Proteomic Analysis of Low-Temperature Stress Response in Maize (Zea mays L.) at the Seedling Stage
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatments
2.2. Protein Extraction and Quality Control
2.3. Filter-Aided Sample Preparation (FASP)
2.4. High-pH Reverse-Phase Peptide Fractionation
2.5. DDA Library Construction and DIA Quantification
2.6. Quantitative Analysis and Data Quality Assessment
2.7. Weighted Gene Co-Expression Network Analysis (WGCNA)
2.8. Data Processing and Bioinformatics
2.9. Hub Gene Validation by qPCR
| Gene | Forward Primer | Reversed Primer |
|---|---|---|
| Zm00001d016659 | CGGATCTCGCCGCCTCTCCC | GCTCGGCGCCGGCGAAGAT |
| LOC100276453 | GCCGCTAGGGTTCCGGCGAG | CAGCAGCAGTCACACCCGGG |
| LOC103645840 | GCCGTCCCCGACCACTACCG | GCTCGCCAACGTCGACTGCTC |
| LOC103633583 | GAAAACCTATAGCTTGCAGCG | GCTCGCCAACGTCGACTGCTC |
3. Results
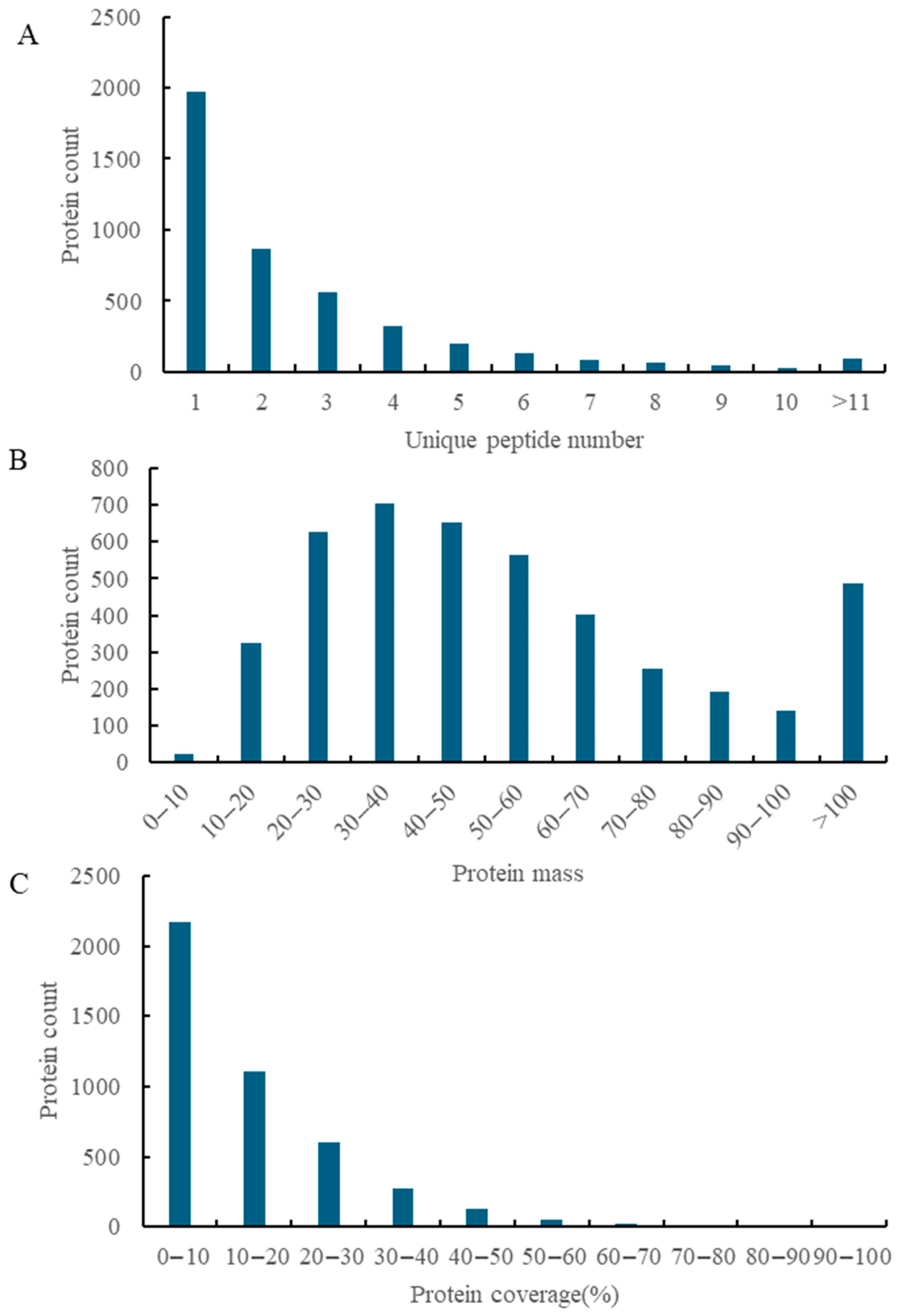
3.1. Statistical Overview of Protein Identification
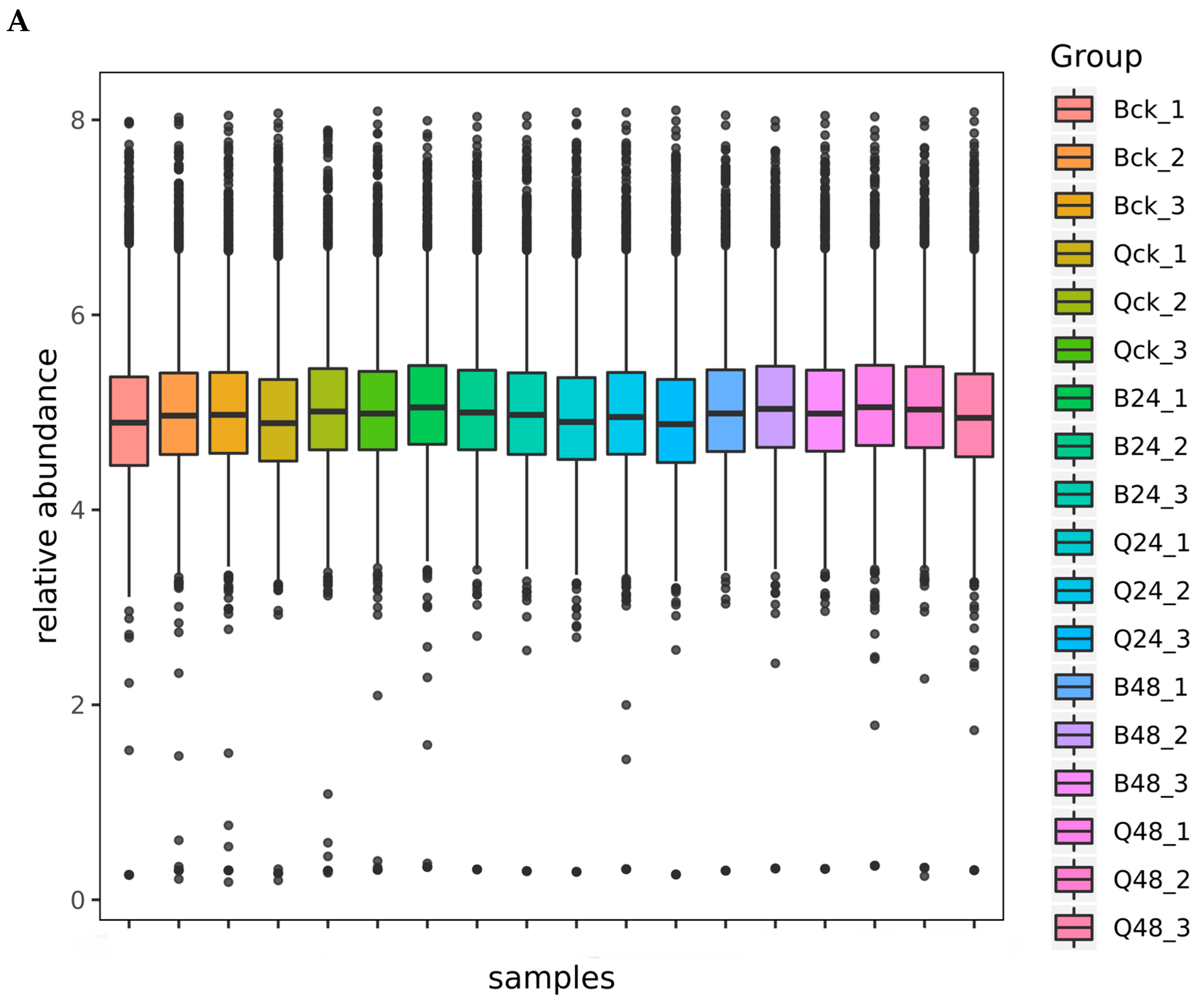
3.2. Data Quality Control
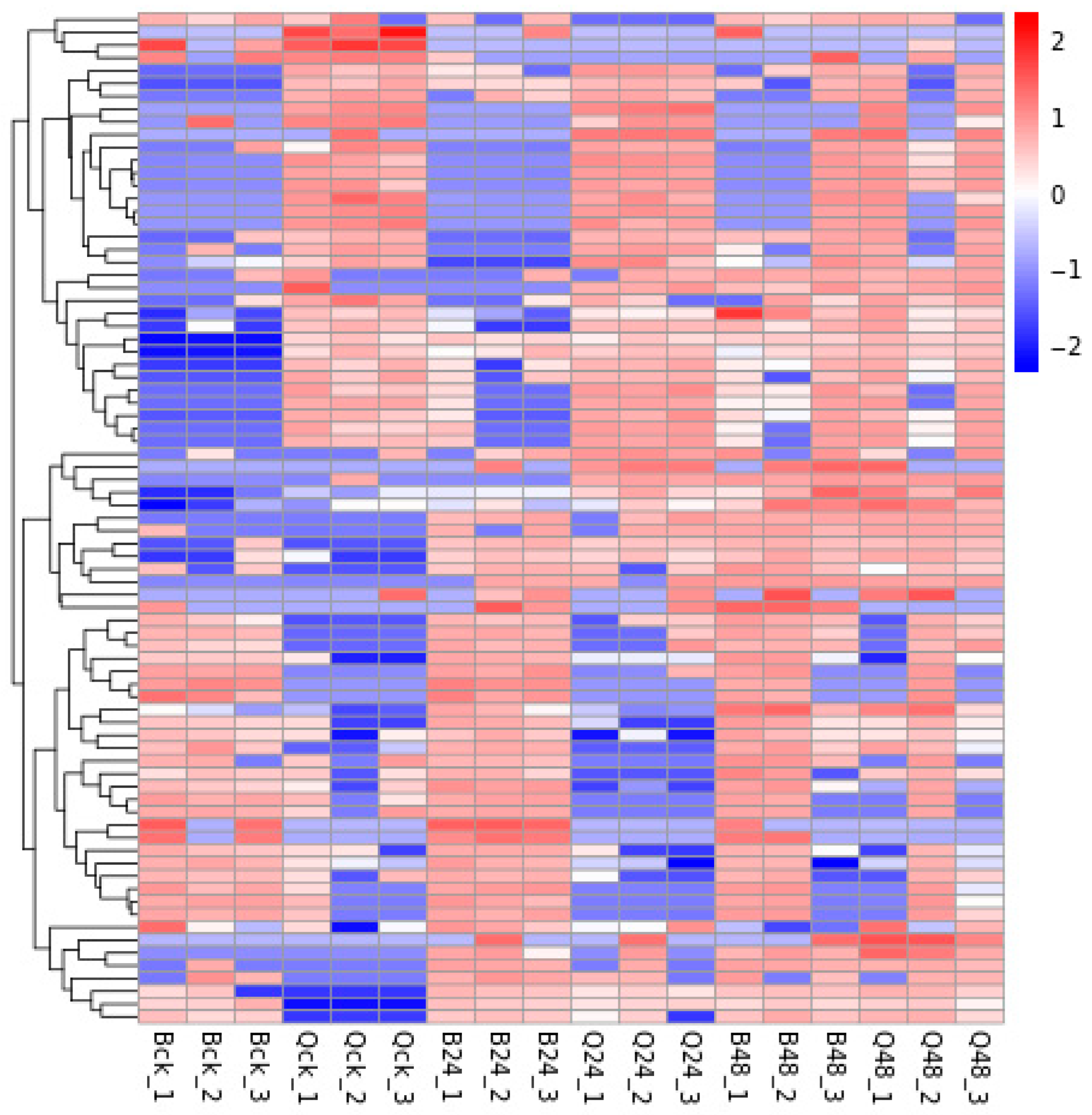
3.3. Hierarchical Clustering Analysis of DEPs
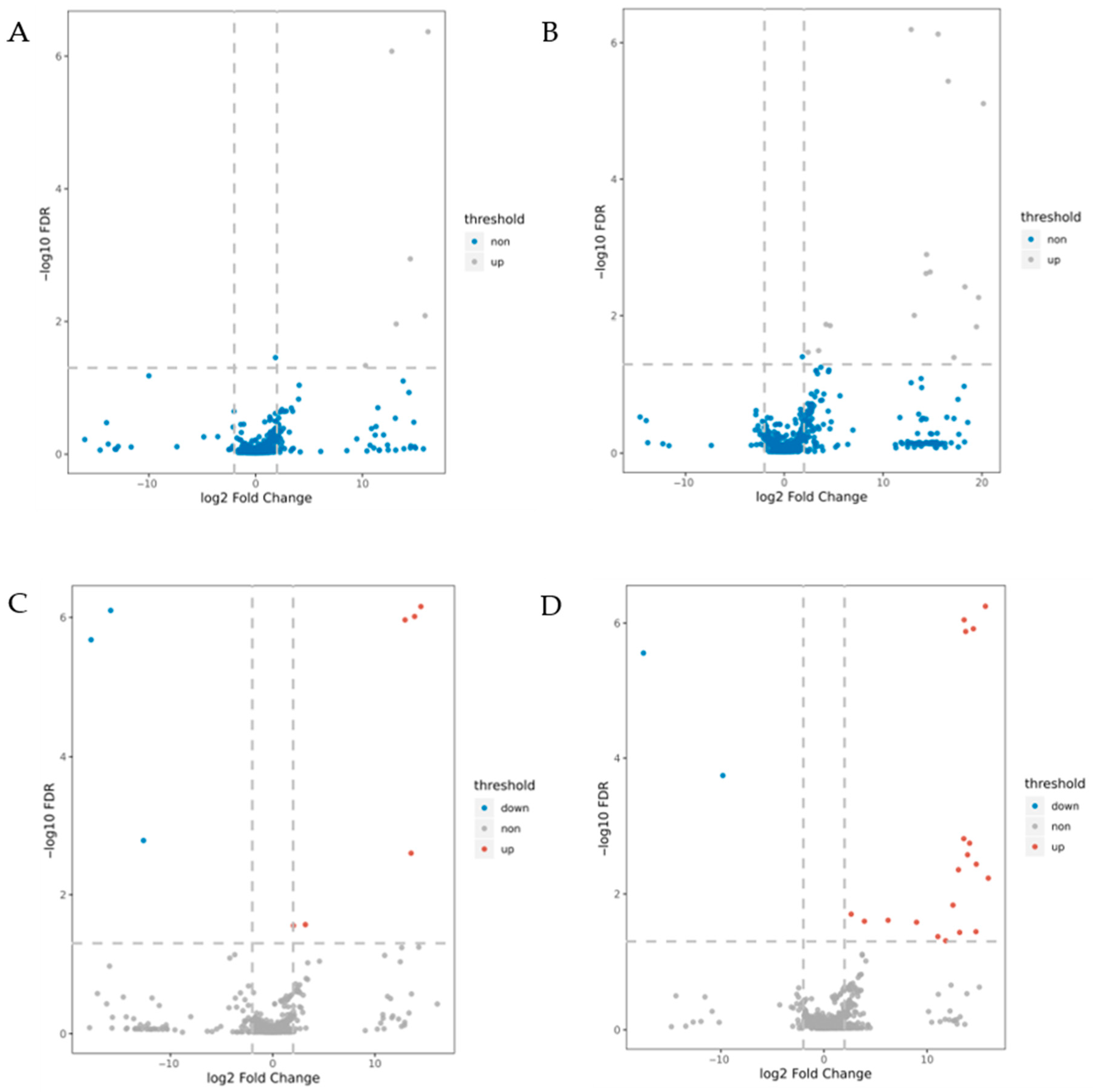
3.4. Identification of DEPs Under Low-Temperature Stress
3.5. GO Annotation and Enrichment Analysis
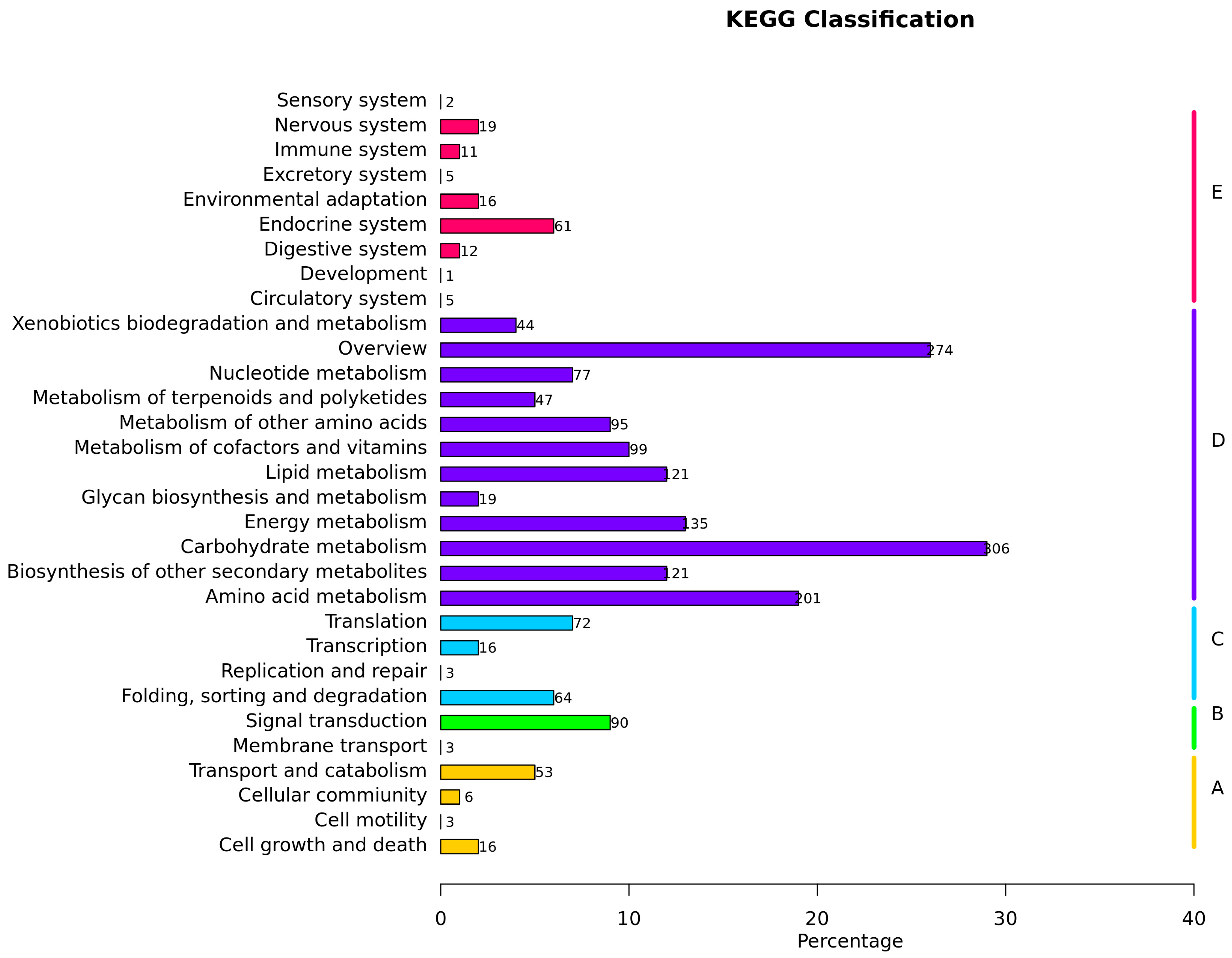
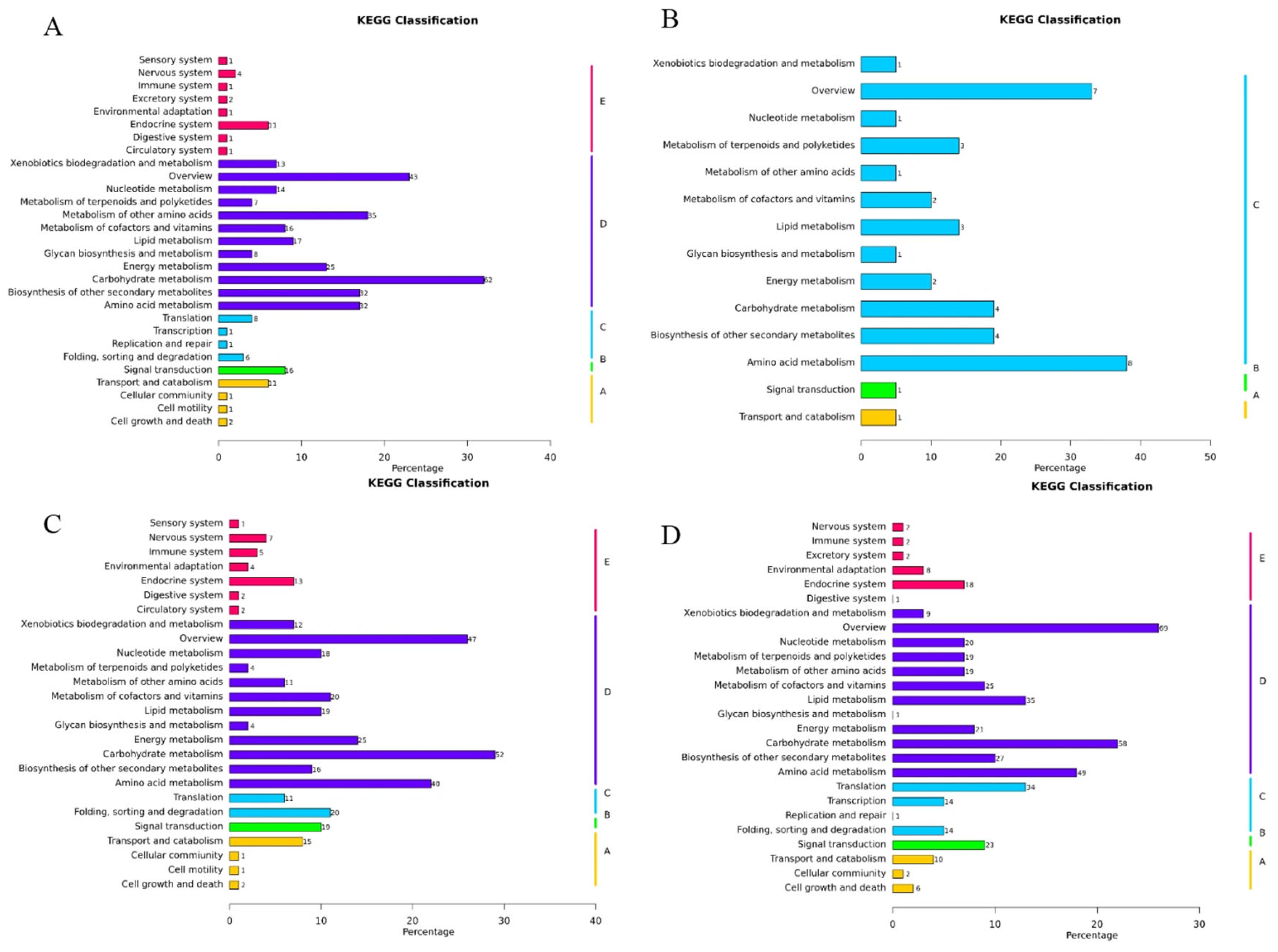
3.6. KEGG Pathway Analysis
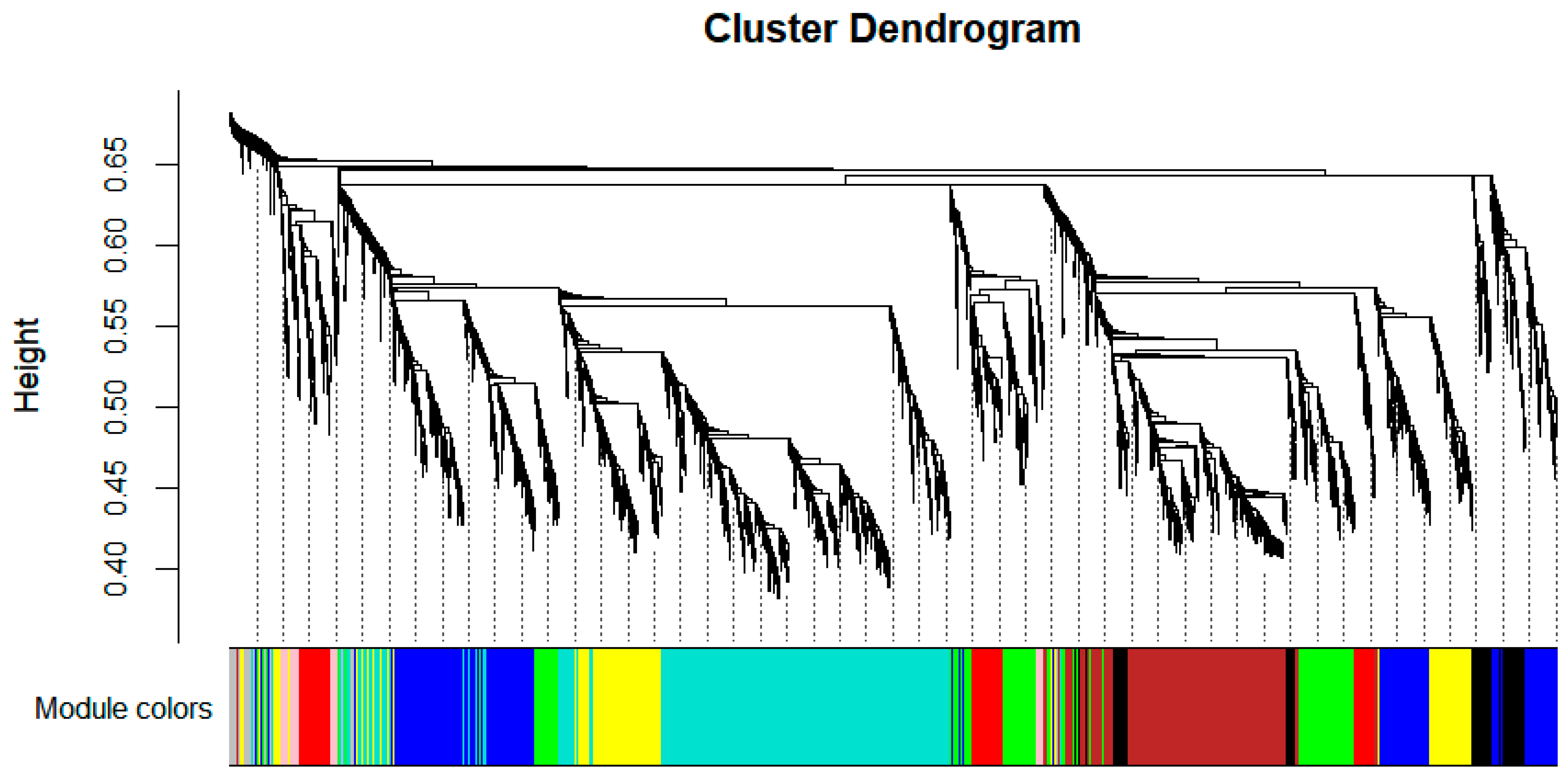
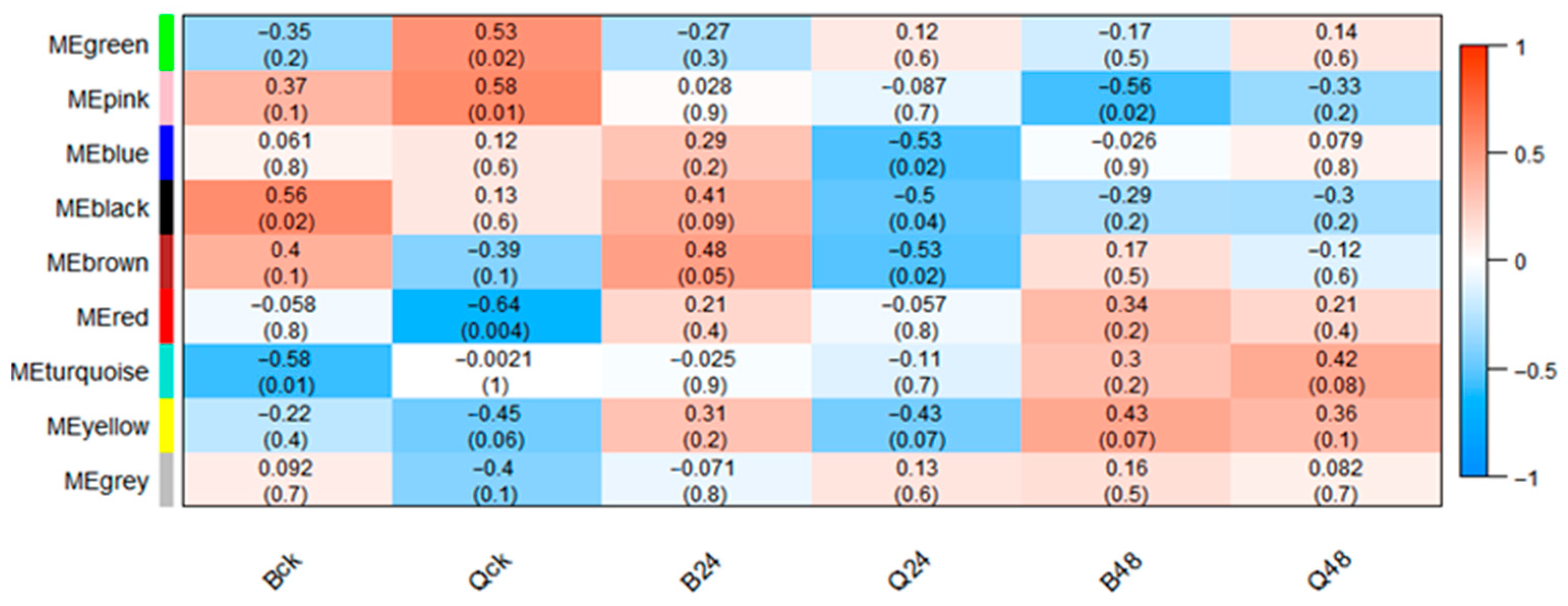
3.7. WGCNA
3.7.1. Module Partitioning and Specific Module Identification
3.7.2. Functional Enrichment Analysis of Representative Modules
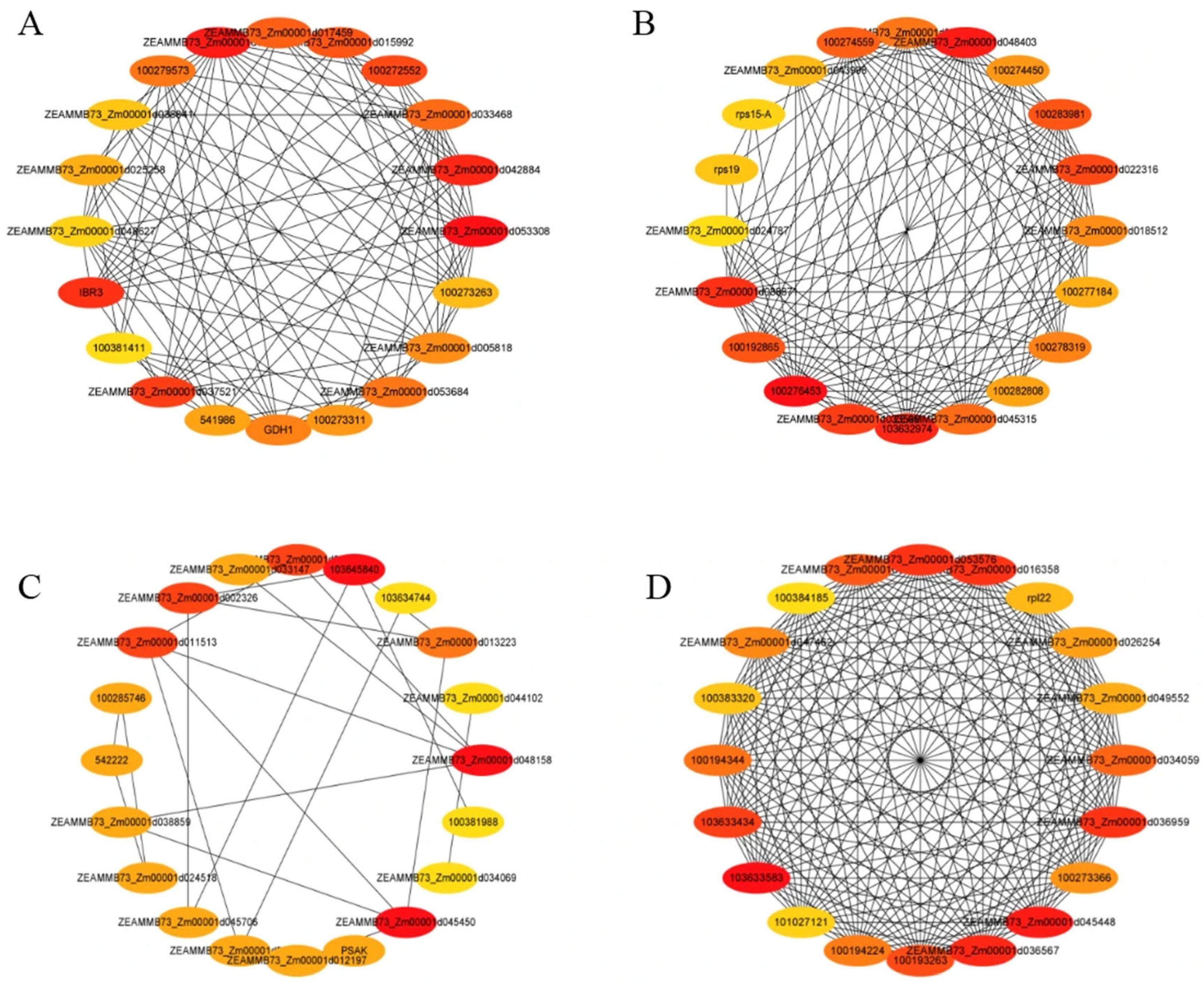
3.7.3. Hub Protein Analysis of Representative Modules
3.8. Hub Gene Validation by qPCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ritonga, F.N.; Chen, S. Physiological and molecular mechanism involved in cold stress tolerance in plants. Plants 2020, 28, 560. [Google Scholar] [CrossRef] [PubMed]
- Bittner, A.; Griebel, T.; van Buer, J.; Juszczak-Debosz, I.; Baier, M. Determining the ROS and the antioxidant status of leaves during cold acclimation. Methods Mol. Biol. 2020, 2156, 241–254. [Google Scholar] [PubMed]
- Wu, Y.; Cai, X.; Tang, Y. Outcomes of low-temperature stress on biological alterations within pothos (Epipremnum aureum) leaves. Life 2022, 12, 1432. [Google Scholar] [CrossRef]
- Guan, Y.; Hwarari, D.; Korboe, H.M.; Ahmad, B.; Cao, Y.; Movahedi, A. Low temperature stress-induced perception and molecular signaling pathways in plants. Environ. Exp. Bot. 2023, 207, 105190. [Google Scholar] [CrossRef]
- Li, P.; Zheng, T.; Domingues, D.S.; Liu, Y.; Ahmad, S. Low-temperature stress in plants: Molecular responses, tolerance mechanisms, plant biodesign and breeding applications. Front. Plant Sci. 2024, 15, 1411636. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Regulatory networks underlying plant responses and adaptation to cold stress. Annu. Rev. Genet. 2024, 58, 43–65. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Cai, L.; Liu, C.; Chen, Z.; Guan, S.; Ma, W.; Pan, G. Low-temperature stress affects reactive oxygen species, osmotic adjustment substances, and antioxidants in rice (Oryza sativa L.) at the reproductive stage. Sci. Rep. 2022, 12, 6224. [Google Scholar] [CrossRef]
- Ashraf, M.A.; Rahman, A. Cellular protein trafficking: A new player in low-temperature response pathway. Plants 2022, 11, 933. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, W.; Li, Y.; Nie, H.; Cui, L.; Li, R.; Tan, L.; Peng, L.; Li, C.; Luo, J.; et al. FERONIA coordinates plant growth and salt tolerance via the phosphorylation of phyB. Nat. Plants 2023, 9, 645–660. [Google Scholar] [CrossRef]
- Aslam, B.; Basit, M.; Nisar, M.A.; Khurshid, M.; Rasool, M.H. Proteomics: Technologies and their applications. J. Chromatogr. Sci. 2017, 55, 182–196. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Sanchez, J.C.; Gooley, A.A.; Appel, R.D.; Humphery-Smith, I.; Hochstrasser, D.F.; Williams, K.L. Progress with proteome projects: Why all proteins expressed by a genome should be identified and how to do it. Biotechnol. Genet. Eng. Rev. 1996, 13, 19–50. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Ma, P.; Wu, Y.; Zhou, Y.; Zhang, G. Quantitative proteomic analysis of the response to cold stress in jojoba, a tropical woody crop. Int. J. Mol. Sci. 2019, 20, 243. [Google Scholar] [CrossRef] [PubMed]
- Kosová, K.; Vítámvás, P.; Prášil, I.T.; Renaut, J. Plant proteome changes under abiotic stress—Contribution of proteomics studies to understanding plant stress response. J. Proteom. 2011, 74, 1301–1322. [Google Scholar] [CrossRef]
- Singh, R.; Jwa, N.S. Understanding the responses of rice to environmental stress using proteomics. J. Proteome Res. 2013, 12, 4652–4669. [Google Scholar] [CrossRef]
- John Martin, J.J.; Song, Y.; Hou, M.; Zhou, L.; Liu, X.; Li, X.; Fu, D.; Li, Q.; Cao, H.; Li, R. Multi-Omics approaches in oil palm research: a comprehensive review of metabolomics, proteomics, and transcriptomics based on low-temperature stress. Int. J. Mol. Sci. 2024, 25, 7695. [Google Scholar] [CrossRef]
- Xing, J.; Tan, J.; Feng, H.; Zhou, Z.; Deng, M.; Luo, H.; Deng, Z. Integrative proteome and phosphoproteome profiling of early cold response in maize seedlings. Int. J. Mol. Sci. 2022, 23, 6493. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, J.; Wang, X.; Li, R.; Chen, B. Proteomic response of hybrid wild rice to cold stress at the seedling stage. PLoS ONE 2018, 13, e0198675. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, Y.; Khan, A.; Ullah, N.; Li, Z.; Zaheer, S.; Zhou, R.; Zhang, Z. Quantitative proteomics-based analysis reveals molecular mechanisms of chilling tolerance in grafted cotton seedlings. Agronomy 2022, 12, 1152. [Google Scholar] [CrossRef]
- Li, H.; Luo, W.; Ji, R.; Xu, Y.; Xu, G.; Qiu, S.; Tang, H. A comparative proteomic study of cold responses in potato leaves. Heliyon 2021, 7, e06002. [Google Scholar] [CrossRef]
- Bibi-Triki, S.; Husson, G.; Maucourt, B.; Vuilleumier, S.; Carapito, C.; Bringel, F. N-terminome and proteogenomic analysis of the Methylobacterium extorquens DM4 reference strain for dichloromethane utilization. J. Proteom. 2018, 179, 131–139. [Google Scholar] [CrossRef]
- Tang, X.; Wang, H.; Long, Q. Proteomics analysis of rice root under low temperature stress at seedling stage. Acta Agric. Boreali-Sin. 2019, 34, 7. [Google Scholar]
- Long, J.; Xing, W.; Wang, Y.; Wu, Z.; Li, W.; Zou, Y.; Sun, J.; Zhang, F.; Pi, Z. Comparative proteomic analysis on chloroplast proteins provides new insights into the effects of low temperature in sugar beet. Bot. Stud. 2022, 63, 18. [Google Scholar] [CrossRef]
- Jan, S.; Rustgi, S.; Barmukh, R.; Shikari, A.B.; Leske, B.; Bekuma, A.; Sharma, D.; Ma, W.; Kumar, U.; Kumar, U.; et al. Advances and opportunities in unraveling cold-tolerance mechanisms in the world’s primary staple food crops. Plant Genome 2024, 17, e20402. [Google Scholar] [CrossRef]
- Yu, T.; Zhang, J.; Cao, J.; Li, X.; Li, S.; Liu, C.; Wang, L. Metabolic insight into cold stress response in two contrasting maize lines. Life 2022, 12, 282. [Google Scholar] [CrossRef] [PubMed]
- Salam, U.; Ullah, S.; Tang, Z.H. An overview of the role of primary and secondary metabolites against different environmental stress factors. Life 2023, 13, 706. [Google Scholar] [CrossRef]
- Weng, Q.; Zhao, Y.; Yanan, Z.; Song, X.; Yuan, J.; Liu, Y. Identification of salt stress-responsive proteins in maize (Zea may) seedlings using itraq-based proteomic technique. Iran. J. Biotechnol. 2021, 19, e2512. [Google Scholar]
- Liao, C.; Peng, Y.; Ma, W.; Liu, R.; Li, C.; Li, X. Proteomic analysis revealed nitrogen-mediated metabolic, developmental, and hormonal regulation of maize (Zea mays L.) ear growth. J. Exp. Bot. 2012, 63, 5275–5288. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.J.; Lan, Y.Q.; Wang, S.H. Research progress in omics of plant’s response to low temperature stress. Shangdong Agric. Sci. 2020, 52, 142–148. [Google Scholar]
- Yu, H.; Kong, X.; Huang, H.; Wu, W.; Park, J.; Yun, D.J.; Lee, B.H.; Shi, H.; Zhu, J.K. STCH4/REIL2 confers cold stress tolerance in arabidopsis by promoting rrna processing and cbf protein translation. Cell Rep. 2020, 30, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, Y.; Lu, S.; Chen, H.; Wu, J.; Zhu, X.; Zou, B.; Hua, J. HsfA1d promotes hypocotyl elongation under chilling via enhancing expression of ribosomal protein genes in Arabidopsis. New Phytol. 2021, 231, 646–660. [Google Scholar] [CrossRef]
- Zhang, J.; Liang, L.; Xiao, J.; Xie, Y.; Zhu, L.; Xue, X.; Xu, L.; Zhou, P.; Ran, J.; Huang, Z.; et al. Genome-wide identification of Polyamine Oxidase (PAO) family genes: Roles of CaPAO2 and CaPAO4 in the cold tolerance of pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2022, 23, 9999. [Google Scholar] [CrossRef]
- Franzoni, G.; Spadafora, N.D.; Sirangelo, T.M.; Ferrante, A.; Rogers, H.J. Biochemical and molecular changes in peach fruit exposed to cold stress conditions. Mol. Hortic. 2023, 3, 24. [Google Scholar] [CrossRef]
- Pacheco, J.M.; Ranocha, P.; Kasulin, L.; Fusari, C.M.; Servi, L.; Aptekmann, A.A.; Gabarain, V.B.; Peralta, J.M.; Borassi, C.; Marzol, E.; et al. Apoplastic class III peroxidases PRX62 and PRX69 promote Arabidopsis root hair growth at low temperature. Nat. Commun. 2022, 13, 1310. [Google Scholar] [CrossRef]
- Ramaiah, M.; Jain, A.; Baldwin, J.C.; Karthikeyan, A.S.; Raghothama, K.G. Characterization of the phosphate starvation-induced glycerol-3-phosphate permease gene family in Arabidopsis. Plant Physiol. 2011, 157, 279–291. [Google Scholar] [CrossRef]
- Carman, P.J.; Barrie, K.R.; Rebowski, G.; Dominguez, R. Structures of the free and capped ends of the actin filament. Science 2023, 380, 1287–1292. [Google Scholar] [CrossRef]
- Kim, J.S.; Kidokoro, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulatory networks in plant responses to drought and cold stress. Plant Physiol. 2024, 195, 170–189. [Google Scholar] [CrossRef] [PubMed]
- Seydel, C.; Kitashova, A.; Fürtauer, L.; Nägele, T. Temperature-induced dynamics of plant carbohydrate metabolism. Physiol. Plant. 2022, 174, e13602. [Google Scholar] [CrossRef]
- Klimm, F.; Schmier, S.; Bohn, H.F.; Kleiser, S.; Thielen, M.; Speck, T. Biomechanics of tendrils and adhesive pads of the climbing passion flower Passiflora discophora. J. Exp. Bot. 2022, 73, 1190–1203. [Google Scholar] [CrossRef]
- Shahryar, N.; Maali-Amiri, R. Metabolic acclimation of tetraploid and hexaploid wheats by cold stress-induced carbohydrate accumulation. J. Plant Physiol. 2016, 204, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Orzechowski, S.; Sitnicka, D.; Grabowska, A.; Compart, J.; Fettke, J.; Zdunek-Zastocka, E. Effect of short-term cold treatment on carbohydrate metabolism in potato leaves. Int. J. Mol. Sci. 2021, 22, 7203. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Shi, Y.; Guo, L.; Fu, D.; Li, M.; Zhang, X.; Li, Z.; Zhuang, J.; Yang, X.; Zuo, J. A natural variant of COOL1 gene enhances cold tolerance for high-latitude adaptation in maize. Cell 2025, 188, 1315–1329.e13. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, C.; Li, M.; Li, Y.; Yan, Z.; Feng, G.; Liu, D. Integrated transcriptomics and metabolomics analysis reveals key regulatory network that response to cold stress in common Bean (Phaseolus vulgaris L.). BMC Plant Biol. 2023, 23, 85. [Google Scholar] [CrossRef]
- Jiang, H.; Shi, Y.; Liu, J.; Li, Z.; Fu, D.; Wu, S.; Li, M.; Yang, Z.; Shi, Y.; Lai, J.; et al. Natural polymorphism of ZmICE1 contributes to amino acid metabolism that impacts cold tolerance in maize. Nat. Plants 2022, 8, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Xie, S.; Nie, L.; Zheng, Y.; Wang, J.; Huang, J.; Zhao, M.; Zhu, S.; Hou, J.; Chen, G.; et al. Comparative proteomics reveals cold acclimation machinery through enhanced carbohydrate and amino acid metabolism in wucai (Brassica campestris L.). Plants 2019, 8, 474. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Ban, Q.; Mao, J.; Lin, M.; Zhu, X.; Xia, Y.; Cao, X.; Zhang, X.; Li, Y. Integrated metabolomic and transcriptomic analysis reveals that amino acid biosynthesis may determine differences in cold-tolerant and cold-sensitive tea cultivars. Int. J. Mol. Sci. 2023, 24, 1907. [Google Scholar] [CrossRef]
- Ling, R.; Chen, G.; Tang, X.; Liu, N.; Zhou, Y.; Chen, D. Acetyl-CoA synthetase 2(ACSS2): A review with a focus on metabolism and tumor development. Discov. Oncol. 2022, 13, 58. [Google Scholar] [CrossRef]
- Zhang, Y.; Chang, M.; Xue, Q.; Wang, H.; Liu, Y.; Wei, H.; Li, J. Intermittent cold stimulation acclimates broilers to acute cold stress by affecting cardiac lipid metabolism. Anim. Biosci. 2025, 38, 775–787. [Google Scholar] [CrossRef]
- Xu, F.Q.; Xue, H.W. The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ. 2019, 42, 2931–2944. [Google Scholar] [CrossRef]
- Sánchez-Bel, P.; Egea, I.; Sánchez-Ballesta, M.T.; Martinez-Madrid, C.; Fernandez-Garcia, N.; Romojaro, F.; Olmos, E.; Estrella, E.; Bolarín, M.C.; Flores, F.B. Understanding the mechanisms of chilling injury in bell pepper fruits using the proteomic approach. J. Proteom. 2012, 75, 5463–5478. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, X.; Liu, C.; Duncan, S.; Hang, R.; Sun, J.; Luo, L.; Ding, Y.; Cao, X. AtPRMT3-RPS2B promotes ribosome biogenesis and coordinates growth and cold adaptation trade-off. Nat. Commun. 2024, 15, 8693. [Google Scholar] [CrossRef]
- Cai, J.; Li, P.; Luo, X.; Chang, T.; Li, J.; Zhao, Y.; Xu, Y. Selection of appropriate reference genes for the detection of rhythmic gene expression via quantitative real-time PCR in Tibetan hulless barley. PLoS ONE 2018, 13, e0190559. [Google Scholar] [CrossRef] [PubMed]


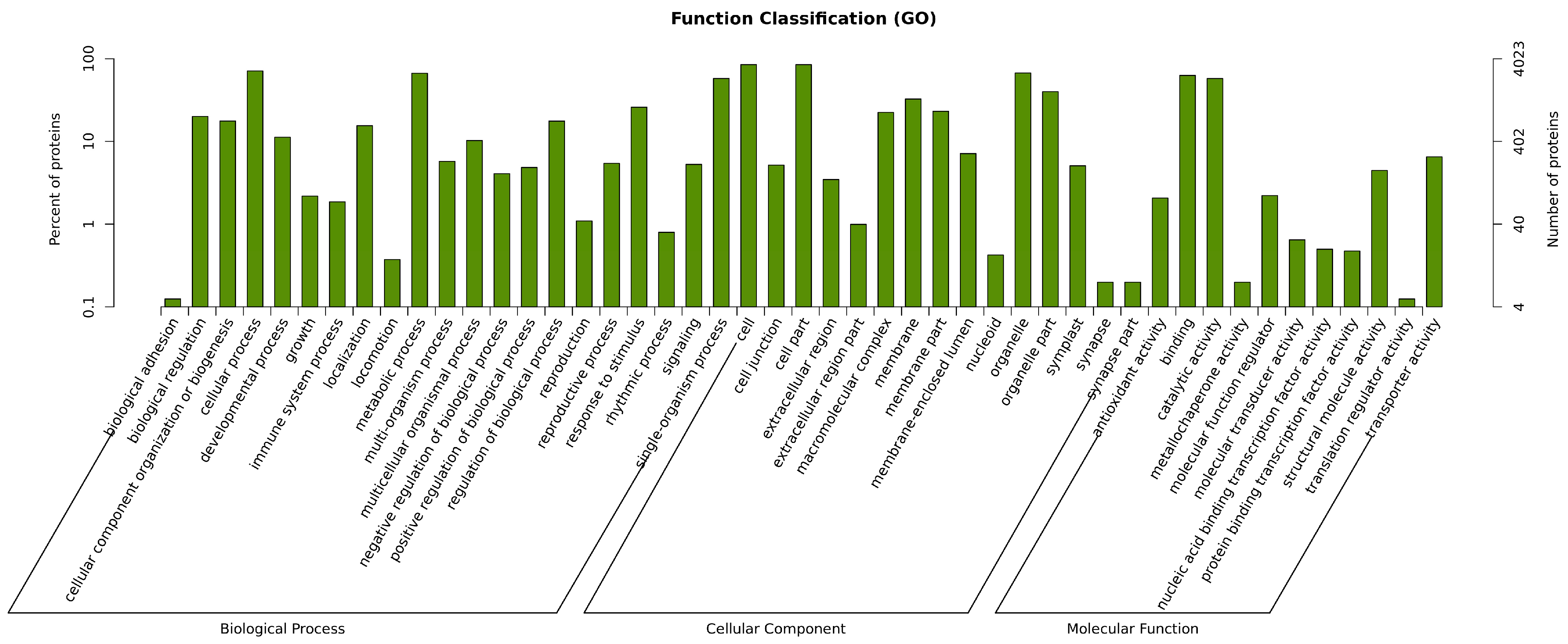
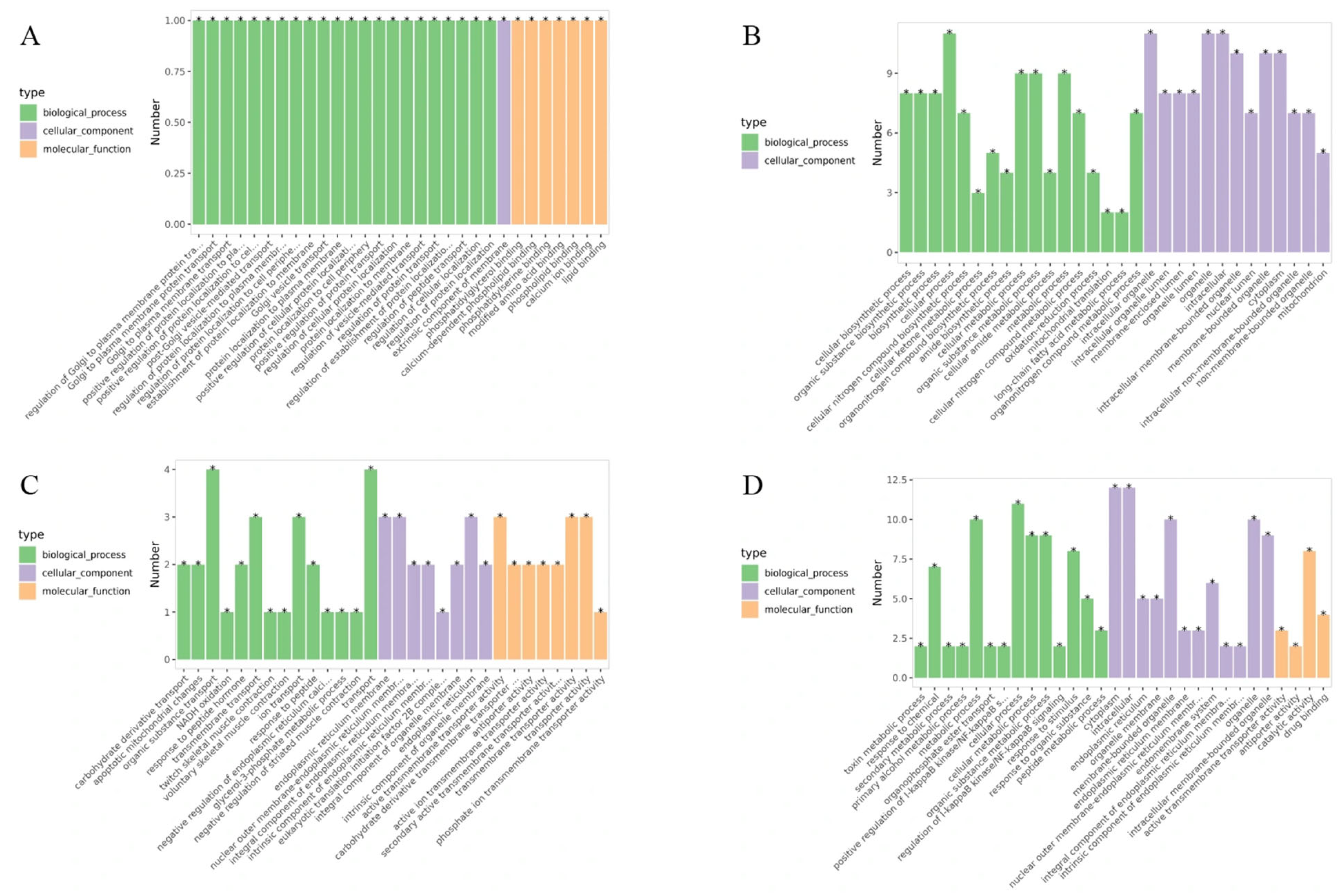

| Type | GO Annotation | GO_ID | p-Value | Count |
|---|---|---|---|---|
| Biological process | Regulation of Golgi to plasma membrane protein transport | GO:0042996 | 0.0003 | 1 |
| Golgi to plasma membrane protein transport | GO:0043001 | 0.0010 | 1 | |
| Golgi to plasma membrane transport | GO:0006893 | 0.0015 | 1 | |
| Positive regulation of protein localization to plasma membrane | GO:1903078 | 0.0016 | 1 | |
| Positive regulation of protein localization to cell periphery | GO:1904377 | 0.0018 | 1 | |
| Post-Golgi vesicle-mediated transport | GO:0006892 | 0.0024 | 1 | |
| Regulation of protein localization to plasma membrane | GO:1903076 | 0.0028 | 1 | |
| Regulation of protein localization to cell periphery | GO:1904375 | 0.0035 | 1 | |
| Establishment of protein localization to membrane | GO:0090150 | 0.0057 | 1 | |
| Golgi vesicle transport | GO:0048193 | 0.0064 | 1 | |
| Protein localization to plasma membrane | GO:0072659 | 0.0072 | 1 | |
| Positive regulation of cellular protein localization | GO:1903829 | 0.0076 | 1 | |
| Protein localization to cell periphery | GO:1990778 | 0.0088 | 1 | |
| Positive regulation of protein transport | GO:0051222 | 0.0112 | 1 | |
| Regulation of cellular protein localization | GO:1903827 | 0.0138 | 1 | |
| Protein localization to membrane | GO:0072657 | 0.0138 | 1 | |
| Regulation of vesicle-mediated transport | GO:0060627 | 0.0150 | 1 | |
| Regulation of protein transport | GO:0051223 | 0.0183 | 1 | |
| Regulation of establishment of protein localization | GO:0070201 | 0.0191 | 1 | |
| Regulation of peptide transport | GO:0090087 | 0.0194 | 1 | |
| Regulation of cellular localization | GO:0060341 | 0.0207 | 1 | |
| Regulation of protein localization | GO:0032880 | 0.0267 | 1 | |
| Cellular component | Extrinsic component of membrane | GO:0019898 | 0.0076 | 1 |
| Molecular function | Phosphatidylglycerol binding | GO:1901611 | 0.0003 | 1 |
| Calcium-dependent phospholipid binding | GO:0005544 | 0.0014 | 1 | |
| Phosphatidylserine binding | GO:0001786 | 0.0016 | 1 | |
| Modified amino acid binding | GO:0072341 | 0.0027 | 1 | |
| Phospholipid binding | GO:0005543 | 0.0105 | 1 | |
| Calcium ion binding | GO:0005509 | 0.0154 | 1 | |
| Lipid binding | GO:0008289 | 0.0183 | 1 |
| Type | GO Annotation | GO_ID | p-Value | Count |
|---|---|---|---|---|
| Biological process | Cellular biosynthetic process | GO:0044249 | 5.06 × 10−6 | 8 |
| Organic substance biosynthetic process | GO:1901576 | 5.83 × 10−6 | 8 | |
| Biosynthetic process | GO:0009058 | 6.87 × 10−6 | 8 | |
| Cellular process | GO:0009987 | 7.44 × 10−6 | 11 | |
| Cellular nitrogen compound biosynthetic process | GO:0044271 | 1.70 × 10−5 | 7 | |
| Cellular ketone metabolic process | GO:0042180 | 2.05 × 10−5 | 3 | |
| Organonitrogen compound biosynthetic process | GO:1901566 | 2.87 × 10−5 | 5 | |
| Amide biosynthetic process | GO:0043604 | 2.88 × 10−5 | 4 | |
| Cellular metabolic process | GO:0044237 | 5.96 × 10−5 | 9 | |
| Organic substance metabolic process | GO:0071704 | 7.78 × 10−5 | 9 | |
| Cellular amide metabolic process | GO:0043603 | 9.74 × 10−5 | 4 | |
| Metabolic process | GO:0008152 | 0.0001 | 9 | |
| Cellular nitrogen compound metabolic process | GO:0034641 | 0.0001 | 7 | |
| Oxidation–reduction process | GO:0055114 | 0.0001 | 4 | |
| Mitochondrial translation | GO:0032543 | 0.0002 | 2 | |
| Long-chain fatty acid metabolic process | GO:0001676 | 0.0002 | 2 | |
| Organonitrogen compound metabolic process | GO:1901564 | 0.0003 | 7 | |
| Cellular component | Intracellular organelle | GO:0043229 | 9.69 × 10−7 | 11 |
| Intracellular organelle lumen | GO:0070013 | 1.19 × 10−6 | 8 | |
| Membrane-enclosed lumen | GO:0031974 | 1.19 × 10−6 | 8 | |
| Organelle lumen | GO:0043233 | 1.19 × 10−6 | 8 | |
| Organelle | GO:0043226 | 1.32 × 10−6 | 11 | |
| Intracellular | GO:0005622 | 3.99 × 10−6 | 11 | |
| Intracellular membrane-bounded organelle | GO:0043231 | 6.54 × 10−6 | 10 | |
| Nuclear lumen | GO:0031981 | 1.16 × 10−5 | 7 | |
| Membrane-bounded organelle | GO:0043227 | 1.29 × 10−5 | 10 | |
| Cytoplasm | GO:0005737 | 1.35 × 10−5 | 10 | |
| Intracellular non-membrane-bounded organelle | GO:0043232 | 2.20 × 10−5 | 7 | |
| Non-membrane-bounded organelle | GO:0043228 | 2.26 × 10−5 | 7 | |
| Mitochondrion | GO:0005739 | 6.98 × 10−5 | 5 |
| Type | GO Annotation | GO_ID | p-Value | Count |
|---|---|---|---|---|
| Biological process | Carbohydrate derivative transport | GO:1901264 | 5.88 × 10−5 | 2 |
| Apoptotic mitochondrial changes | GO:0008637 | 0.0001 | 2 | |
| Organic substance transport | GO:0071702 | 0.0002 | 4 | |
| NADH oxidation | GO:0006116 | 0.0010 | 1 | |
| Response to peptide hormone | GO:0043434 | 0.0010 | 2 | |
| Transmembrane transport | GO:0055085 | 0.0010 | 3 | |
| Twitch skeletal muscle contraction | GO:0014721 | 0.0011 | 1 | |
| Voluntary skeletal muscle contraction | GO:0003010 | 0.0011 | 1 | |
| Ion transport | GO:0006811 | 0.0013 | 3 | |
| Response to peptide | GO:1901652 | 0.0015 | 2 | |
| Negative regulation of endoplasmic reticulum calcium ion concentration | GO:0032471 | 0.0015 | 1 | |
| Glycerol-3-phosphate metabolic process | GO:0006072 | 0.0016 | 1 | |
| Negative regulation of striated muscle contraction | GO:0045988 | 0.0016 | 1 | |
| transport | GO:0006810 | 0.0018 | 4 | |
| Cellular component | Endoplasmic reticulum membrane | GO:0005789 | 3.79 × 10−5 | 3 |
| Nuclear outer membrane–endoplasmic reticulum membrane network | GO:0042175 | 4.25 × 10−5 | 3 | |
| Integral component of endoplasmic reticulum membrane | GO:0030176 | 0.0002 | 2 | |
| Intrinsic component of endoplasmic reticulum membrane | GO:0031227 | 0.0002 | 2 | |
| Eukaryotic translation initiation factor 2B complex | GO:0005851 | 0.0010 | 1 | |
| Integral component of organelle membrane | GO:0031301 | 0.0011 | 2 | |
| Endoplasmic reticulum | GO:0005783 | 0.0013 | 3 | |
| Intrinsic component of organelle membrane | GO:0031300 | 0.0014 | 2 | |
| Molecular function | Active transmembrane transporter activity | GO:0022804 | 1.26 × 10−5 | 3 |
| Carbohydrate derivative transmembrane transporter activity | GO:1901505 | 1.84 × 10−5 | 2 | |
| Antiporter activity | GO:0015297 | 6.38 × 10−5 | 2 | |
| Active ion transmembrane transporter activity | GO:0022853 | 0.0005 | 2 | |
| Secondary active transmembrane transporter activity | GO:0015291 | 0.0005 | 2 | |
| Transmembrane transporter activity | GO:0022857 | 0.0005 | 3 | |
| Transporter activity | GO:0005215 | 0.0007 | 3 | |
| Phosphate ion transmembrane transporter activity | GO:0015114 | 0.0015 | 1 |
| Type | GO Annotation | GO_ID | p-Value | Count |
|---|---|---|---|---|
| Biological process | Toxin metabolic process | GO:0009404 | 3.4371 × 10−5 | 2 |
| Response to chemical | GO:0042221 | 0.0001 | 7 | |
| Secondary metabolic process | GO:0019748 | 0.0002 | 2 | |
| Primary alcohol metabolic process | GO:0034308 | 0.0002 | 2 | |
| Metabolic process | GO:0008152 | 0.0003 | 10 | |
| Organophosphate ester transport | GO:0015748 | 0.0007 | 2 | |
| Positive regulation of I-kappaB kinase/NF-kappaB signaling | GO:0043123 | 0.0008 | 2 | |
| Cellular process | GO:0009987 | 0.0009 | 11 | |
| Cellular metabolic process | GO:0044237 | 0.0011 | 9 | |
| Organic substance metabolic process | GO:0071704 | 0.0014 | 9 | |
| Regulation of I-kappaB kinase/NF-kappaB signaling | GO:0043122 | 0.0016 | 2 | |
| Response to stimulus | GO:0050896 | 0.0018 | 8 | |
| Response to organic substance | GO:0010033 | 0.0019 | 5 | |
| Peptide metabolic process | GO:0006518 | 0.0019 | 3 | |
| Cellular component | Cytoplasm | GO:0005737 | 5.83372 × 10−6 | 12 |
| Intracellular | GO:0005622 | 5.78814 × 10−5 | 12 | |
| Endoplasmic reticulum | GO:0005783 | 0.0002 | 5 | |
| Organelle membrane | GO:0031090 | 0.0005 | 5 | |
| Membrane-bounded organelle | GO:0043227 | 0.0005 | 10 | |
| Endoplasmic reticulum membrane | GO:0005789 | 0.0006 | 3 | |
| Nuclear outer membrane–endoplasmic reticulum Membrane network | GO:0042175 | 0.0007 | 3 | |
| Endomembrane system | GO:0012505 | 0.0011 | 6 | |
| Integral component of endoplasmic reticulum membrane | GO:0030176 | 0.0012 | 2 | |
| Intrinsic component of endoplasmic reticulum membrane | GO:0031227 | 0.0013 | 2 | |
| Organelle | GO:0043226 | 0.0013 | 10 | |
| Intracellular membrane-bounded organelle | GO:0043231 | 0.0018 | 9 | |
| Molecular function | Active transmembrane transporter activity | GO:0022804 | 0.0002 | 3 |
| Antiporter activity | GO:0015297 | 0.0004 | 2 | |
| Catalytic activity | GO:0003824 | 0.0009 | 8 | |
| Drug binding | GO:0008144 | 0.0021 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, T.; Zhang, J.; Ma, X.; Cao, S.; Li, W.; Yang, G. Proteomic Analysis of Low-Temperature Stress Response in Maize (Zea mays L.) at the Seedling Stage. Curr. Issues Mol. Biol. 2025, 47, 784. https://doi.org/10.3390/cimb47090784
Yu T, Zhang J, Ma X, Cao S, Li W, Yang G. Proteomic Analysis of Low-Temperature Stress Response in Maize (Zea mays L.) at the Seedling Stage. Current Issues in Molecular Biology. 2025; 47(9):784. https://doi.org/10.3390/cimb47090784
Chicago/Turabian StyleYu, Tao, Jianguo Zhang, Xuena Ma, Shiliang Cao, Wenyue Li, and Gengbin Yang. 2025. "Proteomic Analysis of Low-Temperature Stress Response in Maize (Zea mays L.) at the Seedling Stage" Current Issues in Molecular Biology 47, no. 9: 784. https://doi.org/10.3390/cimb47090784
APA StyleYu, T., Zhang, J., Ma, X., Cao, S., Li, W., & Yang, G. (2025). Proteomic Analysis of Low-Temperature Stress Response in Maize (Zea mays L.) at the Seedling Stage. Current Issues in Molecular Biology, 47(9), 784. https://doi.org/10.3390/cimb47090784





