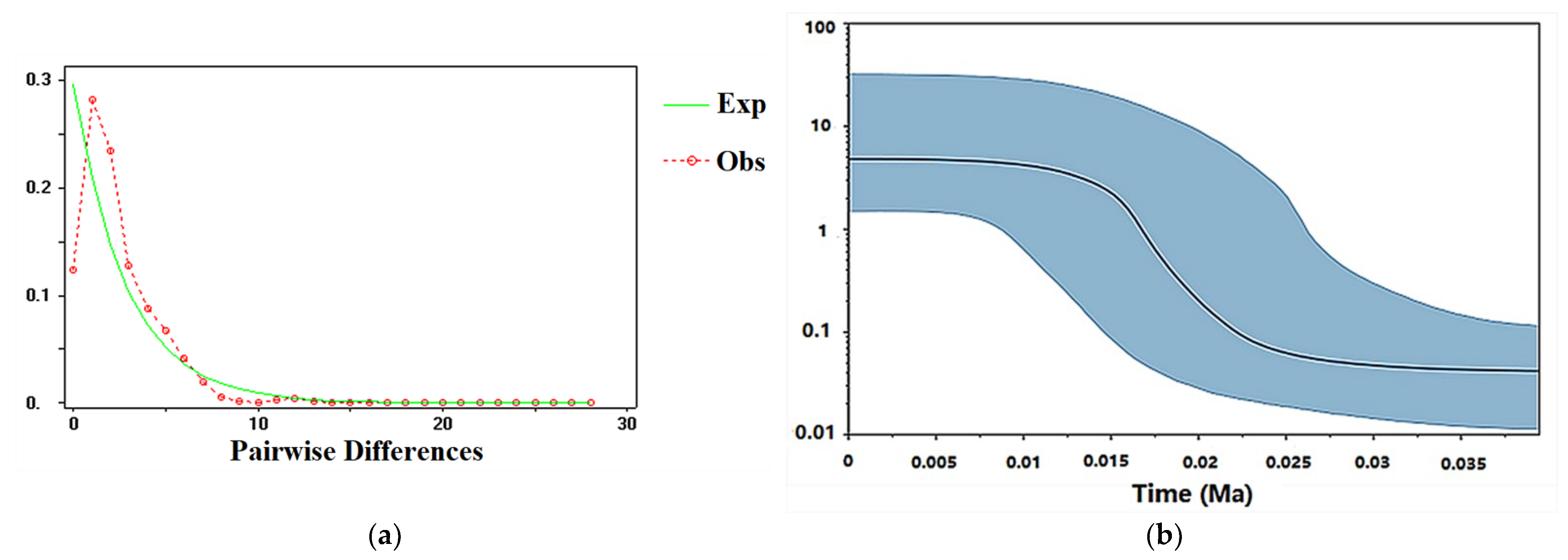
3.6. Intraspecific Divergence Time and Historical Demographic Reconstruction
3.7. Visualization of Dispersal Corridors and Gene Flow Estimation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ling, C.; Yanling, S. The development of biogeography. J. Zool. China 2005, 4, 111–120. [Google Scholar]
- Sanmartín, I. Historical Biogeography: Evolution in Time and Space. Evol. Educ. Outreach 2012, 5, 555–568. [Google Scholar] [CrossRef] [Green Version]
- Avise, J.C. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Hewitt, G.M. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G.M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2004, 359, 183–195. [Google Scholar] [CrossRef] [Green Version]
- Manel, S.; Schwartz, M.K.; Luikart, G.; Taberlet, P. Landscape genetics: Combining landscape ecology and population genetics. Trends Ecol. Evol. 2003, 18, 189–197. [Google Scholar] [CrossRef]
- Liu, Y.; Qiu, Y.; Wang, X.; Yang, H.; Hayashi, M.; Wei, C. Morphological variation, genetic differentiation and phylogeography of the East Asia cicada Hyalessa maculaticollis (Hemiptera: Cicadidae). Syst. Entomol. 2018, 43, 308329. [Google Scholar] [CrossRef]
- Qu, Y.H.; Ericson, P.G.P.; Quan, Q.; Song, G.; Zhang, R.; Gao, B.; Lei, F. Long-term isolation and stability explain high genetic diversity in the Eastern Himalaya. Mol. Ecol. 2014, 23, 705–720. [Google Scholar] [CrossRef]
- Soltis, D.E.; Morris, A.B.; Mclachlan, J.S.; Manos, P.S.; Soltis, P.S. Comparative phylogeography of unglaciated eastern North America. Mol. Ecol. 2006, 15, 4261–4293. [Google Scholar] [CrossRef]
- Brown, J.M.; Abrahamson, W.G.; Way, P.A. Mitochondrial DNA phylogeography of host races of the goldenrod ball gallmaker, Eurosta solidaginis (Diptera: Tephritidae). Evolution 1996, 50, 777–786. [Google Scholar] [CrossRef]
- Austin, J.D.; Lougheed, S.C.; Boag, P.T. Discordant temporal and geographic patterns in maternal lineages of eastern north American frogs, Rana catesbeiana (Ranidae) and Pseudacris crucifer (Hylidae). Mol. Phylogenetics Evol. 2004, 32, 799–816. [Google Scholar] [CrossRef]
- Runck, A.M.; Cook, J.A. Postglacial expansion of the southern red-backed vole (Clethrionomys gapperi) in North America. Mol. Ecol. 2005, 14, 1445–1456. [Google Scholar] [CrossRef] [PubMed]
- Church, S.A.; Kraus, J.M.; Mitchell, J.C.; Church, D.R.; Taylor, D.R. Evidence for multiple Pleistocene refugia in the postglacial expansion of the eastern tiger salamander, Ambystoma tigrinum tigrinum. Evolution 2007, 57, 372–383. [Google Scholar] [CrossRef]
- Zamudio, K.R.; Savage, W.K. Historical isolation, range expansion, and secondary contact of two highly divergent mictochondrial lineages in spotted salamanders (Ambystoma maculatum). Evolution 2003, 57, 1631–1652. [Google Scholar] [CrossRef] [PubMed]
- Bozáňová, J.; Iampor, F.; Mamos, T.; Grabowski, M.; Čiamporová-Zat’ovičová, Z. East Meets West in the Western Carpathians: Contact Zone Between Alpine and Carpathian Caddisfly Lineages Evidenced by COI Barcodes. Res.Sq. 2021. [Google Scholar] [CrossRef]
- Che, J.; Zhou, R.W.; Hu, R.S.; Yan, R.; Papenfuss, R.J.; Wake, R.B.; Zhang, Y.P. Spiny frogs (Paini) illuminate the history of the Himalayan region and Southeast Asia. Proc. Natl. Acad. Sci. USA 2010, 107, 13765–13770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieswetter, C.M.; Schneider, C.J. Phylogeography in the northern Andes: Complex history and cryptic diversity in a cloud forest frog, Pristimantis w-nigrum (Craugastoridae). Mol. Phylogenet. Evol. 2013, 69, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Zhou, K.; Zhang, S.L.; Shu, Y.L.; Zhang, J.H.; Li, E.; Wang, M.S.; Yan, P.; Wu, H.L. Effects of dispersal barriers and geographic distance on the genetic structure of a narrowly distributed frog in a spatially structured landscape. J. Zool. 2019, 309, 295–309. [Google Scholar] [CrossRef]
- Liu, L.; Hao, Z.Z.; Liu, Y.Y.; Wei, X.X.; Cun, Y.Z.; Wang, X.Q. Phylogeography of Pinus armandii and Its Relatives: Heterogeneous Contributions of Geography and Climate Changes to the Genetic Differentiation and Diversification of Chinese White Pines. PLoS ONE 2014, 9, e85920. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.M.; Moeller, M.; Zhang, X.M.; Hollingsworth, M.L.; Liu, J.; Mill, R.R.; Gibby, M.; Li, D.Z. High variation and strong phylogeographic pattern among cpDNA haplotypes in Taxus wallichiana (Taxaceae) in China and North Vietnam. Mol. Ecol. 2007, 16, 4684–4698. [Google Scholar] [CrossRef]
- Yu, D.; Chen, M.; Tang, Q.; Li, X.; Liu, H. Geological events and Pliocene climate fluctuations explain the phylogeographical pattern of the cold water fish Rhynchocypris oxycephalus (Cypriniformes: Cyprinidae) in China. BMC Evol. Biol. 2014, 14, 225. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.T.; Zheng, F.S.; Qin, J.; Lu, M.X.; Du, Y.Z. Genetic Structure of Water Chestnut Beetle: Providing Evidence for Origin of Water Chestnut. PLoS ONE 2016, 11, e0159557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Shi, M.; Chen, X. Population genetic structure of Chilo suppressalis (Walker) (Lepidoptera: Crambidae): Strong subdivision in China inferred from microsatellite markers and mtDNA gene sequences. Mol. Ecol. 2008, 17, 2880–2897. [Google Scholar] [CrossRef] [PubMed]
- Hawlitschek, O.; Hendrich, L.; Espeland, M.; Toussaint, E.F.; Genner, M.J.; Balke, M. Pleistocene climate change promoted rapid diversification of aquatic invertebrates in Southeast Australia. BMC Evol. Biol. 2012, 12, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zhao, M.; Wei, S.; Luo, Z.; Wu, H. Geologic events coupled with Pleistocene climatic oscillations drove genetic variation of Omei treefrog (Rhacophorus omeimontis) in southern China. BMC Evol. Biol. 2015, 15, 289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, P.; Bermike, O.; Bigelow, N.; Brigham-Grette, J.; Velichko, A. Last Interglacial Arctic warmth confirms polar amplification of climate change. Quat. Sci. Rev. 2006, 25, 1383–1400. [Google Scholar]
- Capron, E.; Govin, A.; Stone, E.J.; Masson-Delmotte, V.; Mulitza, S.; Otto-Bliesner, B.L.; Rasmussen, T.L.; Sime, L.C.; Waelbroeck, C.; Wolff, E.W. Temporal and spatial structure of multi-millennial temperature changes at high latitudes during the Last Interglacial. Quat. Sci. Rev. 2014, 103, 116–133. [Google Scholar] [CrossRef] [Green Version]
- Hearty, P.J.; Hollin, J.T.; Neumann, A.C.; O’Leary, M.J.; Mcculloch, M. Global sea-level fluctuations during the last Interglaciation (MIS 5e). Quat. Sci. Rev. 2007, 26, 2090–2112. [Google Scholar] [CrossRef]
- Kopp, R.E.; Simons, F.J.; Mitrovica, J.X.; Maloof, A.C.; Oppenheimer, M. Global and Local Sea Level During the Last Interglacial: A Probabilistic Assessment. Nature 2009, 462, 863–867. [Google Scholar] [CrossRef] [Green Version]
- Köhler, P.; Bintanja, R.; Fischer, H.; Joos, F.; Knutti, R.; Lohmann, G.; Masson-Delmotte, V. What caused Earth’s temperature variations during the last 800,000 years? Data-based evidence on radiative forcing and constraints on climate sensitivity. Quat. Sci. Rev. 2010, 29, 129–145. [Google Scholar] [CrossRef] [Green Version]
- Peltier, W.R. Postglacial coastal evolution: Ice-ocean-solid earth interactions in a period of rapid climate change. Spec. Pap.-Geol. Soc. Am. 2007, 426, 5. [Google Scholar]
- Ye, Z.; Zhu, G.P.; Chen, P.; Zhang, D.; Bu, W. Molecular data and ecological niche modelling reveal the Pleistocene history of a semi-aquatic bug (Microvelia douglasi douglasi) in East Asia. Mol. Ecol. 2014, 23, 3080–3096. [Google Scholar] [CrossRef] [PubMed]
- Sandweiss, D.H.; Maasch, K.A.; Anderson, D.G. Transitions in the mid-holocene. Science 1999, 283, 499–500. [Google Scholar] [CrossRef]
- Mayewski, P.A.; Rohling, E.E.; Stager, J.C.; Karlén, W.; Maasch, K.A.; Meeker, L.D.; Meyerson, E.A.; Gasse, F.; Kreveld, S.V.; Holmgren, K.; et al. Holocene climate variability. Quat. Res. 2004, 62, 243–255. [Google Scholar] [CrossRef]
- Breukelen, M.R.V.; Vonhof, H.B.; Hellstrom, J.C.; Wester, W.C.G.; Kroon, D. Fossil dripwater in stalagmites reveals Holocene temperature and rainfall variation in Amazonia. Earth Planet. Sci. Lett. 2008, 275, 54–60. [Google Scholar] [CrossRef]
- Gomes, V.H.; Mayle, F.E.; Gosling, W.D.; Vieira, I.C.; Salomão, R.P.; Steege, H. Modelling the distribution of Amazonian tree species in response to long-term climate change during the Mid-Late Holocene. J. Biogeogr. 2020, 47, 1530–1540. [Google Scholar] [CrossRef]
- Jiménez-Mejías, P.; Fernández-Mazuecos, M.; Amat, M.E.; Vargas, P. Narrow endemics in European mountains: High genetic diversity within the monospecific genus Pseudomisopates (Plantaginaceae) despite isolation since the late Pleistocene. J. Biogeogr. 2015, 42, 1455–1468. [Google Scholar] [CrossRef]
- Zheng, H.X.; Yan, Y.; Wang, H.; Bai, P.; Zhang, X. Sex-biased olfactory gene expression in the antennae of the adzuki bean beetle, Callosobruchus chinensis. Bull. Insectol. 2021, 74, 115–122. [Google Scholar]
- Yamane, T. Biorational control methods for protection of stored grain legumes against bruchid beetles. Agric. Sci. 2013, 4, 762–766. [Google Scholar] [CrossRef] [Green Version]
- Mainali, B.P.; Kim, H.J.; Park, C.G.; Yoon, Y.N.; Lee, Y.H.; Park, I.H.; Kang, H.W.; Bae, S.D. Interactive effects of temperature and relative humidity on oviposition and development of Callosobruchus chinensis (L.) on azuki bean. J. Stored Prod. Res. 2015, 63, 47–50. [Google Scholar] [CrossRef]
- Cheng, J.X. The spread ways and control measures of insect pests during seed storage. Agric. Tech. Serv. 2009, 12, 1. [Google Scholar]
- Wang, Y.Z.; Li, B.Y.; Hoffmann, A.A.; Cao, L.J.; Gong, Y.J.; Song, W.; Zhu, J.Y.; Wei, S.J. Patterns of genetic variation among geographic and host-plant associated populations of the peach fruit moth Carposina sasakii (Lepidoptera: Carposinidae). BMC Evol. Biol. 2017, 17, 265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuda, M.; Wasano, N.; Kondo, N.; Horng, S.B.; Chou, L.Y.; Tateishi, Y. Habitat-related mtDNA polymorphism in the stored-bean pest Callosobruchus chinensis (Coleoptera: Bruchidae). Bull. Entomol. Res. 2004, 94, 75–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, C.X.; Li, W.C.; Zhu, Z.D.; Li, D.D.; Sun, S.L.; Wang, X.M. Genetic differentiation and diversity of Callosobruchus chinensis collections from China. Bull. Entomol. Res. 2016, 106, 124–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Yang, H.; Ding, H.; Yang, S.; Wang, P. Population genetic structure of the land snail Camaena cicatricosa (Stylommatophora, Camaenidae) in China inferred from mitochondrial genes and ITS2 sequences. Sci. Rep. 2017, 7, 15590. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit, Version 7.1.7. Available online: http://www.mbio.ncsu.edu/bioedit/bioedit.html (accessed on 25 November 2012).
- Sudhir, K.; Glen, S.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar]
- Martins, J.; Solomon, S.E.; Mikheyev, A.S.; Mueller, U.G.; Ortiz, A.; Bacci, M. Nuclear mitochondrial-like sequences in ants: Evidence from Atta cephalotes (Formicidae: Attini). Insect Mol. Biol. 2007, 16, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Cristiano, M.P.; Fernandes-Salomăo, T.M.; Yotoko, K.S. Nuclear mitochondrial DNA: An Achilles’ heel of molecular systematics, phylogenetics, and phylogeographic studies of stingless bees. Apidologie 2012, 43, 527–538. [Google Scholar] [CrossRef] [Green Version]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.L.; Bennett, J.R.; French, C.M. SDMtoolbox 2.0: The next generation Python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. PeerJ 2017, 5, e4095. [Google Scholar] [CrossRef] [Green Version]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under linux and windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GENALEX 6, genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Cheng, L.; Connor, T.R.; Siren, J.; Aanensen, D.M.; Corander, J. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol. Biol. Evol. 2013, 30, 1224–1228. [Google Scholar] [CrossRef]
- Manni, F.; Guérard, E.; Heyer, E. Geographic patterns of (genetic, morphologic, linguistic) variation: How barriers can be detected by “Monmonier’s algorithm”. Hum. Biol. 2004, 76, 173–190. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchene, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; Drummond, A.J.; et al. BEAST 2.5: An Advanced Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [Green Version]
- Hidalgo-Galiana, A.; Ribera, I. Late Miocene diversification of the genus Hydrochus (Coleoptera, Hydrochidae) in the west Mediterranean area. Mol. Phylogenet. Evol. 2011, 59, 377–385. [Google Scholar] [CrossRef]
- Pons, J.; Ribera, I.; Bertranpetit, J.; Balke, M. Nucleotide substitution rates for the full set of mitochondrial protein-coding genes in Coleoptera. Mol. Phylogenet. Evol. 2010, 56, 796–807. [Google Scholar] [CrossRef] [Green Version]
- Ribera, I.; Faille, A. A new microphthalmic stygobitic Graptodytes Seidlitz from Morocco, with a molecular phylogeny of the genus. Zootaxa 2010, 2641, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Brower, A.V. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial-DNA evolution. Proc. Natl. Acad. Sci. USA 1994, 91, 6491–6495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribera, I.; Fresneda, J.; Bucur, R.; Izquierdo, A.; Vogler, A.P.; Salgado, J.M.; Cieslak, A. Ancient origin of a Western Mediterranean radiation of subterranean beetles. BMC Evol. Biol. 2010, 10, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadopoulou, A.; Anastasiou, I.; Keskin, B.; Vogler, A.P. Comparative phylogeography of tenebrionid beetles in the Aegean archipelago: The effect of dispersal ability and habitat preference. Mol. Ecol. 2009, 18, 2503–2517. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [Green Version]
- Kumar, L.S.; Singh, J. Population genetic structure of banana corm weevil Cosmopolites sordidus (Germar) in India. J. Asia-Pac. Entomol. 2018, 21, 1222–1232. [Google Scholar] [CrossRef]
- Mama Racky, N.; Cheikh, T.; Sembene, M. Haplotype Structure and Phylogeographic Evolution of West African Populations of Sitophilus zeamais (Coleoptera, Curculionidae). J. Stored Prod. Res. 2017, 5, 3. [Google Scholar] [CrossRef]
- Kébé, K.; Alvarez, N.; Tuda, M.; Arnqvist, G.; Fox, C.W.; Sembène, M.; Espíndola, A. Global phylogeography of the insect pest Callosobruchus maculatus (Coleoptera:Bruchinae) relates to the history of its main host, Vigna unguiculata. J. Biogeogr. 2017, 44, 2515–2526. [Google Scholar] [CrossRef]
- Ye, Z.; Yuan, J.; Li, M.; Damgaard, J.; Chen, P.P.; Zheng, C.Z.; Yu, H.B.; Fu, S.Y.; Bu, W.J. Geological effects influence population genetic connectivity more than Pleistocene glaciations in the water strider Metrocoris sichuanensis (Insecta: Hemiptera: Gerridae). J. Biogeogr. 2018, 45, 690–701. [Google Scholar] [CrossRef]
- Hong, B. Genetic Structure, Suitability and Demographic History of Scythropus yasumatsui Populations in China. Ph.D Thesis, Northwest A&F University, Shaanxi, China, 2019. [Google Scholar]
- Tsafack, N.; Xie, Y.Z.; Wang, X.P.; Fattorini, S. Influence of Climate and Local Habitat Characteristics on Carabid Beetle Abundance and Diversity in Northern Chinese Steppes. Insects 2020, 11, 19. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Wang, Q.X.; Chang, Q.; Xiang, J.I.; Zhou, K. The divergence of two independent lineages of an endemic Chinese gecko, Gekko swinhonis, launched by the Qinling orogenic belt. Mol. Ecol. 2010, 19, 2490–2500. [Google Scholar] [CrossRef]
- Huang, Z.S.; Yu, F.L.; Gong, H.S.; Song, Y.L.; Zeng, Z.G.; Zhang, Q. Phylogeographical structure and demographic expansion in the endemic alpine stream salamander (Hynobiidae: Batrachuperus) of the Qinling Mountains. Sci. Rep. 2017, 7, 1871. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.H.; Cheng, F.Y.; Zhou, S.L. Genetic Structure of the Tree Peony (Paeonia rockii) and the Qinling Mountains as a Geographic Barrier Driving the Fragmentation of a Large Population. PLoS ONE 2012, 7, e34955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.Y.; Guo, J.J.; Liu, N.; Zhang, R.Z. Genetic structure of the invasive Colorado potato beetle Leptinotarsa decemlineata populations in China. J. Integr. Agric. 2020, 19, 350–359. [Google Scholar] [CrossRef]
- Liu, Y.X.; Dietrich, C.H.; Wei, C. Genetic divergence, population differentiation and phylogeography of the cicada Subpsaltria yangi based on molecular and acoustic data: An example of the early stage of speciation? BMC Evol. Biol. 2019, 19, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.X.; Yang, Z.F.; Zhang, G.Y.; Yu, Q.Q.; Wei, C. Cicada parasitic moths from China (Lepidoptera: Epipyropidae): Morphology, identity, biology, and biogeography. Syst. Biodivers. 2018, 16, 417–427. [Google Scholar] [CrossRef]
- Zhang, B.J.; Wu, P.; Zhao, X.N.; Wang, Y.B.; Gao, X.D. Changes in vegetation condition in areas with different gradients (1980–2010) on the Loess Plateau, China. Environ. Earth Sci. 2012, 68, 2427–2438. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, Y.Q.; Luo, L.Z.; Jin, M.; Gong, Z.J.; Jiang, Y.L.; Li, T. Genetic diversity and population structure of Sitodiplosis mosellana in northern China. PLoS ONE 2013, 8, e78415. [Google Scholar] [CrossRef] [Green Version]
- Fang, F.; Ji, Y.; Zhao, Q.; Wang, Y.; Gao, W.; Chu, K.; Sun, H. Phylogeography of the Chinese endemic freshwater crab Sinopotamon acutum (Brachyura, Potamidae). Zool. Scr. 2015, 44, 653–666. [Google Scholar] [CrossRef]
- Hu, L.J.; Uchiyama, K.; Shen, H.L.; Saito, Y.; Tsuda, Y.; Ide, Y. Nuclear DNA Microsatellites Reveal Genetic Variation but a Lack of Phylogeographical Structure in an Endangered Species, Fraxinus mandshurica, Across North-east China. Ann. Bot. 2008, 102, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Qu, Y.H.; Zhang, R.Y.; Quan, Q.; Song, G.; Li, S.H.; Lei, F.M. Incomplete lineage sorting or secondary admixture: Disentangling historical divergence from recent gene flow in the Vinous-throated parrotbill (Paradoxornis webbianus). Mol. Ecol. 2012, 21, 6117–6133. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Liu, R.R.; Wang, L.Y.; Qiu, Q.; Chen, K.M.; Liu, J.Q. Phylogeographic analyses suggest that deciduous species (Ostryopsis davidiana Decne., Betulaceae) survived in northern China during the Last Glacial Maximum. J. Biogeogr. 2009, 36, 2148–2155. [Google Scholar] [CrossRef]
- Gilles, J.; Litrico, I.; Tillard, E.; Duvallet, G. Genetic Structure and Gene Flow Along an Altitudinal Gradient Among Two Stomoxyine species (Diptera: Muscidae) on La Réunion Island. J. Med. Entomol. 2007, 44, 433–439. [Google Scholar] [CrossRef]
- Li, J.J.; Huang, S.H.; Liu, Z.Y.; Bi, Y.X. Climate-Driven Range Shifts of Brown Seaweed Sargassum horneri in the Northwest Pacific. Front. Mar. Sci. 2020, 7, 570881. [Google Scholar] [CrossRef]
- Engelhardt, E.K.; Neuschulz, E.L.; Hof, C. Ignoring biotic interactions overestimates climate change effects: The potential response of the spotted nutcracker to changes in climate and resource plants. J. Biogeogr. 2020, 47, 143–154. [Google Scholar] [CrossRef] [Green Version]
- Maharjan, R.; Yi, H.; Young, Y.; Jang, Y.; Kim, Y.; Bae, S. Effects of low temperatures on the survival and development of Callosobruchus chinensis (L.) (Coleoptera: Bruchidae) under different storage durations. J. Asia-Pac. Entomol. 2017, 20, 893–900. [Google Scholar] [CrossRef]
- Maharjan, R.; Ahn, J.; Park, C.; Yoon, Y.; Jang, Y.; Kang, H.; Bae, S. Effects of temperature on development of the Azuki bean weevil, Callosobruchus chinensis (Coleoptera: Bruchidae) on two leguminous seeds. J. Stored Prod. Res. 2017, 72, 90–99. [Google Scholar] [CrossRef]
- Sanjo, J.V.; Nameer, P.O. The expanding distribution of the Indian Peafowl (Pavo cristatus) as an indicator of changing climate in Kerala, southern India: A modelling study using MaxEnt. Ecol. Indic. 2020, 110, 105930. [Google Scholar]
- Li, X.; Xu, D.; Jin, Y.; Zhuo, Z.; Wang, R. Predicting the current and future distributions of Brontispa longissima (Coleoptera: Chrysomelidae) under climate change in China. Glob. Ecol. Conserv. 2020, 25, e01444. [Google Scholar] [CrossRef]
- Qu, Y.H.; Luo, X.; Zhang, R.Y.; Song, G.; Zou, F.S.; Lei, F.M. Lineage diversification and historical demography of a montane bird Garrulax elliotii—Implications for the Pleistocene evolutionary history of the eastern Himalayas. BMC Evol. Biol. 2011, 11, 174. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.H.; Wang, I.J.; Comes, H.P.; Peng, H.; Qiu, Y.X. Contributions of historical and contemporary geographic and environmental factors to phylogeographic structure in a Tertiary relict species, Emmenopterys henryi (Rubiaceae). Sci. Rep. 2016, 6, 24041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, S.J.; Cao, L.J.; Gong, Y.J.; Shi, B.C.; Wang, S.; Zhang, F.; Guo, X.J.; Wang, Y.M.; Chen, X.X. Population genetic structure and approximate Bayesian computation analyses reveal the southern origin and northward dispersal of the oriental fruit moth Grapholita molesta (Lepidoptera: Tortricidae) in its native range. Mol. Ecol. 2015, 24, 4094–4111. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Yuan, J.J.; Zhen, Y.H.; Damgaard, J.; Yamada, K.; Zhu, X.X.; Jiang, K.; Yang, X.; Wang, W.W.; Bu, W.J.; et al. Local environmental selection and lineage admixture act as significant mechanisms in the adaptation of the widespread East Asian pond skater Gerris latiabdominis to heterogeneous landscapes. J. Biogeogr. 2020, 47, 1154–1165. [Google Scholar] [CrossRef]
- Li, J.; Cao, J.J.; Niu, J.Q.; Liu, X.X.; Zhang, Q.W. Identification of the population structure of Myzus persicae (Hemiptera: Aphididae) on peach trees in China using microsatellites. J. Insect Sci. 2015, 15, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fountain, T.; Duvaux, L.; Horsburgh, G.; Reinhardt, K.; Butlin, R.K. Human-facilitated metapopulation dynamics in an emerging pest species, cimex lectularius. Mol. Ecol. 2014, 23, 1071–1084. [Google Scholar] [CrossRef] [Green Version]
- Thomas, E.; van Zonneveld, M.; Loo, J.; Hodgkin, T.; Galluzzi, G.; van Etten, J. Present spatial diversity patterns of Theobroma cacao L. in the Neotropics reflect genetic differentiation in Pleistocene refugia followed by human-influenced dispersal. PLoS ONE 2012, 7, e47676. [Google Scholar] [CrossRef] [Green Version]
- Cui, S.F.; Wang, L.; Qiu, J.P.; Geng, X.Q.; Liu, Z.C. Effects of hypoxia/hypercapnia on the metablism of Callosobruchus chinensis (L.) larvae. J. Stored Prod. Res. 2019, 83, 322–330. [Google Scholar] [CrossRef]
- Damte, T.; Mitiku, G. Pattern of egg distribution by Adzuki bean beetle, Callosobruchus chinensis (L.) (Coleoptera: Chysomelidae) in stored chickpea under natural infestation. J. Stored Prod. Res. 2020, 88, 101683. [Google Scholar] [CrossRef]
- Li, W.Z.; An, J.J.; Fu, G.X.; Wang, Y.H.; Chen, Z.S.; You, X.F.; Yuan, G.H. Repellent Effect of 12 Kinds of Flavoring Powder and Agar on Callosobruchus chinensis Adults. J. Henan Univ. Technol. (China) 2008, 29, 4. [Google Scholar]
- Chen, F.J. Occurrence and Integrated Control of Callosobruchus chinensis in Mountainous Area of Southwest Zhejiang province. Hubei Plant Prot. 2003, 6, 8. [Google Scholar]
- Ou, J. Determination and Analysis of the Complete Mitochondrial Genome of Six Stored Grain Beetles. Ph.D. Dissertation, Shaanxi Normal University, Xi’an, China, 2015. [Google Scholar]
- Wang, Y.; Talekar, N.S.; Chen, B.; Li, Z.Y.; Chen, Z.L. Study on the Influence of Legume Varieties on the Growth and Oviposition Preference of Callosobruchus chinensis (L.). J. Yunnan Agric. Univ. 2010, 1, 6. [Google Scholar]
- Tang, Y.S.; Jiang, Y.H.; Zheng, Y.K. Studies on the Selection of Germplasm Resources Phenotype Varieties of Broad Bean and Pea Resistance to Callosobruchus chinensis. Yunnan Nongye Keji 2014, 5, 4. [Google Scholar]
- Wang, Q. Genetic Differentiation of Callosobruchus chinensis (L.) among Different Geographical Populations. Ph.D Dissertation, Southwest University, Chongqing, China, 2008. [Google Scholar]
- Xu, J.J. Molecular Detection of Different Geographic Populations and Interspecific Relationships of Several Important Bean Wee-vil Species. Ph.D. Dissertation, University of Yangzhou, Yangzhou, China, 2007. [Google Scholar]
- Deng, Y.X. Effects of Environmental Factors on Four Kinds of Stored Product Insect Pests. Ph.D. Dissertation, Southwest Agricultural University, Chongqing, China, 2001. [Google Scholar]
- Deng, Y.X.; Zhu, W.B.; Li, L.S. Preliminary Study on Geographic Population of Callosobruchus chinensis (L.). Grain Storage 1989, 03, 55. [Google Scholar]
- Song, Y.F. Study on Tolerance of Callosobruchus chinensis (Linnaeus) Adults to Low Temperature and Its Physiological and Biochemical Mechanisms. Ph.D. Dissertation, Southwest University, Chongqing, China, 2017. [Google Scholar]
- Leng, T.R.; Yin, Z.C.; Bu, R.; Hao, X.Y.; Gao, X.M.; Yin, F.X. The Harm Factors Detection of Callosobruchus chinensis and Resistance Estimation of Mung Bean Varieties to Callosobruchus chinensis. Liaoning Agric. Sci. 2016, 3, 24–27. [Google Scholar]


| Gene | Primers | Annealing Temperature | Length (bp) |
|---|---|---|---|
| COI | F: AATAAATGATTATTTTCCACTAATCATAAAGACATCGGGA | 56 °C | 1533 |
| R: TTAATTTGTTAGTAGGGGTAATTCGGAGTATCTATG | |||
| COII | F: ATTTTTACTTGAAAAACAATTCTTCTTCAAGAC | 62 °C | 688 |
| R: AAATTTTGATTATTTTAGAAATTCATTTAATAAAATAATTAGGAGT | |||
| Cyt-b | F: ATGAAAATAAATTTTCGAAAAACCCACC | 56 °C | 1140 |
| R: TTAGTGGTAAATGATTTTATCTCATATTTTGTATAAAATTGA | |||
| 12S rRNA | F: AAAAAATTTTATTTTGGTTATTTAATTAGATTTTTCTTGGT | 62 °C | 752 |
| R: GTCTTTCTAGGCACACTTTCCAG |
| Population Code | N | Hap | S | Hd ± SD | Pi ± SD | Tajima’s D | Fu’s Fs |
|---|---|---|---|---|---|---|---|
| SJT | 10 | Hap-1(5), Hap-2(1), Hap-3(1), Hap-4(2), Hap-5(1); | 6 | 0.756 ± 0.130 | 0.00084 ± 0.00017 | 0.45768 | −0.23033 |
| SCQ | 10 | Hap-3(3), Hap-4(2), Hap-6(1), Hap-7(1), Hap-8(1); | 4 | 0.857 ± 0.108 | 0.00056 ± 0.00013 | 0.08124 | −1.69431 |
| SCW | 14 | Hap-4(1), Hap-9(1), Hap-10(1); | 2 | 1.000 ± 0.272 | 0.00048 ± 0.00016 | 0 | −1.2164 |
| SLZ | 10 | Hap-4(6), Hap-11(1), Hap-12(1), Hap-13(1), Hap-14(1); | 10 | 0.667 ± 0.163 | 0.00082 ± 0.00023 | −1.53448 | −0.27358 |
| SXX | 10 | Hap-2(5), Hap-4(1), Hap-15(2), Hap-16(1), Hap-17(1); | 6 | 0.756 ± 0.130 | 0.00066 ± 0.00017 | −0.53927 | −0.78721 |
| SXH | 10 | Hap-1(1), Hap-2(5), Hap-4(3), Hap-18(1); | 6 | 0.711 ± 0.117 | 0.00067 ± 0.00019 | −0.49593 | 0.44029 |
| SYQ | 10 | Hap-3(5), Hap-4(1), Hap-19(1), Hap-20(1); | 3 | 0.643 ± 0.184 | 0.00033 ± 0.00012 | −0.81246 | −1.38724 |
| SSY | 10 | Hap-1(2), Hap-3(1), Hap-4(5), Hap-21(1), Hap-22(1), Hap-23(1); | 8 | 0.800 ± 0.114 | 0.00078 ± 0.00019 | −0.8324 | −1.33144 |
| SD1 | 10 | Hap-3(2), Hap-4(4), Hap-22(4), Hap-24(1); | 5 | 0.764 ± 0.083 | 0.00073 ± 0.00011 | 0.73905 | 0.80985 |
| SD2 | 10 | Hap-3(1), Hap-15(1), Hap-25(1); | 2 | 1.000 ± 0.272 | 0.00048 ± 0.00016 | 0 | −1.2164 |
| HL | 13 | Hap-26(6), Hap-27(5), Hap-28(1), Hap-29(1); | 4 | 0.679 ± 0.089 | 0.00049 ± 0.00007 | 0.25198 | 0.19892 |
| HB1 | 12 | Hap-3(4), Hap-10(1), Hap-19(1), Hap-30(2); | 4 | 0.750 ± 0.139 | 0.00054 ± 0.00010 | −0.12075 | −0.42156 |
| HB2 | 17 | Hap-3(8), Hap-4(1), Hap-20(1), Hap-31(1); | 4 | 0.491 ± 0.175 | 0.00026 ± 0.00011 | −1.71166 | −1.4146 |
| SW | 14 | Hap-3(4), Hap-4(1), Hap-15(2), Hap-32(1), Hap33(1), Hap34(1); | 6 | 0.844 ± 0.103 | 0.00054 ± 0.00013 | −1.18946 | −2.60454 |
| JX | 16 | Hap-3(5), Hap-6(1), Hap-15(2), Hap-35(1), Hap-36(1); | 11 | 0.756 ± 0.130 | 0.00084 ± 0.00034 | −1.76515 | −0.23033 |
| AH | 11 | Hap-4(5), Hap-37(3), Hap-38(1), Hap-39(1), Hap-40(1), Hap-41(1); | 5 | 0.803 ± 0.096 | 0.00057 ± 0.00010 | −0.11051 | −1.92425 |
| TJ | 12 | Hap-3(5), Hap-4(4), Hap-42(1), Hap-43(1); | 3 | 0.709 ± 0.099 | 0.00032 ± 0.00007 | −0.38482 | −0.93979 |
| NM | 12 | Hap-3(1), Hap-44(7), Hap-45(2), Hap-46(1), Hap-47(1); | 9 | 0.667 ± 0.141 | 0.00058 ± 0.00024 | −1.83035 | −0.69537 |
| SC | 11 | Hap-3(2), Hap-9(3), Hap-26(3), Hap-48(1), Hap-49(1); | 5 | 0.844 ± 0.080 | 0.00067 ± 0.00011 | 0.27556 | −0.73268 |
| HJ | 23 | Hap-3(8), Hap-30(1), Hap-50(1), Hap-51(1), Hap-52(1); | 7 | 0.576 ± 0.163 | 0.00055 ± 0.00022 | −1.3042 | −0.83145 |
| JS | 14 | Hap-3(13), Hap-33(2), Hap-53(4), Hap-54(1), Hap-55(1); | 14 | 0.595 ± 0.108 | 0.00065 ± 0.00033 | −2.04699 | 0.51876 |
| GZ | 14 | Hap-4(8), Hap-41(1), Hap-56(1); | 3 | 0.378 ± 0.181 | 0.00021 ± 0.00012 | −1.56222 | −0.45861 |
| ALL | 224 | 58 | 0.876 ± 0.016 | 0.00085 ± 0.00006 | −2.24496 | −26.7717 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Li, M.; Zheng, H.; Dong, T.; Zhang, X. A Phylogeographical Analysis of the Beetle Pest Species Callosobruchus chinensis (Linnaeus, 1758) in China. Insects 2022, 13, 145. https://doi.org/10.3390/insects13020145
Wang F, Li M, Zheng H, Dong T, Zhang X. A Phylogeographical Analysis of the Beetle Pest Species Callosobruchus chinensis (Linnaeus, 1758) in China. Insects. 2022; 13(2):145. https://doi.org/10.3390/insects13020145
Chicago/Turabian StyleWang, Fang, Min Li, Haixia Zheng, Tian Dong, and Xianhong Zhang. 2022. "A Phylogeographical Analysis of the Beetle Pest Species Callosobruchus chinensis (Linnaeus, 1758) in China" Insects 13, no. 2: 145. https://doi.org/10.3390/insects13020145
APA StyleWang, F., Li, M., Zheng, H., Dong, T., & Zhang, X. (2022). A Phylogeographical Analysis of the Beetle Pest Species Callosobruchus chinensis (Linnaeus, 1758) in China. Insects, 13(2), 145. https://doi.org/10.3390/insects13020145





