Diagnostic Value of Artificial Intelligence-Assisted Endoscopic Ultrasound for Pancreatic Cancer: A Systematic Review and Meta-Analysis
Abstract
:1. Introduction
2. Materials and Methods
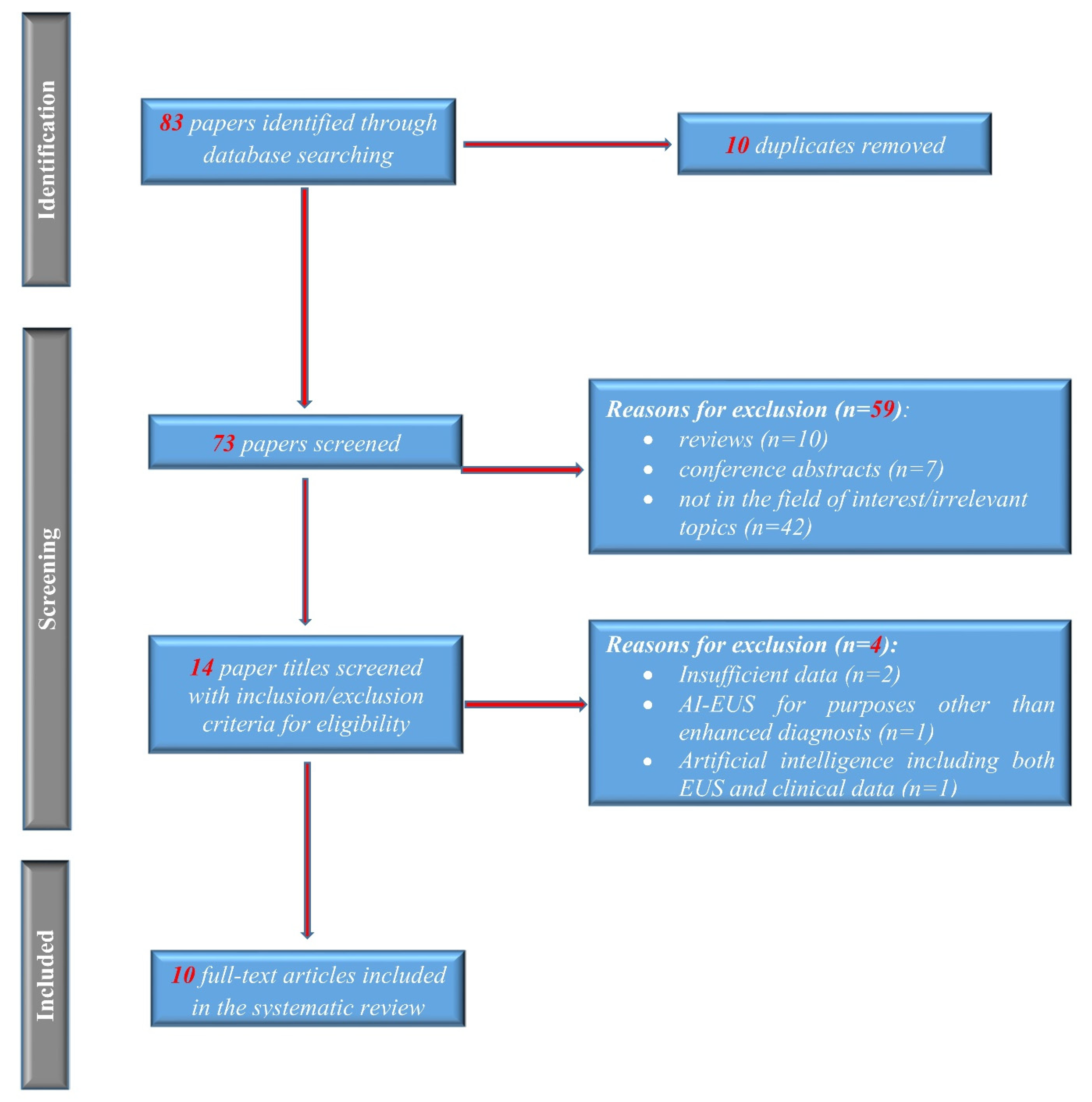
2.1. Search Strategy and Study Selection
2.2. Inclusion and Exclusion Criteria
2.3. Data Extraction
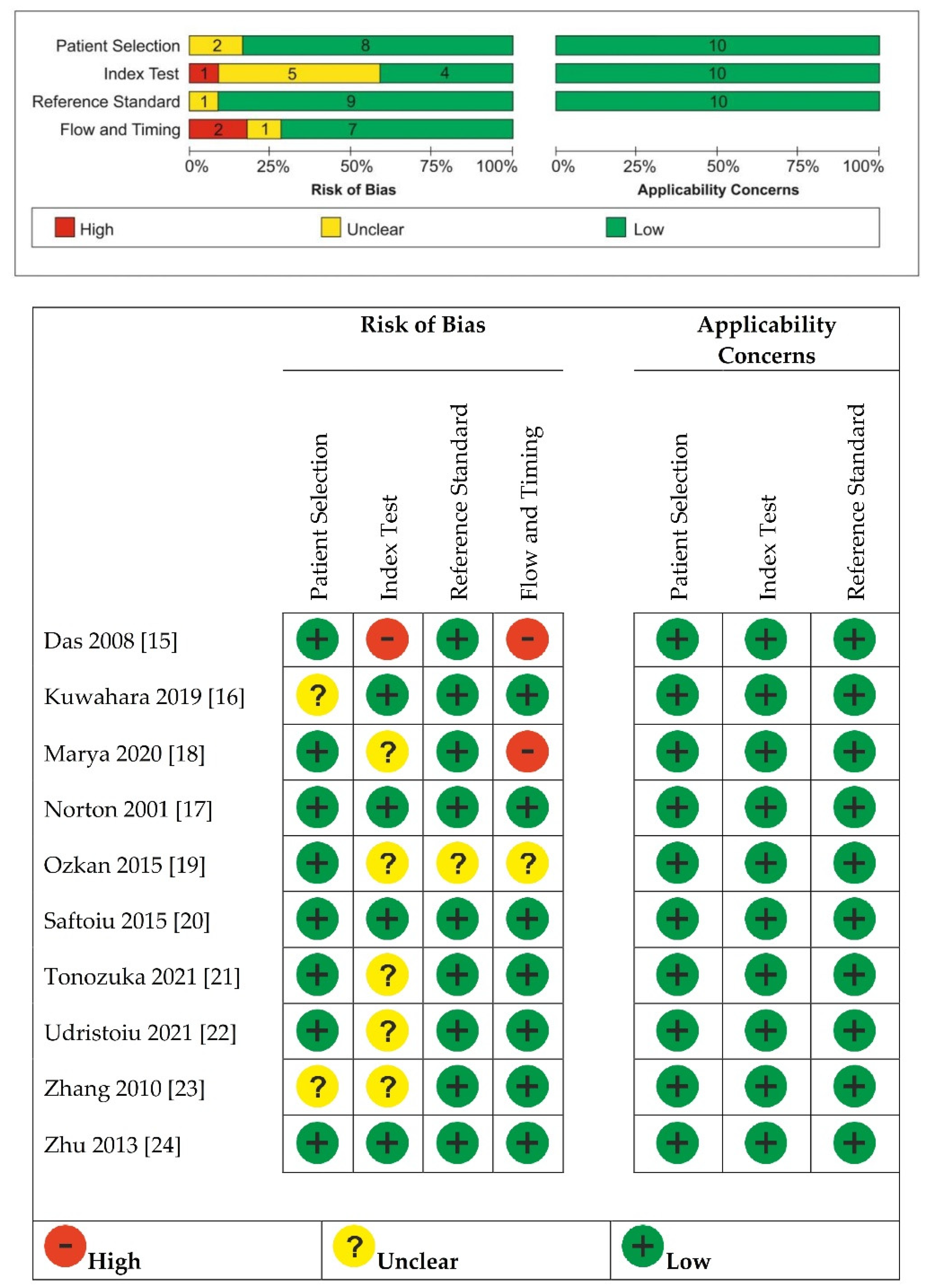
2.4. Quality Assessment of the Studies
2.5. Statistical Methods
3. Results
3.1. Electronic Search Results and Study Characteristics
3.2. Quality of Included Studies
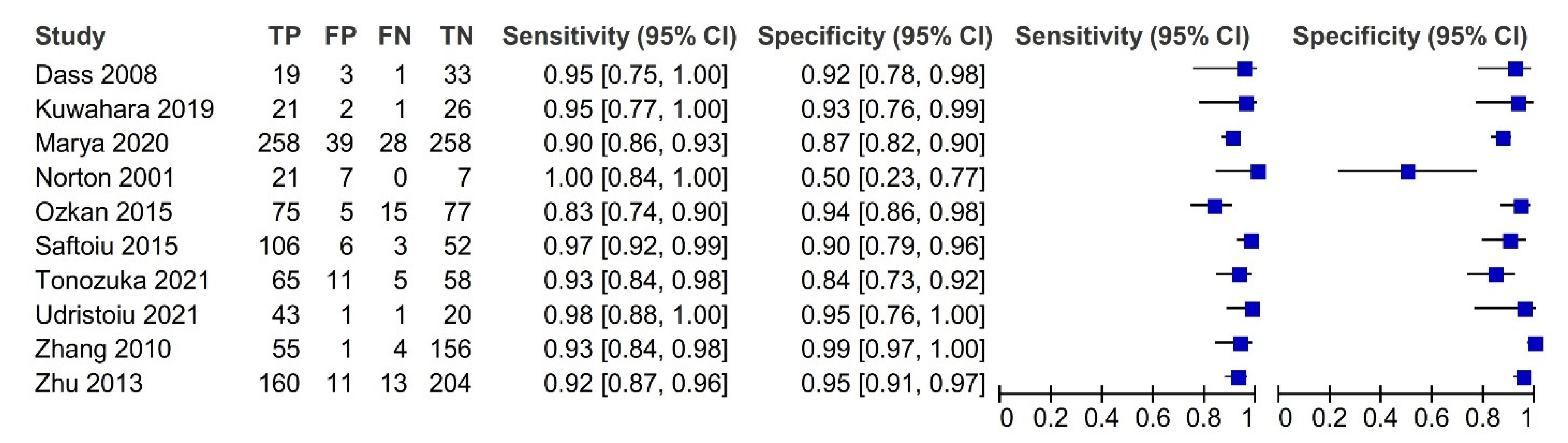
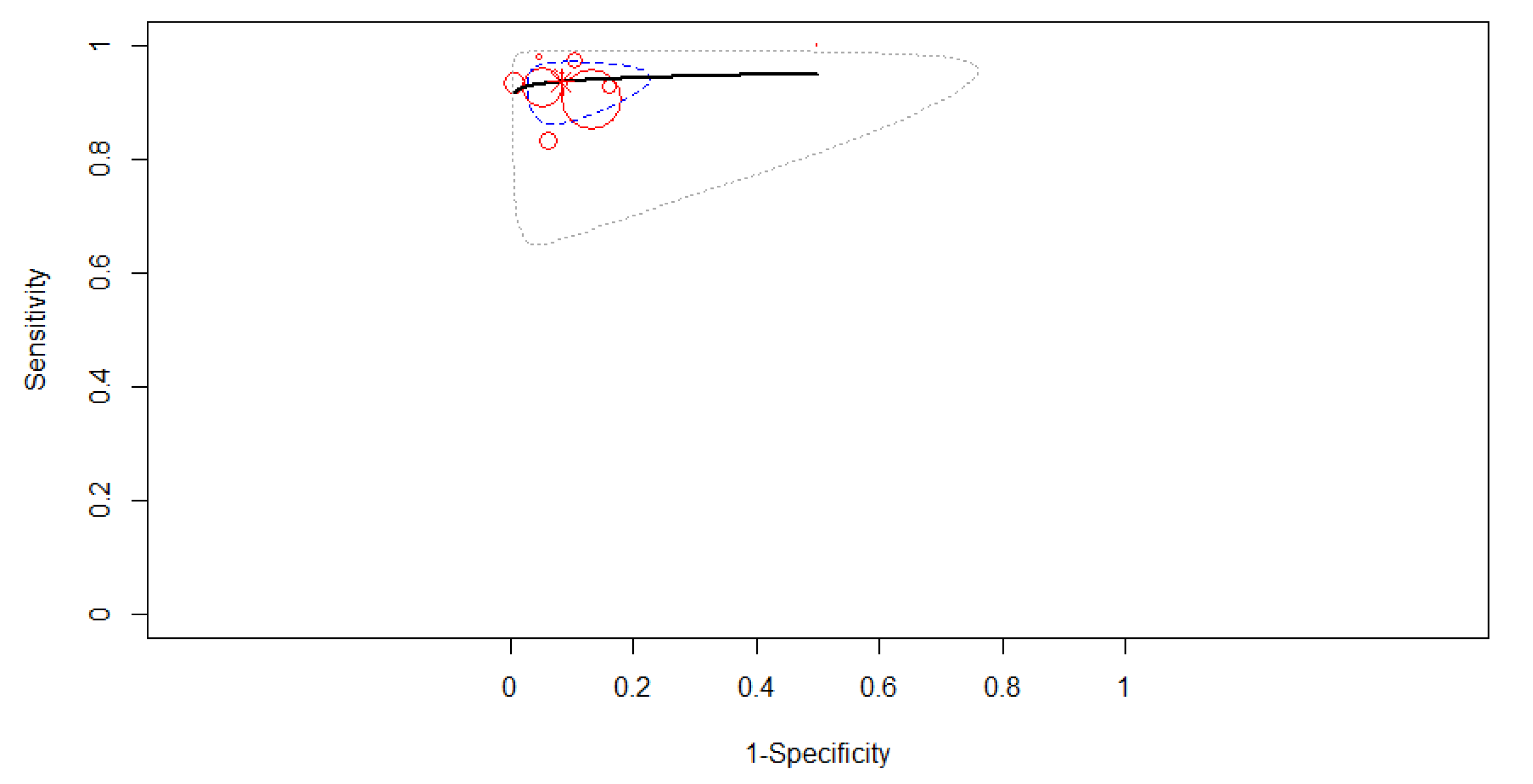
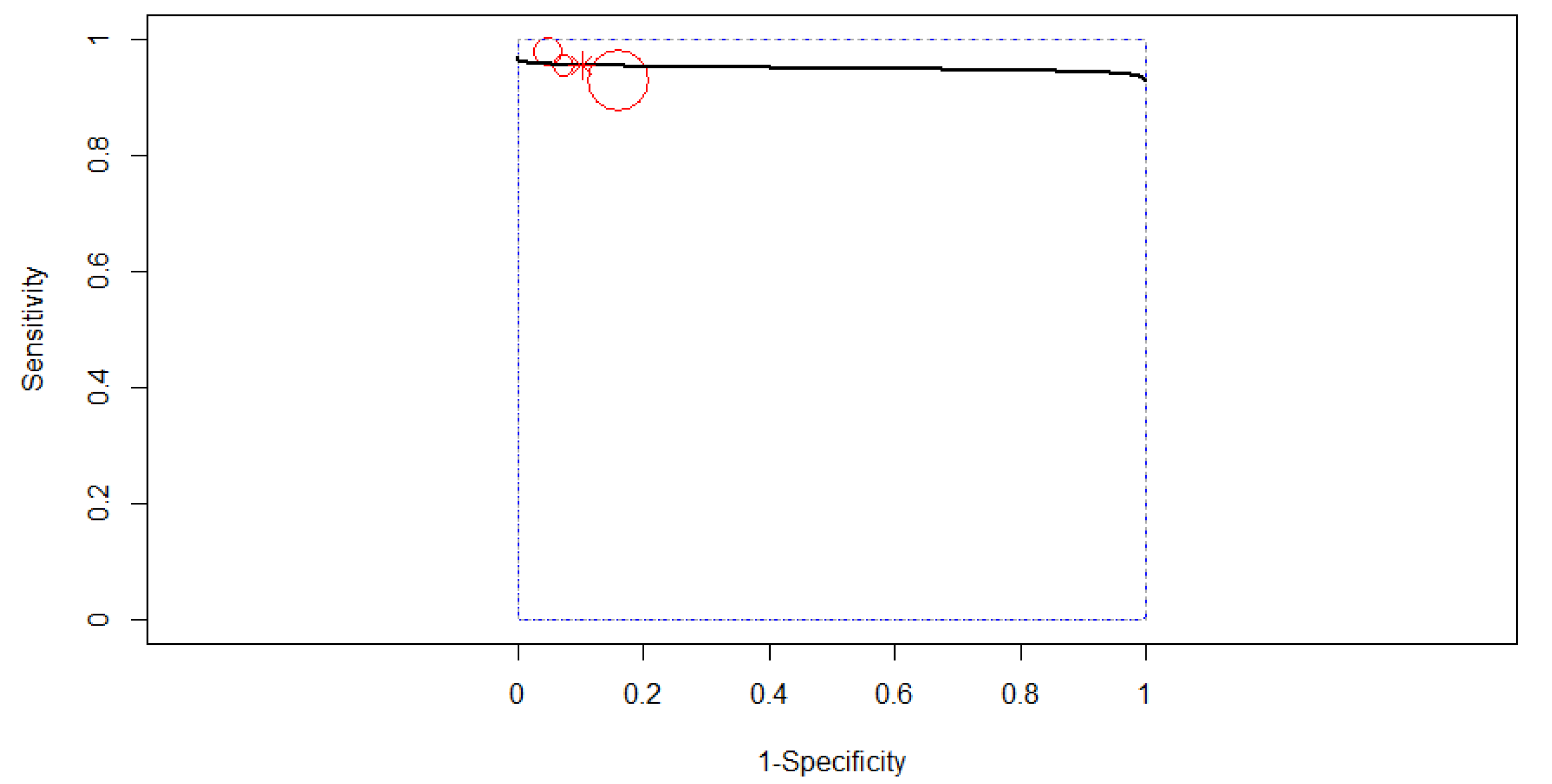
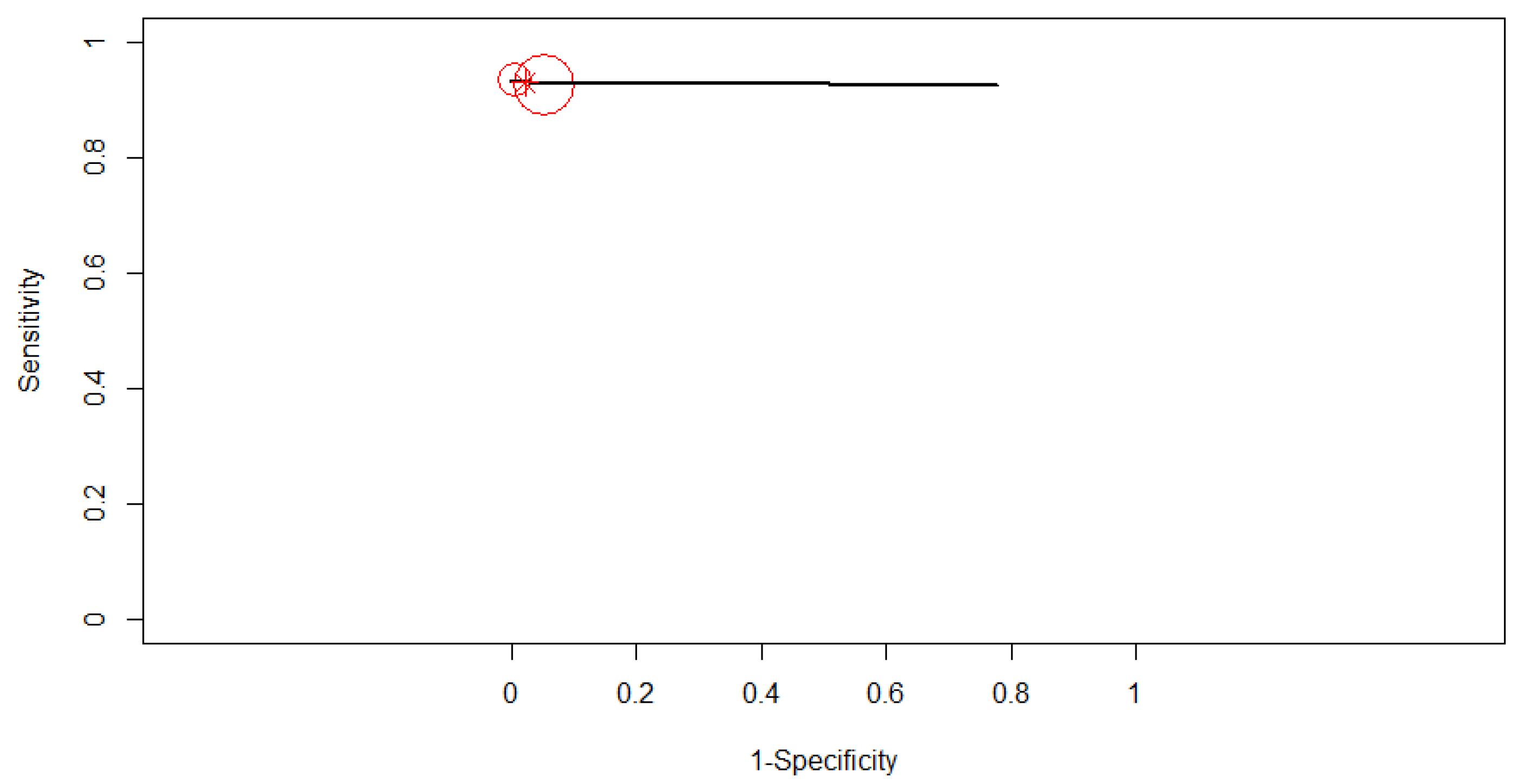
3.3. Diagnostic Accuracy
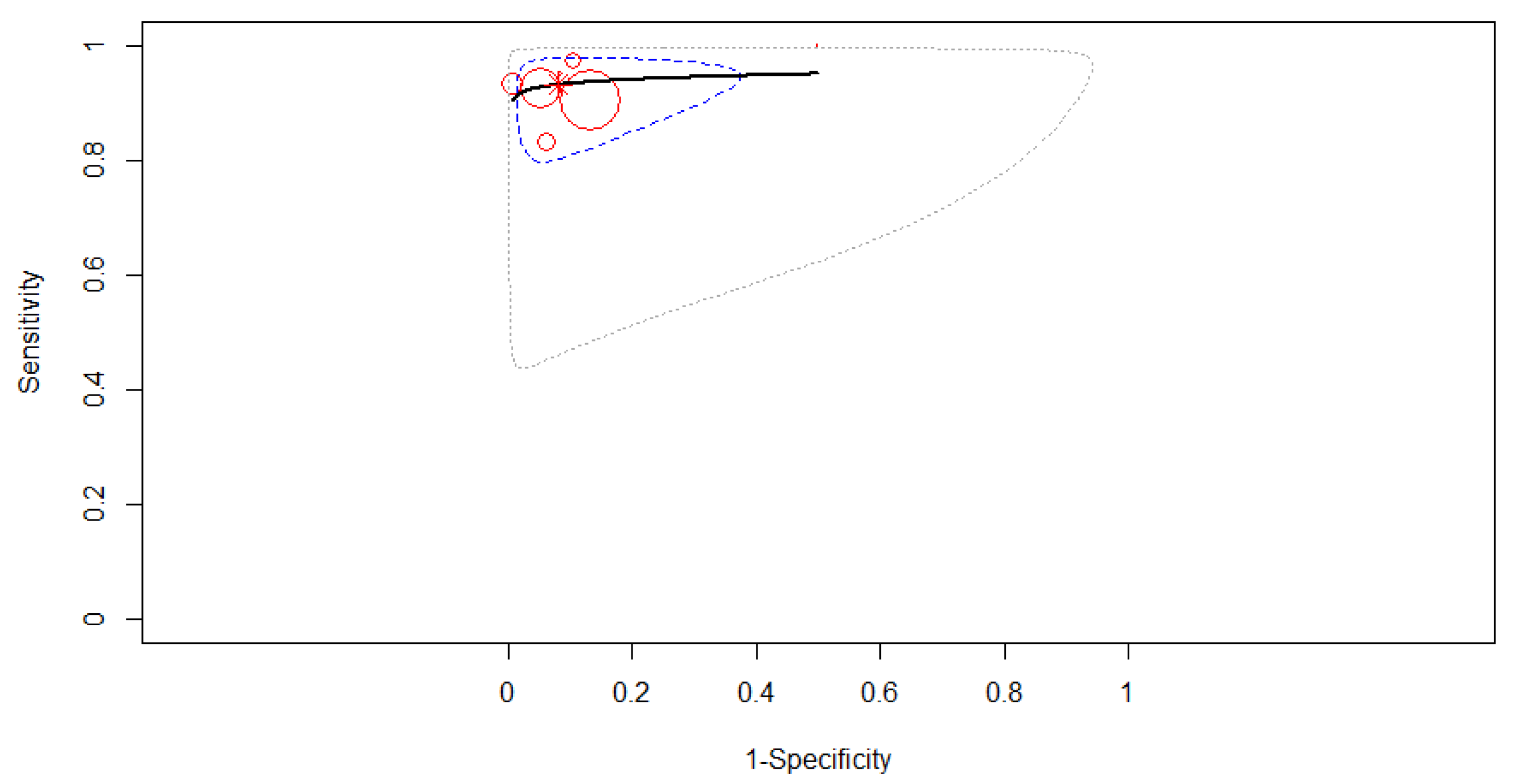
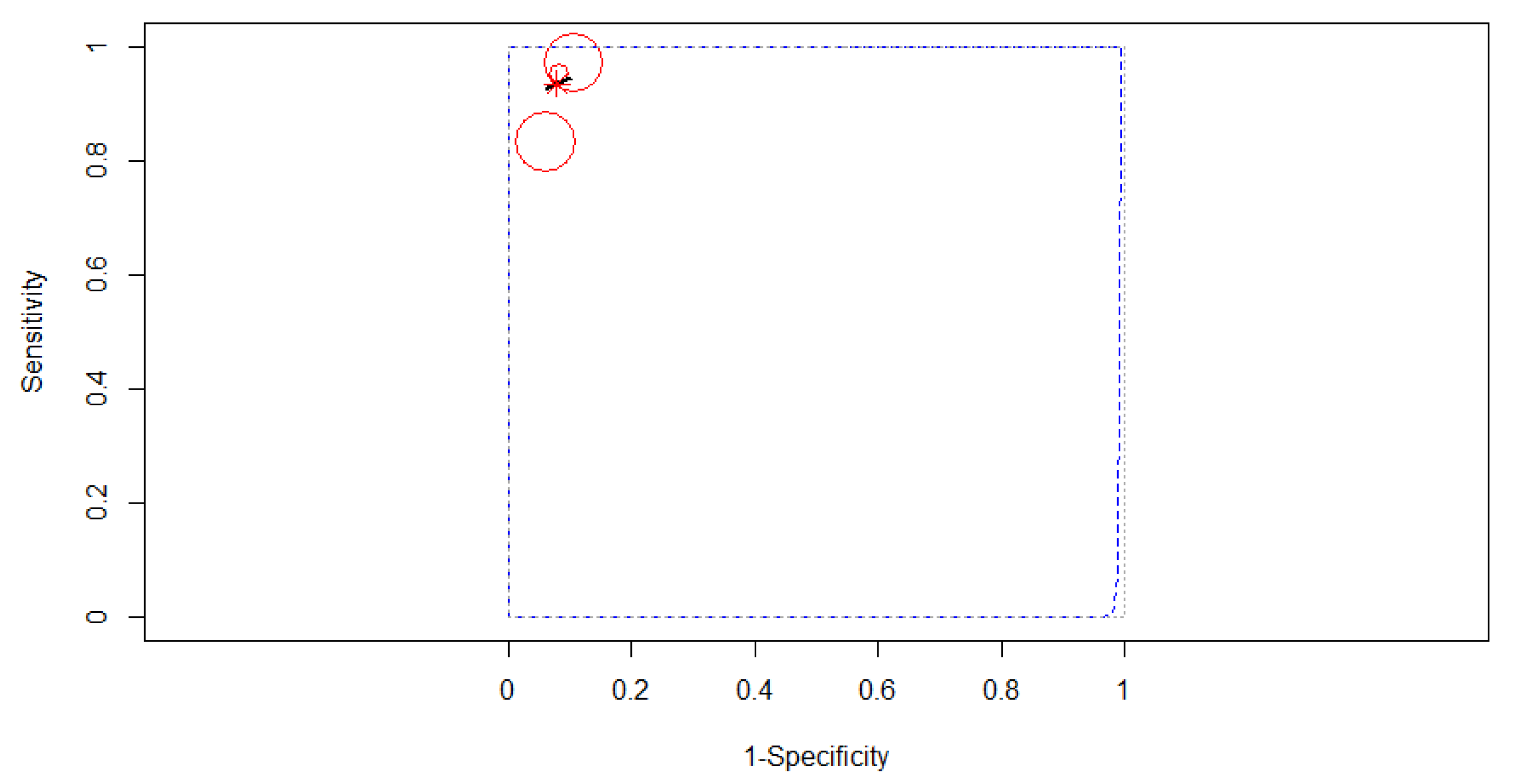
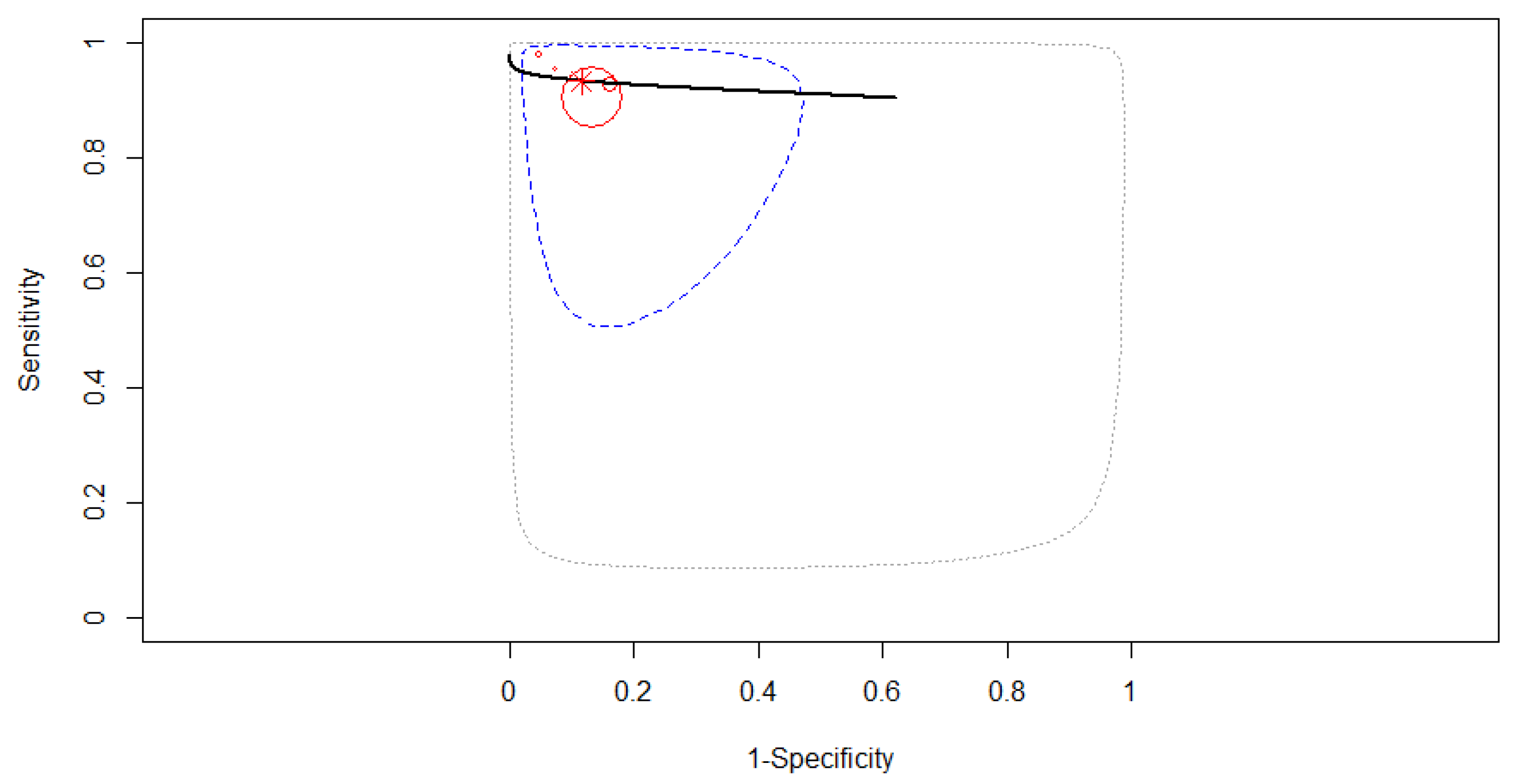
3.4. Subgroup Analysis
3.4.1. Subgroup Analysis Based on the Type of Computer-Aided Diagnosis
3.4.2. Subgroup Analysis Based on the Algorithm of Artificial Intelligence
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Primers 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Surveillance, Epidemiology and End Results Program (SEER). Available online: https://seer.cancer.gov/statfacts/html/pancreas.html (accessed on 1 December 2021).
- Winter, J.M.; Brennan, M.; Tang, L.H.; D’Angelica, M.I.; DeMatteo, R.P.; Fong, Y.; Klimstra, D.S.; Jarnagin, W.; Allen, P.J. Survival after resection of pancreatic adenocarcinoma: Results from a single institution over three decades. Ann. Surg. Oncol. 2012, 19, 169–175. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef] [PubMed]
- Ehteshami Bejnordi, B.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.W.M.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women with Breast Cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60. [Google Scholar] [CrossRef] [Green Version]
- Doi, K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef] [Green Version]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61. [Google Scholar] [CrossRef]
- Ma, J.; Wu, F.; Zhu, J.; Xu, D.; Kong, D. A pre-trained convolutional neural network based method for thyroid nodule diagnosis. Ultrasonics 2017, 73, 221–230. [Google Scholar] [CrossRef]
- Mori, Y.; Kudo, S.E.; Misawa, M.; Saito, Y.; Ikematsu, H.; Hotta, K.; Ohtsuka, K.; Urushibara, F.; Kataoka, S.; Ogawa, Y.; et al. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann. Intern. Med. 2018, 169, 357–366. [Google Scholar] [CrossRef]
- Kanesaka, T.; Lee, T.-C.; Uedo, N.; Lin, K.-P.; Chen, H.-Z.; Lee, J.-Y.; Wang, H.-P.; Chang, H.-T. Computer-aided diagnosis for identifying and delineating early gastric cancers in magnifying narrow-band imaging. Gastrointest. Endosc. 2018, 87, 1339–1344. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.H.; Sun, L.-M.; Lin, C.-L.; Hsieh, M.J.; Hsu, C.Y.; Kao, C.H. Development of a prediction model for pancreatic cancer in patients with type 2 diabetes using logistic regression and artificial neural network models. Cancer Manag. Res. 2018, 10, 6317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, L.C.; Park, S.; Kawamoto, S.; Fouladi, D.F.; Shayesteh, S.; Zinreich, E.S.; Graves, J.S.; Horton, K.M.; Hruban, R.H.; Yuille, A.L.; et al. Utility of CT Radiomics Features in Differentiation of Pancreatic Ductal Adenocarcinoma from Normal Pancreatic Tissue. Am. J. Roentgenol. 2019, 213, 349–357. [Google Scholar] [CrossRef] [PubMed]
- McInnes, M.D.; Moher, D.; Thombs, B.D.; McGrath, T.A.; Bossuyt, P.M.; Clifford, T.; Cohen, J.F.; Deeks, J.J.; Gatsonis, C.; Hooft, L.; et al. Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies: The PRISMA-DTA Statement. JAMA 2018, 319, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Nguyen, C.C.; Li, F.; Li, B. Digital image analysis of EUS images accurately differentiates pancreatic cancer from chronic pancreatitis and normal tissue. Gastrointest. Endosc. 2008, 67, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, T.; Hara, K.; Mizuno, N.; Okuno, N.; Matsumoto, S.; Obata, M.; Kurita, Y.; Koda, H.; Toriyama, K.; Onishi, S.; et al. Usefulness of deep learning analysis for the diagnosis of malignancy in intraductal papillary mucinous neoplasms of the pancreas. Clin. Transl. Gastroenterol. 2019, 10, e00045. [Google Scholar] [CrossRef]
- Norton, I.D.; Zheng, Y.; Wiersema, M.S.; Greenleaf, J.; Clain, J.E.; DiMagno, E.P. Neural network analysis of EUS images to differentiate between pancreatic malignancy and pancreatitis. Gastrointest. Endosc. 2001, 54, 625–629. [Google Scholar] [CrossRef]
- Marya, N.B.; Powers, P.D.; Chari, S.T.; Gleeson, F.C.; Leggett, C.L.; Abu Dayyeh, B.K.; Chandrasekhara, V.; Iyer, P.G.; Majumder, S.; Pearson, R.K.; et al. Utilisation of artificial intelligence for the development of an EUS-convolutional neural network model trained to enhance the diagnosis of autoimmune pancreatitis. Gut 2021, 70, 1335–1344. [Google Scholar] [CrossRef]
- Ozkan, M.; Cakiroglu, M.; Kocaman, O.; Kurt, M.; Yilmaz, B.; Can, G.; Korkmaz, U.; Dandıl, E.; Eksi, Z. Age-based computer-aided diagnosis approach for pancreatic cancer on endoscopic ultrasound images. Endosc. Ultrasound 2016, 5, 101–107. [Google Scholar]
- Săftoiu, A.; Vilmann, P.; Dietrich, C.F.; Iglesias-Garcia, J.; Hocke, M.; Seicean, A.; Ignee, A.; Hassan, H.; Streba, C.T.; Ioncică, A.M.; et al. Quantitative contrast-enhanced harmonic EUS in differential diagnosis of focal pancreatic masses (with videos). Gastrointest. Endosc. 2015, 82, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Tonozuka, R.; Itoi, T.; Nagata, N.; Kojima, H.; Sofuni, A.; Tsuchiya, T.; Ishii, K.; Tanaka, R.; Nagakawa, Y.; Mukai, S. Deep learning analysis for the detection of pancreatic cancer on endosonographic images: A pilot study. J. Hepato-Biliary-Pancreat. Sci. 2021, 28, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Udriștoiu, A.L.; Cazacu, I.M.; Gruionu, L.G.; Gruionu, G.; Iacob, A.V.; Burtea, D.E.; Ungureanu, B.S.; Costache, M.I.; Constantin, A.; Popescu, C.F.; et al. Real-time computer-aided diagnosis of focal pancreatic masses from endoscopic ultrasound imaging based on a hybrid convolutional and long short-term memory neural network model. PLoS ONE 2021, 16, e0251701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-M.; Yang, H.; Jin, Z.-D.; Yu, J.-G.; Cai, Z.-Y.; Li, Z.-S. Differential diagnosis of pancreatic cancer from normal tissue with digital imaging processing and pattern recognition based on a support vector machine of EUS images. Gastrointest. Endosc. 2010, 72, 978–985. [Google Scholar] [CrossRef]
- Zhu, M.; Xu, C.; Yu, J.; Wu, Y.; Li, C.; Zhang, M.; Jin, Z.; Li, Z. Differentiation of pancreatic cancer and chronic pancreatitis using computer-aided diagnosis of endoscopic ultrasound (EUS) images: A diagnostic test. PLoS ONE 2013, 8, e63820. [Google Scholar] [CrossRef] [Green Version]
- Maguchi, H. The roles of endoscopic ultrasonography in the diagnosis of pancreatic tumors. J. Hepato-Biliary-Pancreat. Surg. 2004, 11, 1–3. [Google Scholar] [CrossRef]
- Zhang, L.; Sanagapalli, S.; Stoita, A. Challenges in diagnosis of pancreatic cancer. Gastrointest. Endosc. 2018, 24, 2047. [Google Scholar] [CrossRef]
- Goyal, H.; Mann, R.; Gandhi, Z.; Perisetti, A.; Zhang, Z.; Sharma, N.; Saligram, S.; Inamdar, S.; Tharian, B. Application of artificial intelligence in pancreaticobiliary diseases. Ther. Adv. Gastrointest. Endosc. 2021, 14, 2631774521993059. [Google Scholar] [CrossRef]
- Lijmer, G.J.; Bossuyt, P.M.; Heisterkamp, S.H. Exploring sources of heterogeneity in systematic reviews of diagnostic tests. Stat. Med. 2002, 21, 1525–1537. [Google Scholar] [CrossRef]
- Campbell, C.; Ying, Y. Learning with support vector machines. Synth. Lect. Artif. Intell. Mach. Learn. 2011, 5, 1–95. [Google Scholar] [CrossRef]
- Sarker, I.H. Machine learning: Algorithms, real-world applications and research directions. Comput. Sci. 2021, 2, 160. [Google Scholar] [CrossRef] [PubMed]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-G.; Jun, S.; Cho, Y.-W.; Lee, H.; Kim, G.B.; Seo, J.B.; Kim, N. Deep learning in medical imaging: General overview. Korean J. Radiol. 2017, 18, 570–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, H.-C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müller, M.F.; Meyenberger, C.; Bertschinger, P.; Schaer, R.; Marincek, B. Pancreatic tumors: Evaluation with endoscopic US, CT, and MR imaging. Radiology 1994, 190, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, H.; Kitano, M.; Suetomi, Y.; Maekawa, K.; Takeyama, Y.; Kudo, M. Utility of contrast-enhanced endoscopic ultrasonography for diagnosis of small pancreatic carcinomas. Ultrasound Med. Biol. 2008, 34, 525–532. [Google Scholar] [CrossRef] [PubMed]
| No. crt. | Author | Study Design | Comparison | No. of Patients (Overall Data) | No. of Images (Overall Data) | Testing Data | Final Diagnosis | Analysis Target | Type of Computer-Aided Diagnosis (CAD) | Algorithm of AI |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Kuwahara 2019 | retrospective | benign IPMN vs malignant IPMN | 50 | 3970 | no separate testing data | 27 benign IPMN/23 malignant IPMN | B-mode images | deep learning-based CAD | CNN |
| 2 | Das 2008 | retrospective | normal pancreas vs. chronic pancreatitis (CP) vs PDAC | 56 | 319 | 50% of all data | 2 normal pancreas/12 CP/22 PDAC | Texture features from B-mode image | conventional CAD | ANN |
| 3 | Marya 2020 | retrospective | autoimmune pancreatitis vs. normal pancreas vs. CP vs. PDAC | 583 | 1,174,461 | 123 patients | 146 AIP/292 PDAC/72 CP/73NP | B-mode images | conventional CAD | CNN |
| 4 | Norton 2001 | retrospective | CP vs PDAC | 35 | N/A | N/A | 14 CP/21 PDAC | Grey-scale pixels from B-mode image | conventional CAD | Basic Neuronal Network/Machine Learning |
| 5 | Ozkan 2015 | retrospective | PDAC vs. normal pancreas | 172 | 332 | 72 (42 PDAC, 30 normal pancreas) | 202 PDAC/130 normal pancreas (images) | Digital features from B-mode image | conventional CAD | ANN |
| 6 | Saftoiu 2015 | Prospective | CP vs. PDAC | 167 | 15% of pts | 112 PDAC/55 CP | TIC parameters from contrast-enhanced EUS | conventional CAD | ANN | |
| 7 | Tonozuka 2021 | Prospective | normal pancreas vs. CP vs. PDAC | 139 | 1390 | 47 pts, 470 images (25 PDAC, 12 CP, 10 NP) | 76 PDAC/34 CP/29 normal pancreas | B-mode images | deep learning-based CAD | CNN |
| 8 | Udristoiu 2021 | Retrospective | CP vs. PDAC vs. NET | 65 | 3360 | 672 images from 65 pts | 30 PDAC 20 CP/15 NET | Multi parametric (B-mode, contrast, elastography) | deep learning-based CAD | CNN |
| 9 | Zhang 2010 | Retrospective | CP vs. PDAC vs. normal pancreas | 216 | 50% of all data | 153 PDAC/20 normal pancreas/43 CP | Texture features from B-mode image | conventional CAD | SVM | |
| 10 | Zhu 2013 | Retrospective | CP vs. PDAC | 388 | 50% of all data (194; 131 PDAC, 63 CP) | 262 PDAC/126 CP | Texture features from B-mode image | conventional CAD | SVM |
| Number of Studies | Pooled Sensitivity (95% CI) | Pooled Specificity (95% CI) | Pooled DOR (95% CI) | AUC Partial AUC (Restricted to Observed FPRs and Normalized) | |
|---|---|---|---|---|---|
| AI | 10 | 0.92 | 0.9 | 128.9 | 0.95 (0.93) |
| (0.89–0.95) | (0.83–0.94) | (71.2–233.8) | |||
| CNN | 4 | 0.91 | 0.87 | 86.2 | 0.94 (0.81) |
| (0.88–0.94) | (0.83–0.9) | (39.7–187.2) | |||
| ANN | 3 | 0.93 | 0.92 | 141.5 | 0.95 (0.91) |
| (0.78–0.98) | (0.86–0.95) | (55.8–358.9) | |||
| SVM | 2 | 0.93 | 0.98 | 547.9 | 0.93 (0.92) |
| (0.89–0.96) | (0.85–0.99) | (64.3–4669.6) | |||
| Deep Learning | 3 | 0.95 | 0.9 | 161.2 | 0.97 (0.94) |
| (0.89–0.98) | (0.78–0.95) | (36.9–702.3) | |||
| Conventional | 7 | 0.92 | 0.91 | 138.3 | 0.95 (0.93) |
| (0.87–0.95) | (0.85–0.96) | (64.9–294.1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dumitrescu, E.A.; Ungureanu, B.S.; Cazacu, I.M.; Florescu, L.M.; Streba, L.; Croitoru, V.M.; Sur, D.; Croitoru, A.; Turcu-Stiolica, A.; Lungulescu, C.V. Diagnostic Value of Artificial Intelligence-Assisted Endoscopic Ultrasound for Pancreatic Cancer: A Systematic Review and Meta-Analysis. Diagnostics 2022, 12, 309. https://doi.org/10.3390/diagnostics12020309
Dumitrescu EA, Ungureanu BS, Cazacu IM, Florescu LM, Streba L, Croitoru VM, Sur D, Croitoru A, Turcu-Stiolica A, Lungulescu CV. Diagnostic Value of Artificial Intelligence-Assisted Endoscopic Ultrasound for Pancreatic Cancer: A Systematic Review and Meta-Analysis. Diagnostics. 2022; 12(2):309. https://doi.org/10.3390/diagnostics12020309
Chicago/Turabian StyleDumitrescu, Elena Adriana, Bogdan Silviu Ungureanu, Irina M. Cazacu, Lucian Mihai Florescu, Liliana Streba, Vlad M. Croitoru, Daniel Sur, Adina Croitoru, Adina Turcu-Stiolica, and Cristian Virgil Lungulescu. 2022. "Diagnostic Value of Artificial Intelligence-Assisted Endoscopic Ultrasound for Pancreatic Cancer: A Systematic Review and Meta-Analysis" Diagnostics 12, no. 2: 309. https://doi.org/10.3390/diagnostics12020309
APA StyleDumitrescu, E. A., Ungureanu, B. S., Cazacu, I. M., Florescu, L. M., Streba, L., Croitoru, V. M., Sur, D., Croitoru, A., Turcu-Stiolica, A., & Lungulescu, C. V. (2022). Diagnostic Value of Artificial Intelligence-Assisted Endoscopic Ultrasound for Pancreatic Cancer: A Systematic Review and Meta-Analysis. Diagnostics, 12(2), 309. https://doi.org/10.3390/diagnostics12020309









