The Analysis of Solanum lycopersicum Sap Dark Proteome Reveals Ordered and Disordered Protein Abundance
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition of S. lycopersicum Protein Sequences
2.2. Further Classification and Curation of up Dark Protein Database from S. lycopersicum
2.3. Intrinsically Disordered Prediction of up Dataset
2.4. Reanalysis of S. lycopersicum Mass Spectra
2.5. Proteomic Data Processing and Visualization
3. Results
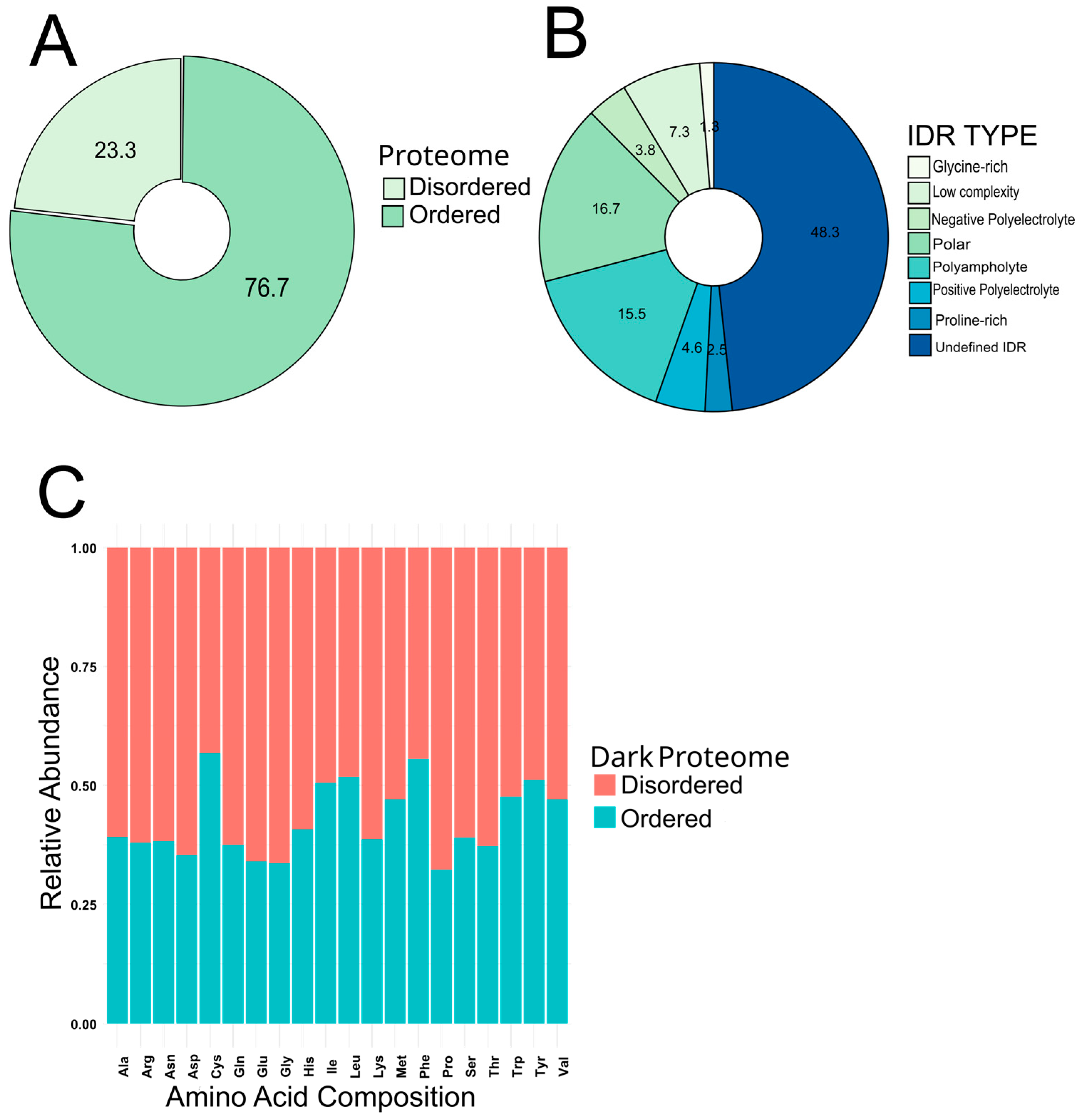
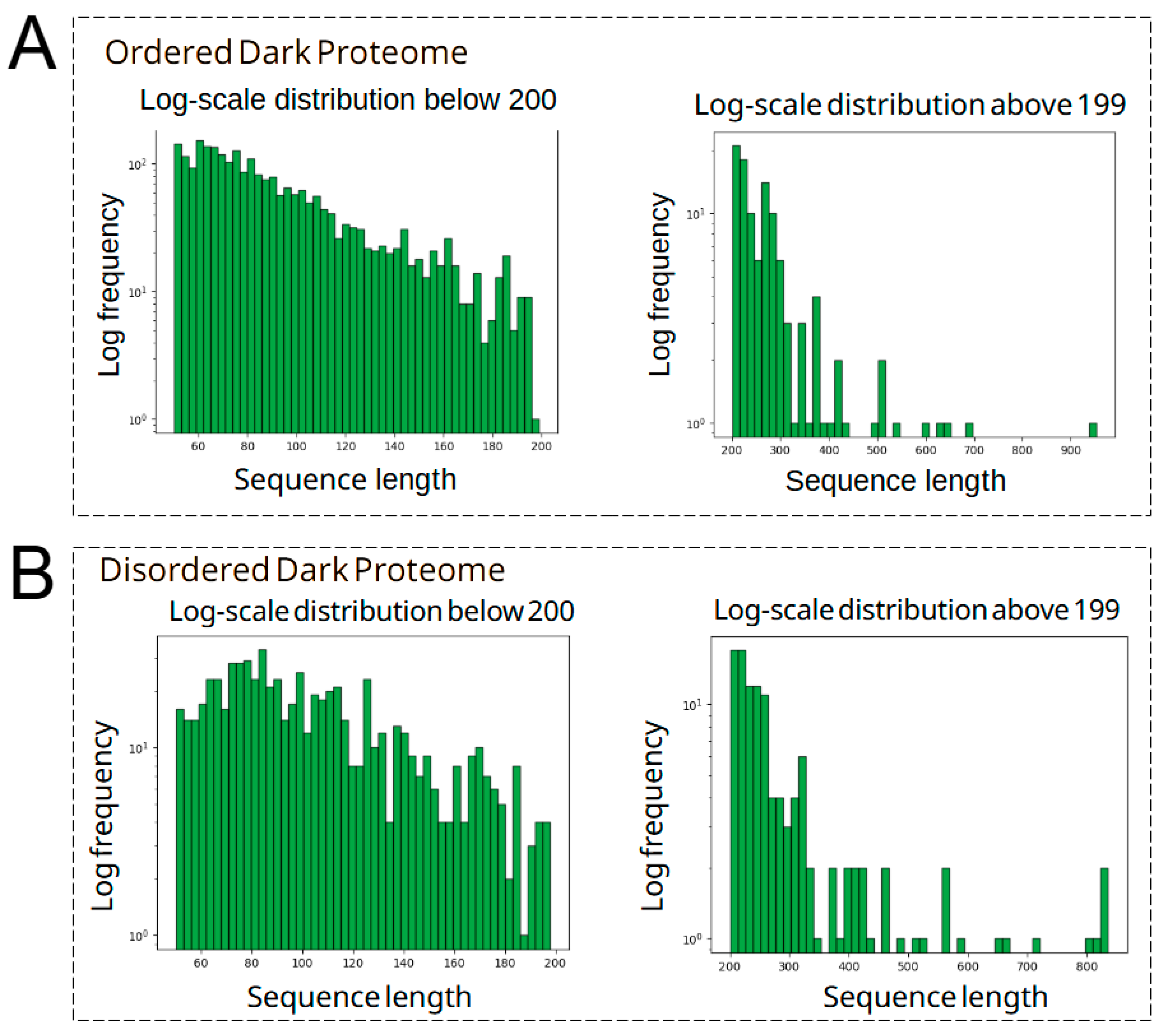
3.1. Identification, Processing, and Curation of Dark Proteins in S. lycopersicum and Their Functional Assesment into Ordered and Disordered Regions
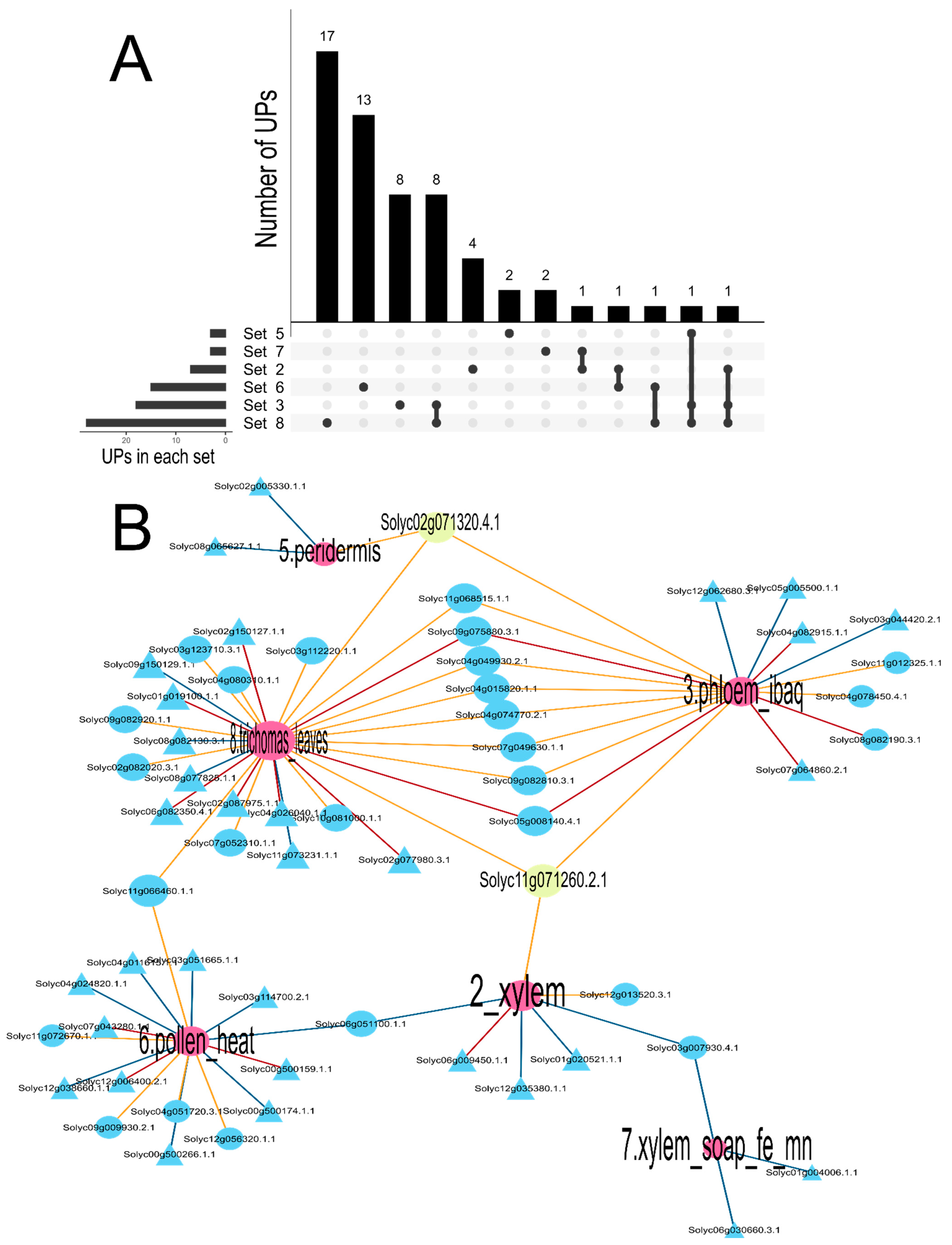
3.2. Experimental Identification of Dark Proteins and Network Analysis of Dark Proteins
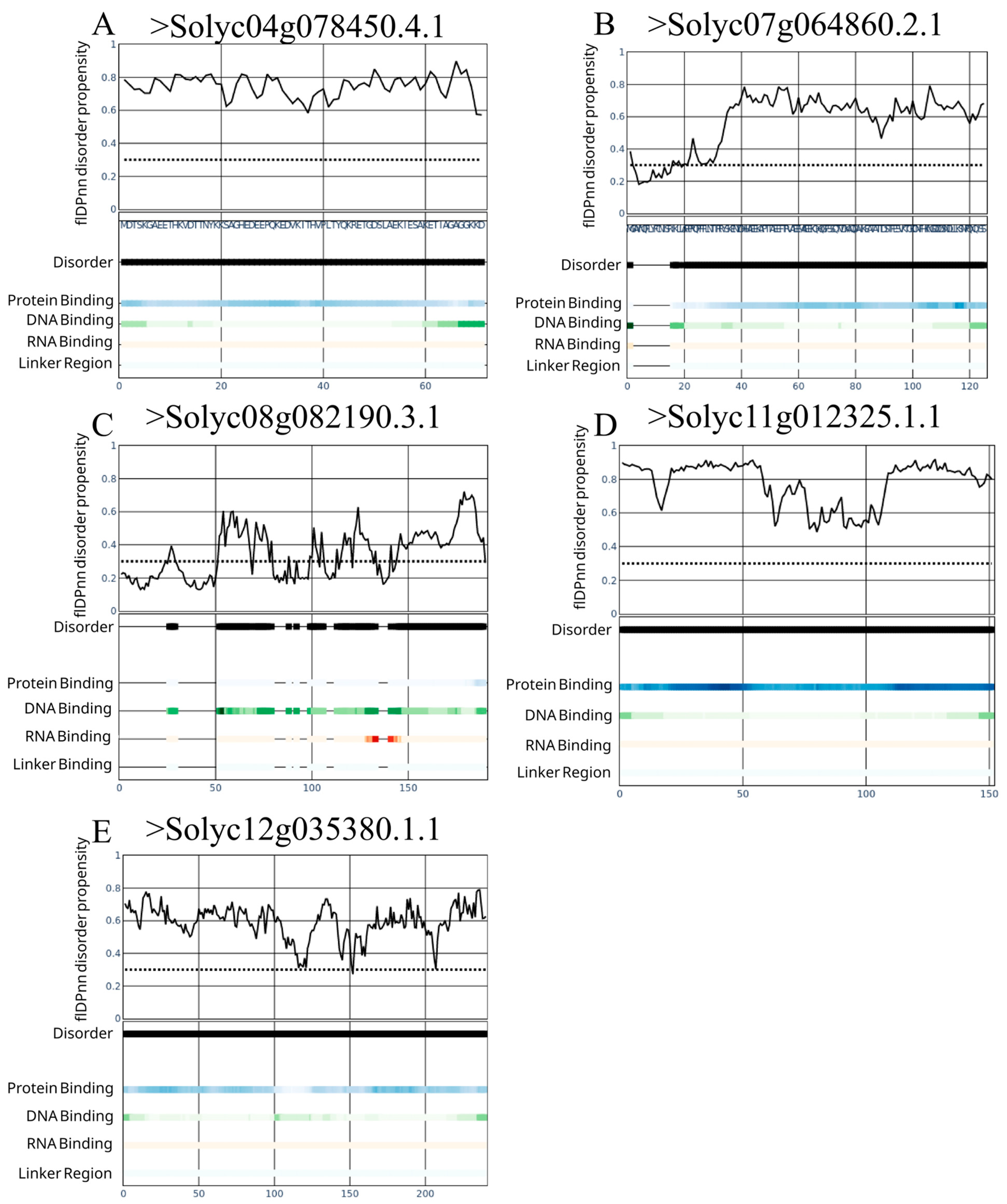
3.3. Identification of Intrinsically Disordered Proteins in the Vascular Transport System of S. lycopersicum
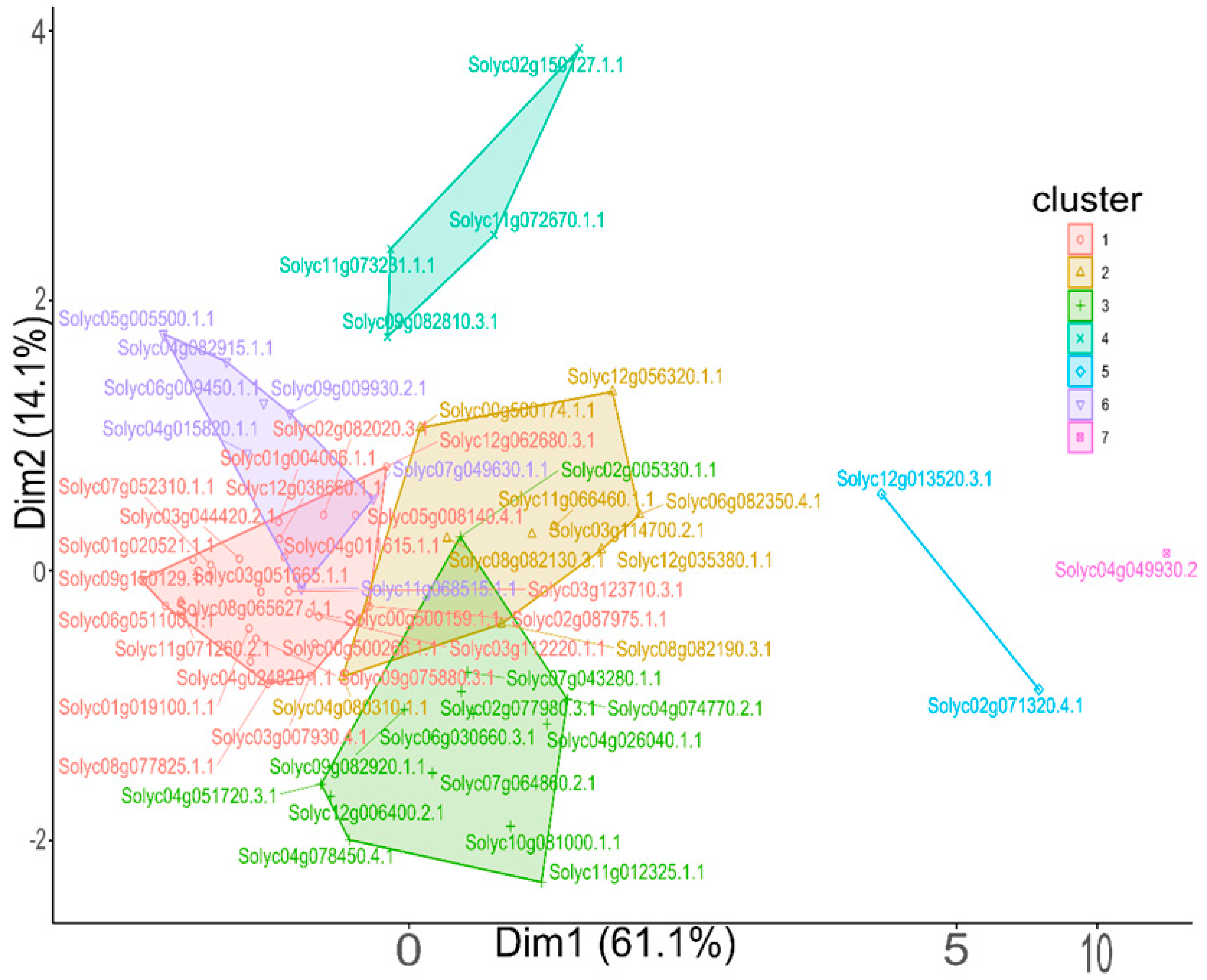
3.4. Clustering Analysis of Dark Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| UP | Unknown Protein |
| IDP | Intrinsically Disordered Proteins |
| IDR | Intrinsically Disordered regions |
| TF | Transcription Factors |
| CRY | Criptochormes |
| LEA | Late Embryogenesis Abundant |
| LFQ | Label-Free Quantification |
| UPDB | Uncharacterized Protein Data Bank |
References
- Sant’Ana, D.V.P.; Lefsrud, M. Tomato proteomics: Tomato as a model for crop proteomics. Sci. Hortic. 2018, 239, 224–233. [Google Scholar] [CrossRef]
- Gerszberg, A.; Hnatuszko-Konka, K.; Kowalczyk, T.; Kononowicz, A.K. Tomato (Solanum lycopersicum L.) in the service of biotechnology. Plant Cell Tissue Organ Cult. 2015, 120, 881–902. [Google Scholar] [CrossRef]
- Karlova, R.; Chapman, N.; David, K.; Angenent, G.C.; Seymour, G.B.; de Maagd, R.A. Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 2014, 65, 4527–4541. [Google Scholar] [CrossRef]
- Kimura, S.; Sinha, N. Tomato (Solanum lycopersicum): A Model Fruit-Bearing Crop. Cold Spring Harb. Protoc. 2008, 2008, pdb.emo105. [Google Scholar] [CrossRef]
- Doganlar, S.; Frary, A.; Tanksley, S.D. The genetic basis of seed-weight variation: Tomato as a model system. Theor. Appl. Genet. 2000, 100, 1267–1273. [Google Scholar] [CrossRef]
- Alexander, L.; Grierson, D. Ethylene biosynthesis and action in tomato: A model for climacteric fruit ripening. J. Exp. Bot. 2002, 53, 2039–2055. [Google Scholar] [CrossRef] [PubMed]
- Rombauts, S. Annotation of the Tomato Genome. In The Tomato Genome; Causse, M., Giovannoni, J., Bouzayen, M., Zouine, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 159–171. [Google Scholar]
- The Tomato Genome Consortium (TGC). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef]
- Hosmani, P.S.; Flores-Gonzalez, M.; van de Geest, H.; Maumus, F.; Bakker, L.V.; Schijlen, E.; van Haarst, J.; Cordewener, J.; Sanchez-Perez, G.; Peters, S.; et al. An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. bioRxiv 2019, 767764. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef]
- Gautam, A.; Pandey, P.; Pandey, A.K. Chapter 20—Proteomics in relation to abiotic stress tolerance in plants. In Plant Life Under Changing Environment; Tripathi, D.K., Pratap Singh, V., Chauhan, D.K., Sharma, S., Prasad, S.M., Dubey, N.K., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 513–541. [Google Scholar]
- Griffiths, W.J.; Wang, Y. Mass spectrometry: From proteomics to metabolomics and lipidomics. Chem. Soc. Rev. 2009, 38, 1882–1896. [Google Scholar] [CrossRef] [PubMed]
- Bitard-Feildel, T.; Callebaut, I. Exploring the dark foldable proteome by considering hydrophobic amino acids topology. Sci. Rep. 2017, 7, 41425. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Nepomnyachiy, S.; Ben-Tal, N.; Kolodny, R. Global view of the protein universe. Proc. Natl. Acad. Sci. USA 2014, 111, 11691–11696. [Google Scholar] [CrossRef]
- Faure, G.; Callebaut, I. Comprehensive Repertoire of Foldable Regions within Whole Genomes. PLoS Comput. Biol. 2013, 9, e1003280. [Google Scholar] [CrossRef]
- Tautz, D.; Domazet-Lošo, T. The evolutionary origin of orphan genes. Nat. Rev. Genet. 2011, 12, 692–702. [Google Scholar] [CrossRef]
- Caetano-Anollés, G.; Caetano-Anollés, D. An Evolutionarily Structured Universe of Protein Architecture. Genome Res. 2003, 13, 1563–1571. [Google Scholar] [CrossRef]
- Perdigão, N.; Heinrich, J.; Stolte, C.; Sabir, K.S.; Buckley, M.J.; Tabor, B.; Signal, B.; Gloss, B.S.; Hammang, C.J.; Rost, B.; et al. Unexpected features of the dark proteome. Proc. Natl. Acad. Sci. USA 2015, 112, 15898–15903. [Google Scholar] [CrossRef] [PubMed]
- Perdigão, N.; Rosa, A.C.; O’Donoghue, S.I. The Dark Proteome Database. BioData Min. 2017, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Perdigão, N.; Rosa, A. Dark Proteome Database: Studies on Dark Proteins. High-Throughput 2019, 8, 8. [Google Scholar] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Scaiewicz, A.; Levitt, M. Unique function words characterize genomic proteins. Proc. Natl. Acad. Sci. USA 2018, 115, 6703–6708. [Google Scholar] [CrossRef]
- Woodcock, S.; Mornon, J.P.; Henrissat, B. Detection of secondary structure elements in proteins by hydrophobic cluster analysis. Protein Eng. 1992, 5, 629–635. [Google Scholar] [CrossRef]
- Zamora-Briseño, J.A.; Pereira-Santana, A.; Reyes-Hernández, S.J.; Cerqueda-García, D.; Castaño, E.; Rodríguez-Zapata, L.C. Towards an understanding of the role of intrinsic protein disorder on plant adaptation to environmental challenges. Cell Stress Chaperones 2021, 26, 141–150. [Google Scholar] [CrossRef]
- Covarrubias, A.A.; Romero-Pérez, P.S.; Cuevas-Velazquez, C.L.; Rendón-Luna, D.F. The functional diversity of structural disorder in plant proteins. Arch. Biochem. Biophys. 2020, 680, 108229. [Google Scholar] [CrossRef] [PubMed]
- Choura, M.; Ebel, C.; Hanin, M. Genomic analysis of intrinsically disordered proteins in cereals: From mining to meaning. Gene 2019, 714, 143984. [Google Scholar] [CrossRef]
- Sun, X.; Jones, W.T.; Rikkerink, E.H.A. GRAS proteins: The versatile roles of intrinsically disordered proteins in plant signalling. Biochem. J. 2012, 442, 1–12. [Google Scholar] [CrossRef]
- Kang, C.Y.; Lian, H.L.; Wang, F.F.; Huang, J.R.; Yang, H.Q. Cryptochromes, Phytochromes, and COP1 Regulate Light-Controlled Stomatal Development in Arabidopsis. Plant Cell 2009, 21, 2624–2641. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.H.; Yang, H.Q. Cryptochrome Signaling in Plants. Photochem. Photobiol. 2007, 83, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Wang, X.; Zhang, L. Structural and Functional Dynamics of Dehydrins: A Plant Protector Protein under Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 3420. [Google Scholar] [CrossRef]
- Banerjee, A.; Roychoudhury, A. Group II late embryogenesis abundant (LEA) proteins: Structural and functional aspects in plant abiotic stress. Plant Growth Regul. 2016, 79, 1–17. [Google Scholar] [CrossRef]
- Kovacs, D.; Agoston, B.; Tompa, P. Disordered plant LEA proteins as molecular chaperones. Plant Signal. Behav. 2008, 3, 710–713. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Bandeira, N.; Perez-Riverol, Y.; Sharma, V.; Carver, J.J.; Mendoza, L.; Kundu, D.J.; Wang, S.; Bandla, C.; Kamatchinathan, S.; et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. 2023, 51, D1539–D1548. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Andreeva, A.; Blum, M.; Chuguransky, S.R.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Llinares-López, F.; Meng-Papaxanthos, L.; et al. The Pfam protein families database: Embracing AI/ML. Nucleic Acids Res. 2025, 53, D523–D534. [Google Scholar] [CrossRef]
- Hu, G.; Katuwawala, A.; Wang, K.; Wu, Z.; Ghadermarzi, S.; Gao, J.; Kurgan, L. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat. Commun. 2021, 12, 4438. [Google Scholar] [CrossRef]
- Necci, M.; Piovesan, D.; Clementel, D.; Dosztányi, Z.; Tosatto, S.C.E. MobiDB-lite 3.0: Fast consensus annotation of intrinsic disorder flavors in proteins. Bioinformatics 2021, 36, 5533–5534. [Google Scholar] [CrossRef]
- Necci, M.; Piovesan, D.; CAID Predictors; DisProt Curators; Tosatto, S.C.E. Critical assessment of protein intrinsic disorder prediction. Nat. Methods 2021, 18, 472–481. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Schaab, C.; Geiger, T.; Stoehr, G.; Cox, J.; Mann, M. Analysis of High Accuracy, Quantitative Proteomics Data in the MaxQB Database. Mol. Cell. Proteom. 2012, 11, M111.014068. [Google Scholar] [CrossRef]
- Kong, A.T.; Leprevost, F.V.; Avtonomov, D.M.; Mellacheruvu, D.; Nesvizhskii, A.I. MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry–based proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hou, X.; Du, T. Responses of tomato fruit water balance and xylem hydraulic property of pedicel and calyx to water deficit and salinity stress. Environ. Exp. Bot. 2023, 206, 105195. [Google Scholar] [CrossRef]
- Braun, D.M. Phloem Loading and Unloading of Sucrose: What a Long, Strange Trip from Source to Sink. Annu. Rev. Plant Biol. 2022, 73, 553–584. [Google Scholar] [CrossRef]
- Sun, C.; Wei, J.; Gu, X.; Wu, M.; Li, M.; Liu, Y.; An, N.; Wu, K.; Wu, S.; Wu, J.; et al. Different multicellular trichome types coordinate herbivore mechanosensing and defense in tomato. Plant Cell 2024, 36, 4952–4969. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Symeonidi, E.; Pang, T.Y.; Denyer, T.; Weidauer, D.; Bezrutczyk, M.; Miras, M.; Zöllner, N.; Hartwig, T.; Wudick, M.M.; et al. Distinct identities of leaf phloem cells revealed by single cell transcriptomics. Plant Cell 2021, 33, 511–530. [Google Scholar] [CrossRef] [PubMed]
- Kuntorini, E.M.; Sari, S.G.; Fariani, R. The morphoanatomy, histochemistry, and phytochemistry of the leaves and fruits of Rhodomyrtus tomentosa. Biodivers. J. Biol. Divers. 2023, 24, 98–105. [Google Scholar] [CrossRef]
- Beckman, C.H. Phenolic-storing cells: Keys to programmed cell death and periderm formation in wilt disease resistance and in general defence responses in plants? Physiol. Mol. Plant Pathol. 2000, 57, 101–110. [Google Scholar] [CrossRef]
- Wright, B.W.; Yi, Z.; Weissman, J.S.; Chen, J. The dark proteome: Translation from noncanonical open reading frames. Trends Cell Biol. 2022, 32, 243–258. [Google Scholar] [CrossRef]
- Kulkarni, P.; Uversky, V.N. Intrinsically Disordered Proteins: The Dark Horse of the Dark Proteome. Proteomics 2018, 18, 1800061. [Google Scholar] [CrossRef]
- Medvedev, K.E.; Pei, J.; Grishin, N.V. DisEnrich: Database of enriched regions in human dark proteome. Bioinformatics 2022, 38, 1870–1876. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, J.; Sun, N.; Tu, C.; Shi, X.; Cheng, H.; Liu, S.; Li, S.; Wang, Y.; Zheng, Y.; et al. Intrinsically Disordered Proteins as Important Players during Desiccation Stress of Soybean Radicles. J. Proteome Res. 2017, 16, 2393–2409. [Google Scholar] [CrossRef]
- Das Laha, S.; Das, D.; Ghosh, T.; Podder, S. Enrichment of intrinsically disordered residues in ohnologs facilitates abiotic stress resilience in Brassica rapa. J. Plant Res. 2023, 136, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, J.; Deng, L.; Liu, H.; Liu, H.; Zhao, W.; Zhao, Y.; Sun, X.; Fan, S.; Wang, H.; et al. An intrinsically disordered region-containing protein mitigates the drought–growth trade-off to boost yields. Plant Physiol. 2023, 192, 274–292. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Velazquez, C.L.; Vellosillo, T.; Guadalupe, K.; Schmidt, H.B.; Yu, F.; Moses, D.; Brophy, J.A.; Cosio-Acosta, D.; Das, A.; Wang, L.; et al. Intrinsically disordered protein biosensor tracks the physical-chemical effects of osmotic stress on cells. Nat. Commun. 2021, 12, 5438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhu, C.; Duan, Y.; Liu, T.; Liu, H.; Su, C.; Lu, Y. The intrinsically disordered region from PP2C phosphatases functions as a conserved CO2 sensor. Nat. Cell Biol. 2022, 24, 1029–1037. [Google Scholar] [CrossRef]
- Orand, T.; Delaforge, E.; Lee, A.; Kragelj, J.; Tengo, M.; Tengo, L.; Blackledge, M.; Boeri Erba, E.; Davis, R.J.; Palencia, A.; et al. Bipartite binding of the intrinsically disordered scaffold protein JIP1 to the kinase JNK1. Proc. Natl. Acad. Sci. USA 2025, 122, e2419915122. [Google Scholar] [CrossRef]
- Hansen, J.C.; Lu, X.; Ross, E.D.; Woody, R.W. Intrinsic Protein Disorder, Amino Acid Composition, and Histone Terminal Domains. J. Biol. Chem. 2006, 281, 1853–1856. [Google Scholar] [CrossRef]
- Wetzler, D.E.; Fuchs Wightman, F.; Bucci, H.A.; Rinaldi, J.; Caramelo, J.J.; Iusem, N.D.; Ricardi, M.M. Conformational plasticity of the intrinsically disordered protein ASR1 modulates its function as a drought stress-responsive gene. PLoS ONE 2018, 13, e0202808. [Google Scholar] [CrossRef]
- Xoconostle-Cázares, B.; Martínez-Navarro, A.C.; Ruiz-Medrano, R. Phloem Long-Distance Trafficking of RNAs and Proteins. eLS 2016, 1–9. [Google Scholar] [CrossRef]
- Kehr, J.; Kragler, F. Long distance RNA movement. New Phytol. 2018, 218, 29–40. [Google Scholar] [CrossRef]
- Tolstyko, E.A.; Lezzhov, A.A.; Morozov, S.Y.; Solovyev, A.G. Phloem transport of structured RNAs: A widening repertoire of trafficking signals and protein factors. Plant Sci. 2020, 299, 110602. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyes-Soria, F.A.; Guillén-Chable, F.; Castaño de la Serna, E.; Sánchez-Teyer, L.F.; Herrera-Alamillo, M.A.; Pereira-Santana, A.; Rodriguez-Zapata, L.C. The Analysis of Solanum lycopersicum Sap Dark Proteome Reveals Ordered and Disordered Protein Abundance. Curr. Issues Mol. Biol. 2025, 47, 769. https://doi.org/10.3390/cimb47090769
Reyes-Soria FA, Guillén-Chable F, Castaño de la Serna E, Sánchez-Teyer LF, Herrera-Alamillo MA, Pereira-Santana A, Rodriguez-Zapata LC. The Analysis of Solanum lycopersicum Sap Dark Proteome Reveals Ordered and Disordered Protein Abundance. Current Issues in Molecular Biology. 2025; 47(9):769. https://doi.org/10.3390/cimb47090769
Chicago/Turabian StyleReyes-Soria, Francisco Antonio, Francisco Guillén-Chable, Enrique Castaño de la Serna, Lorenzo Felipe Sánchez-Teyer, Miguel Angel Herrera-Alamillo, Alejandro Pereira-Santana, and Luis Carlos Rodriguez-Zapata. 2025. "The Analysis of Solanum lycopersicum Sap Dark Proteome Reveals Ordered and Disordered Protein Abundance" Current Issues in Molecular Biology 47, no. 9: 769. https://doi.org/10.3390/cimb47090769
APA StyleReyes-Soria, F. A., Guillén-Chable, F., Castaño de la Serna, E., Sánchez-Teyer, L. F., Herrera-Alamillo, M. A., Pereira-Santana, A., & Rodriguez-Zapata, L. C. (2025). The Analysis of Solanum lycopersicum Sap Dark Proteome Reveals Ordered and Disordered Protein Abundance. Current Issues in Molecular Biology, 47(9), 769. https://doi.org/10.3390/cimb47090769





