Integration of eQTL and GEO Datasets to Identify Genes Associated with Breast Ductal Carcinoma In Situ
Abstract
1. Introduction
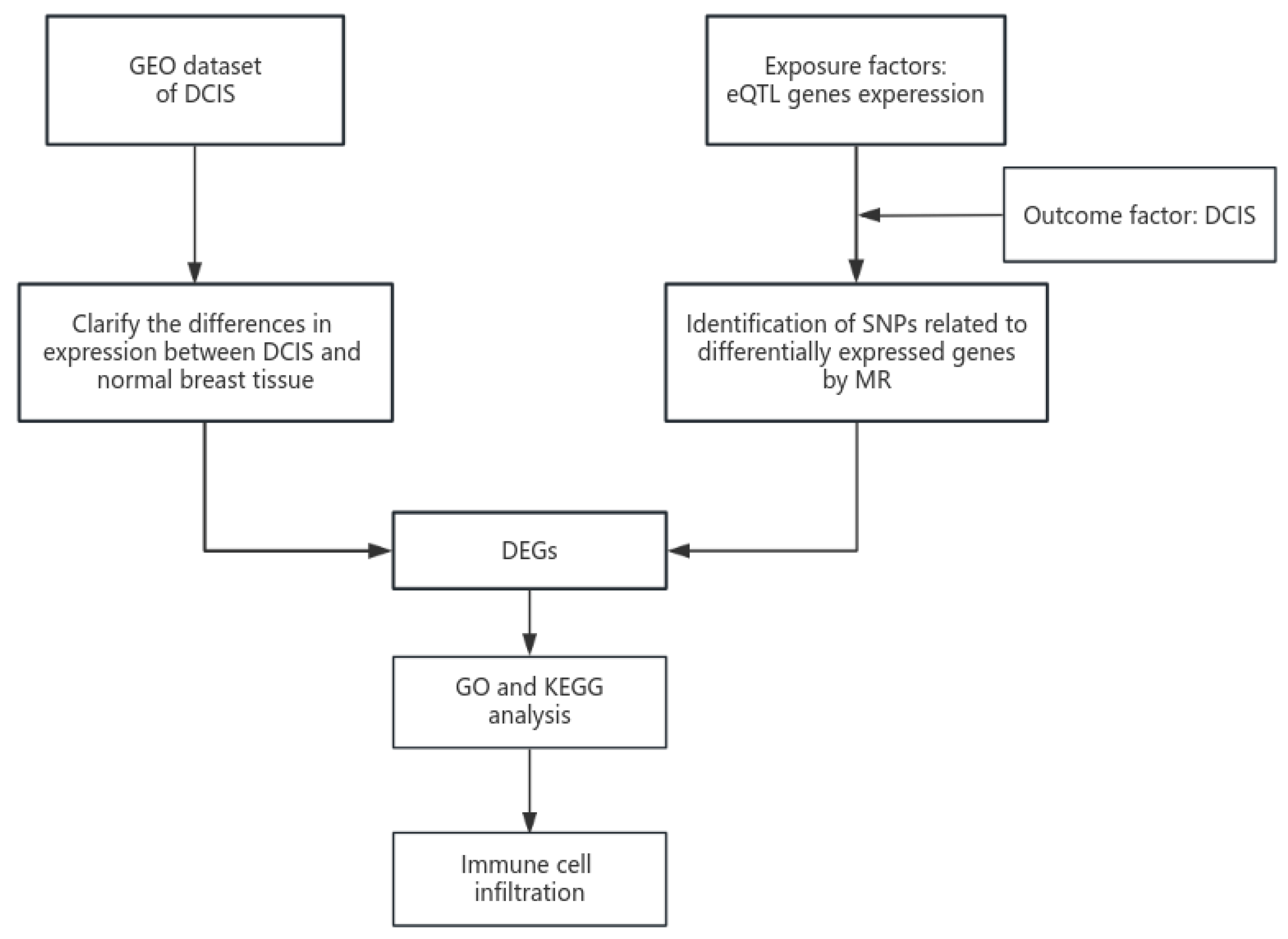
2. Materials and Methods
2.1. GEO Datasets
2.1.1. GEO Dataset Selection
2.1.2. GEO Dataset Statistical Analysis
2.2. eQTL Dataset Processing
2.2.1. eQTL Dataset Acquisition
2.2.2. eQTL Statistical Analysis
2.3. GEO and eQTL Joint Analysis
2.3.1. Acquisition and Validation of Intersection Genes
2.3.2. GO and KEGG Pathway Analyses
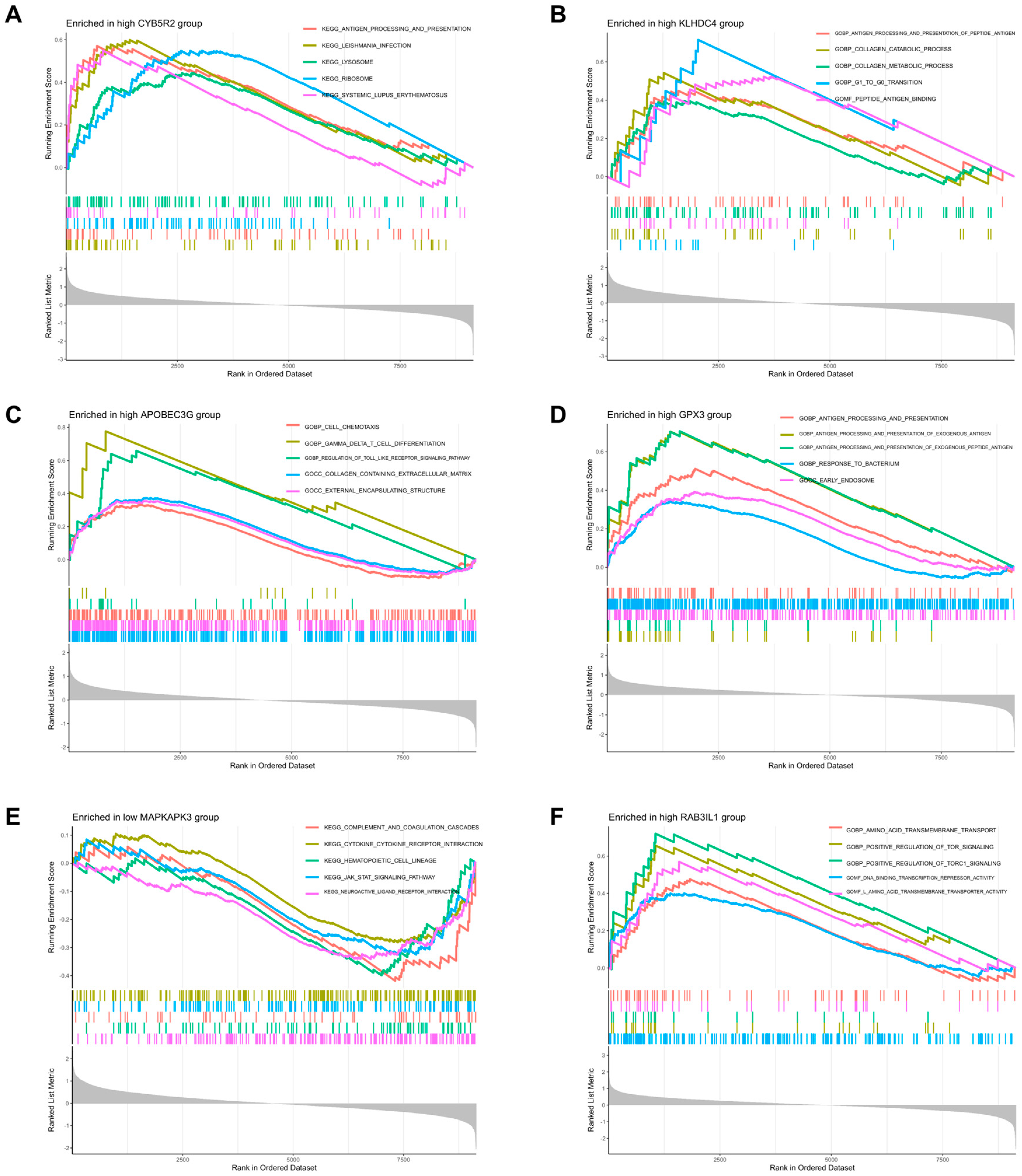
2.3.3. Gene Set Enrichment Analysis (GSEA)
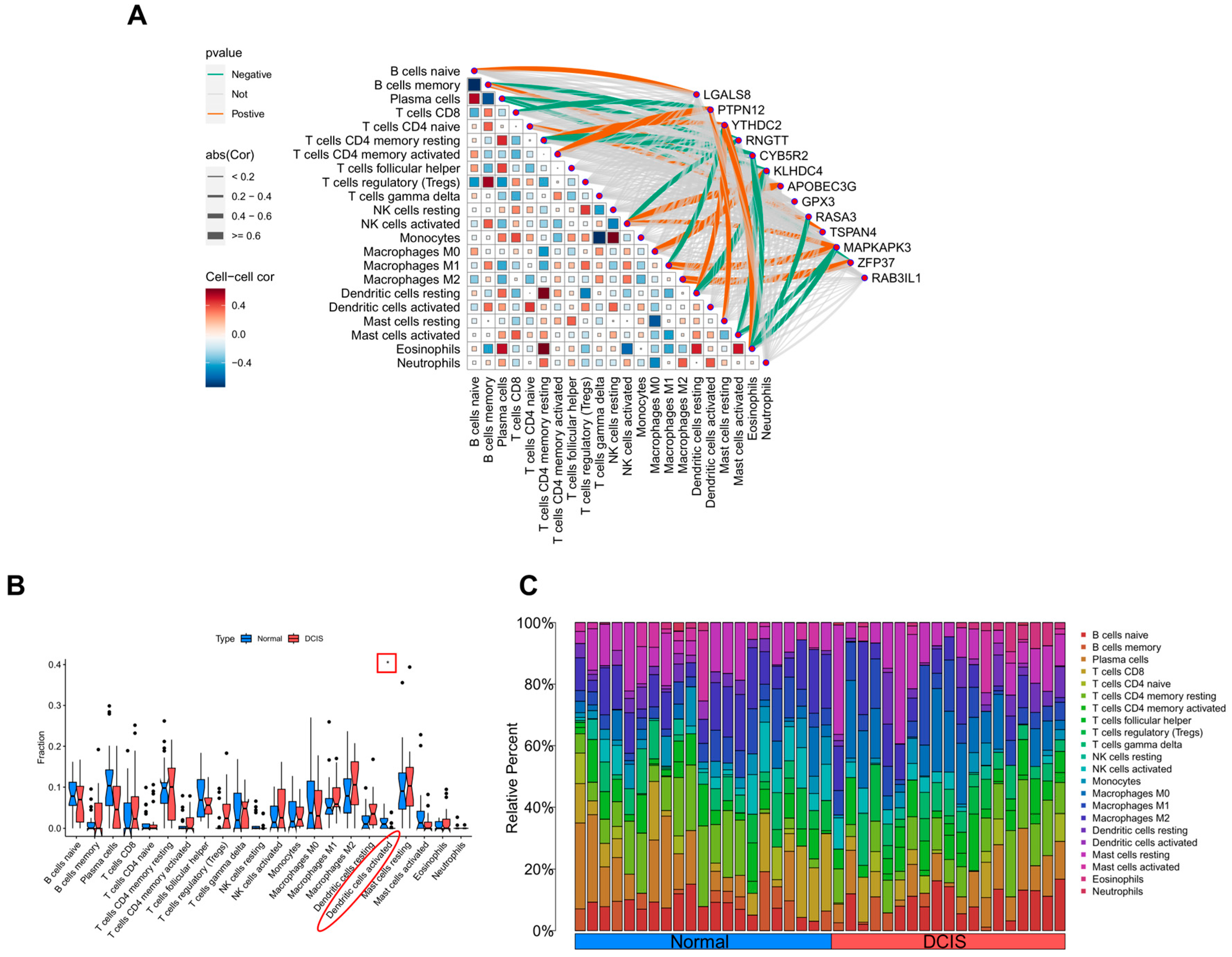
2.3.4. Immune Cell Infiltration
2.4. Data Validation
2.5. Gene Expression Validation
2.6. Functional Experimental Validation
2.6.1. Cell Culture
2.6.2. Cell Transfection
2.6.3. Quantitative Real-Time PCR (qRT-PCR)
2.6.4. Transwewll Invation Assay
2.7. Statistics
3. Results
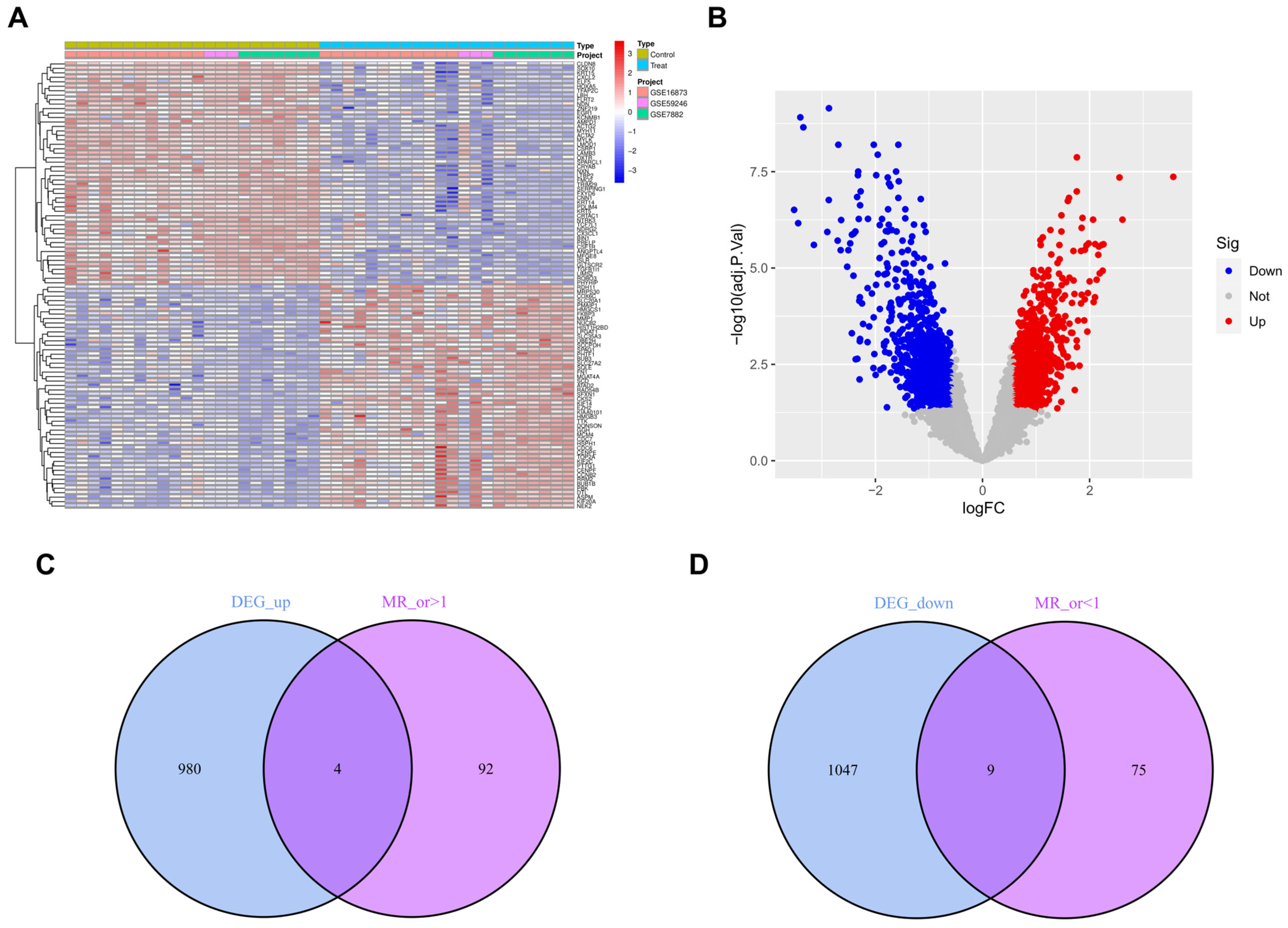
3.1. Outcomes of the GEO and eQTL Datasets
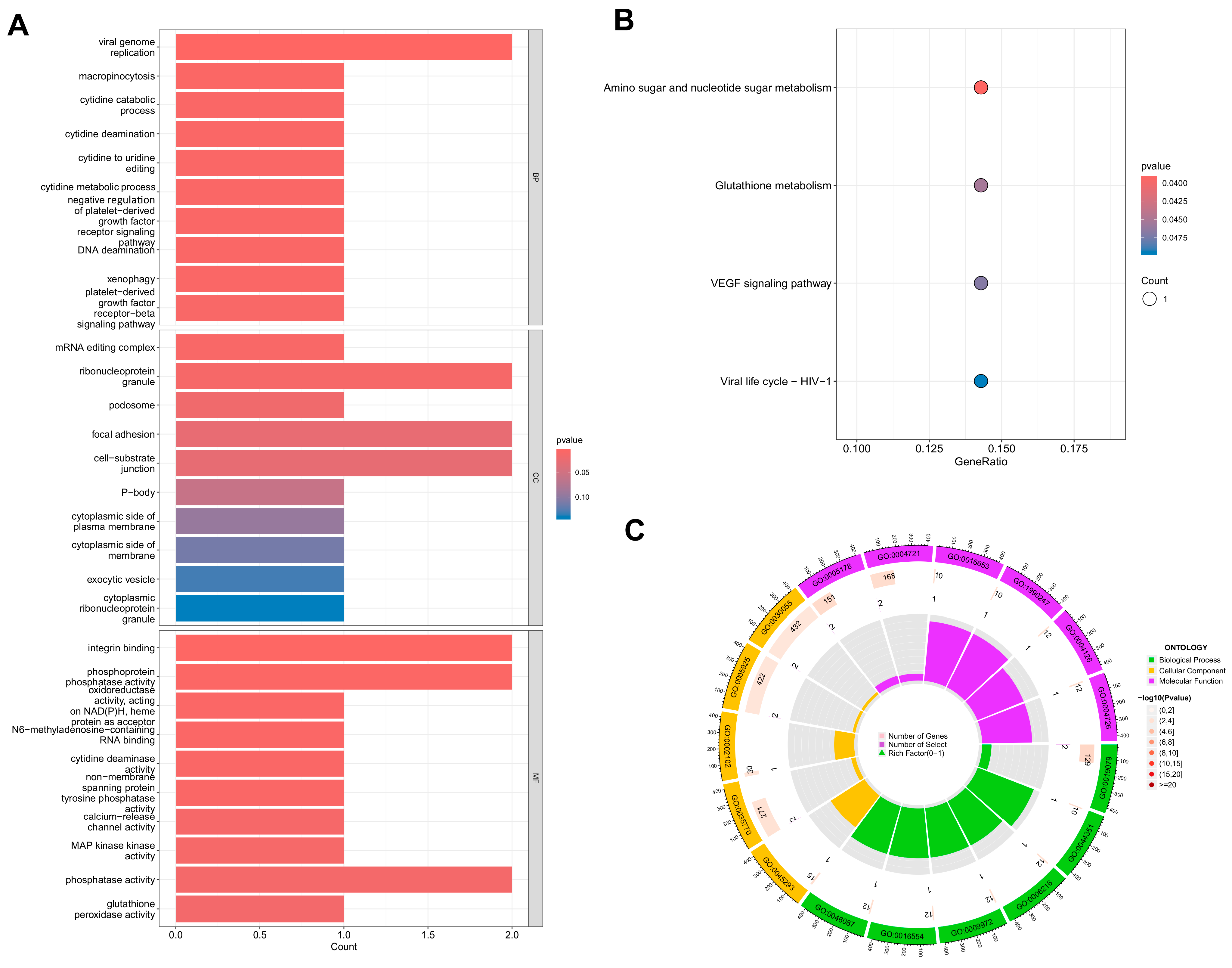
3.2. GO and KEGG
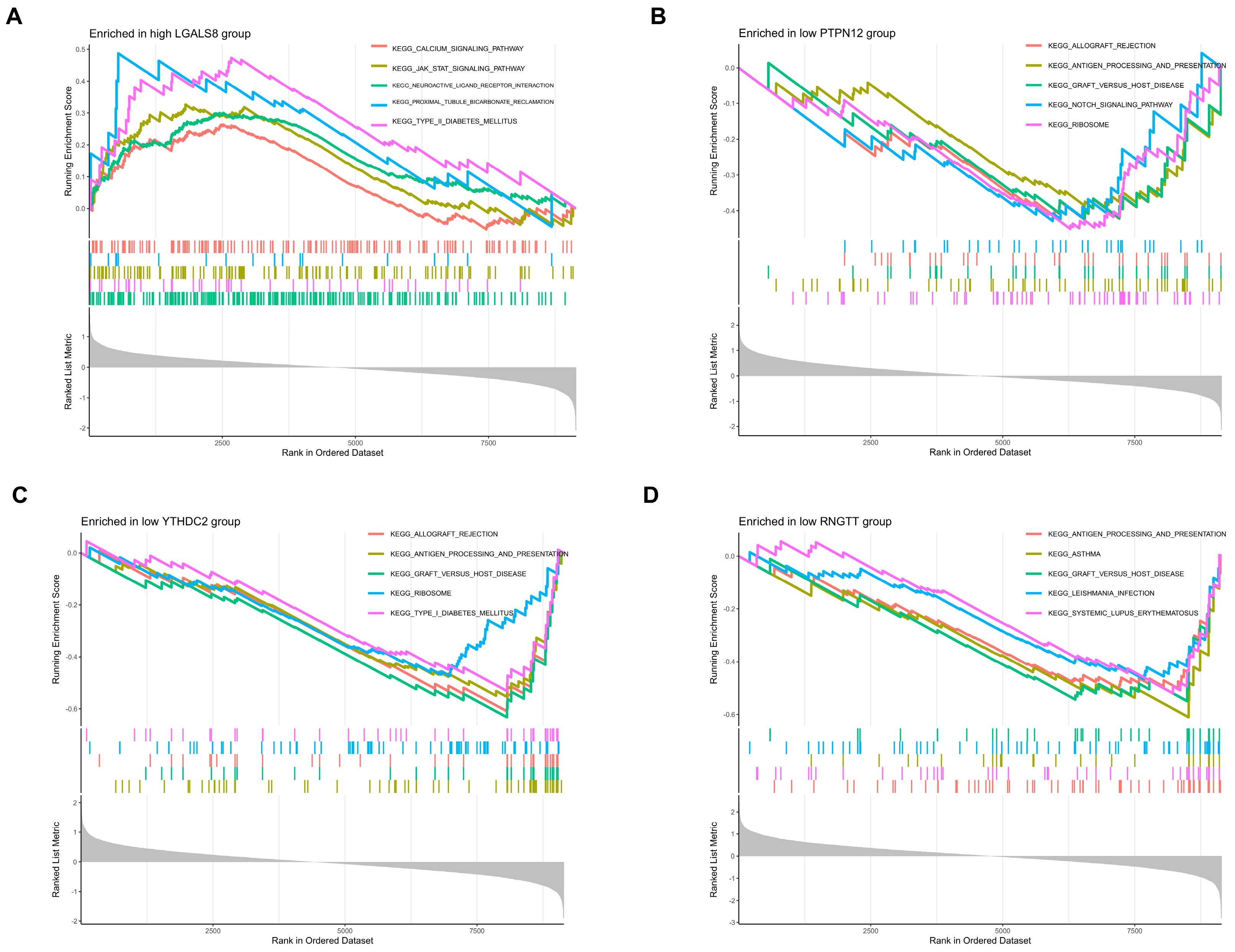
3.3. GSEA
3.4. CIBERSORT Analysis
3.5. Statistical Validation
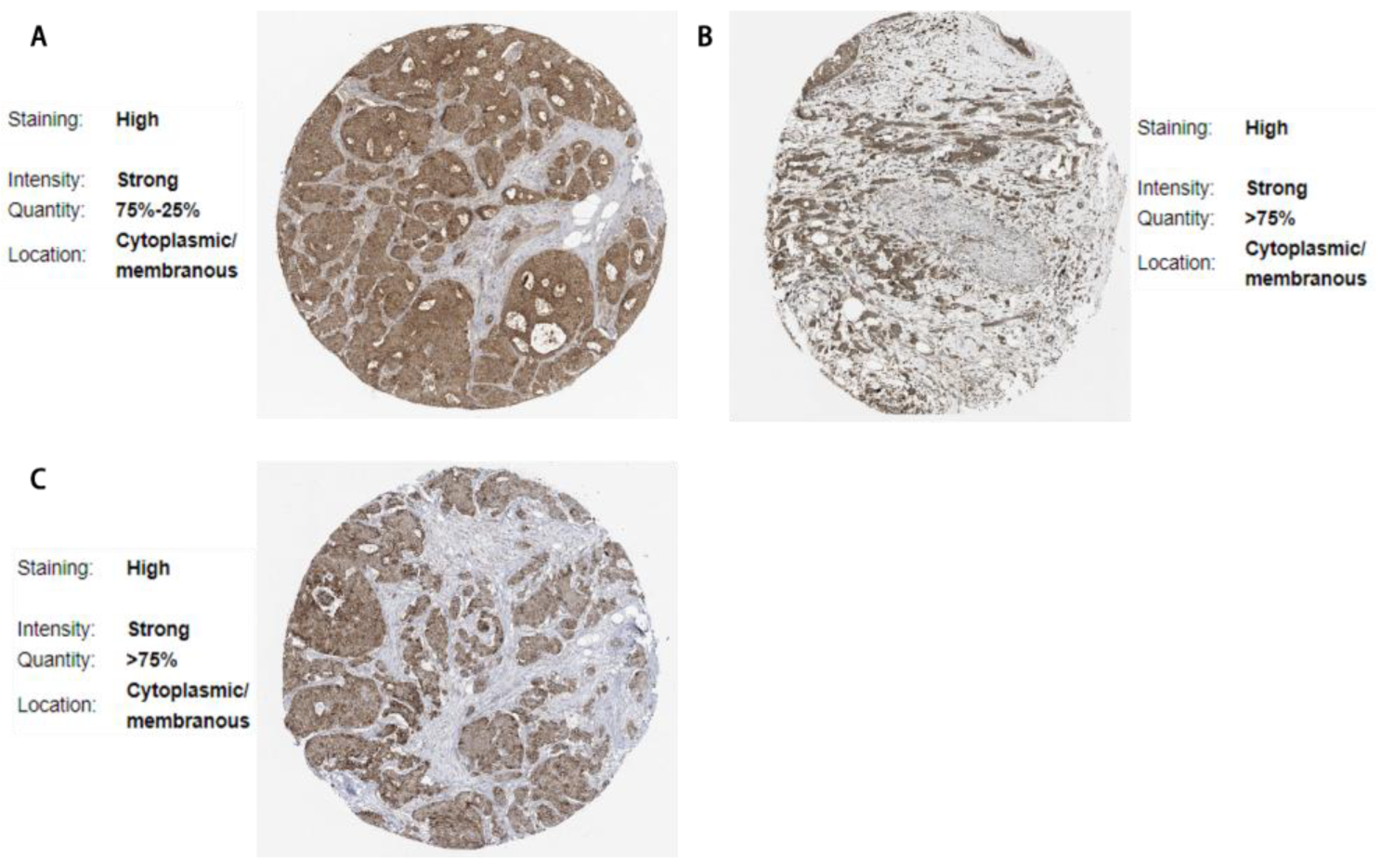
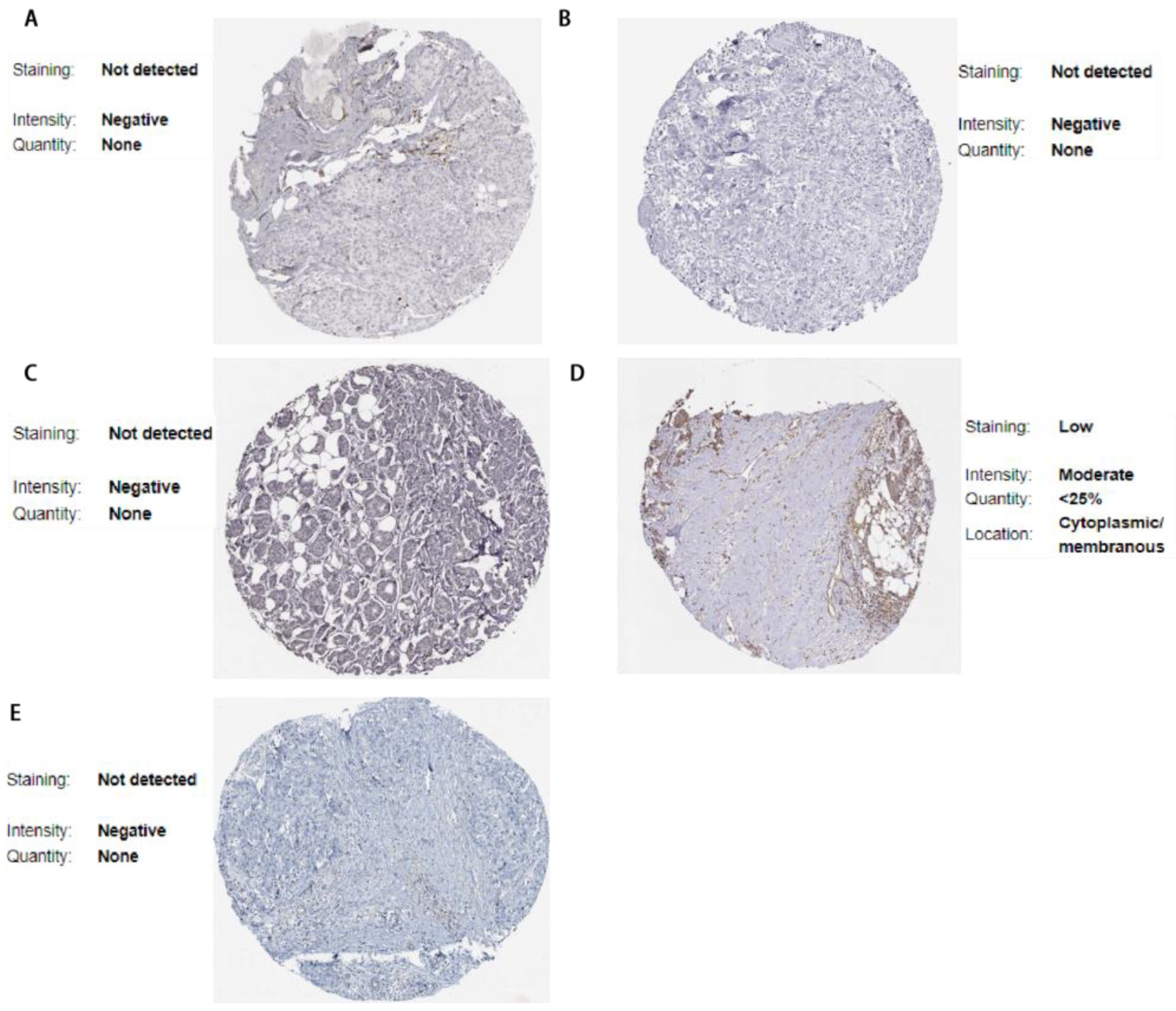
3.6. Validation via the Human Protein Atlas
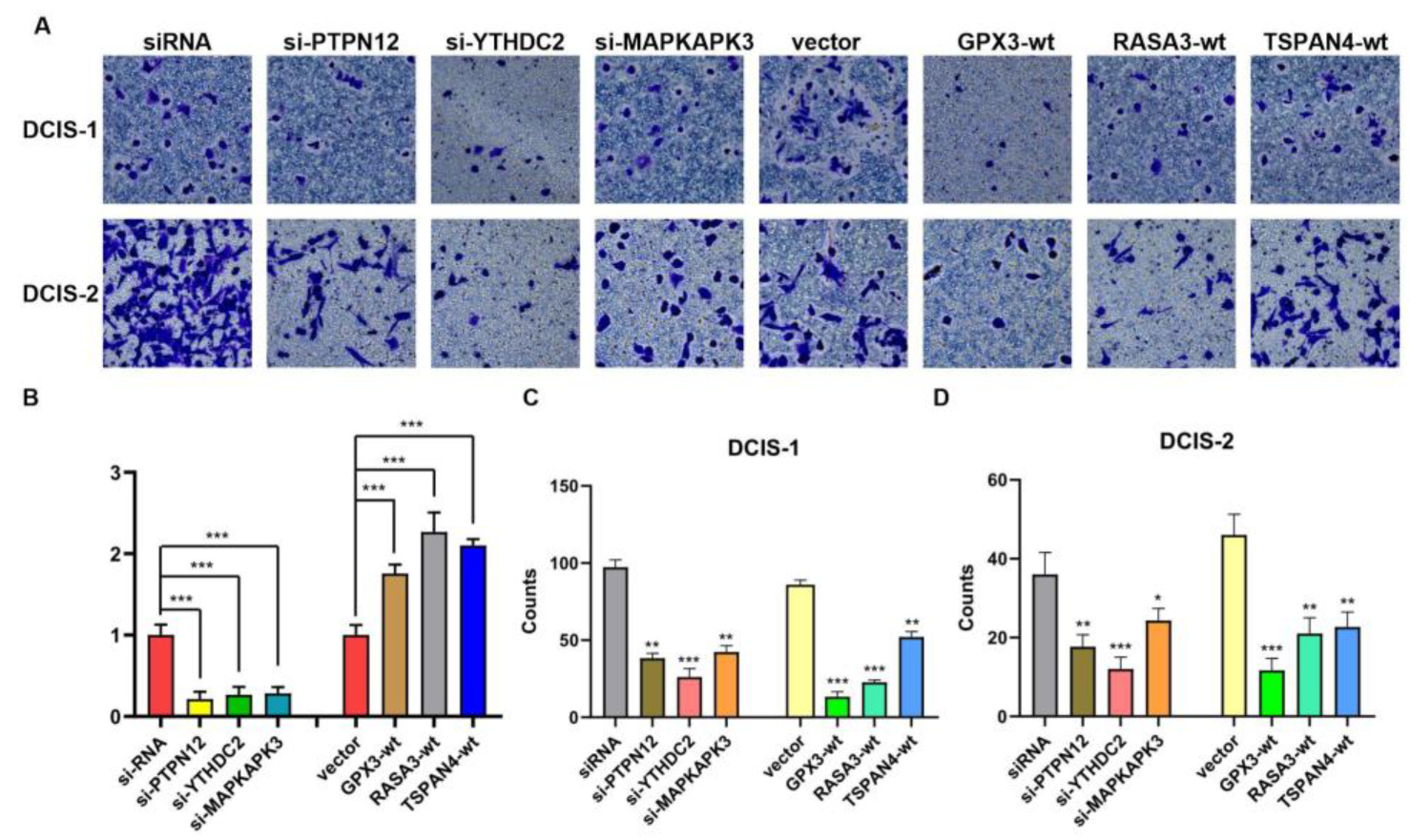
3.7. Results of Cellular Experiments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DCIS | Breast ductal carcinoma in situ |
| MR | Mendelian randomization |
| GEO | Gene Expression Omnibus |
| DEGs | Differentially expressed genes |
| SNPs | Single-nucleotide polymorphisms |
| eQTL | Expression quantitative trait locus |
| GWAS | Genome-Wide association study database |
| GO | Gene ontology analysis |
| KEGG | Kyoto encyclopedia of genes and genomes analysis |
| CIBERSORT | Cell-type identification by estimating relative subsets of RNA transcripts |
| TME | Immune-related tumor microenvironment |
| IDC | Invasive ductal cancer |
| IVs | Instrumental variables |
| LD | Linkage disequilibrium |
| WIV | Weak instrumental variable |
| MRE | MR egger |
| WM | Weighted median |
| IVM | Inverse variance weighted |
| SM | Simple mode |
| OR | Odds Ratio |
| GSEA | Gene Set Enrichment Analysis |
| NES | Normalized enrichment score |
| NOM | Nominal |
| FDR | False discovery rate |
| HPA | Human Protein Atlas |
| siRNAs | Small interfering RNAs |
| qRT-PCR | Quantitative real-time PCR |
| BP | Biological process |
| CC | Cellular component |
| MF | Molecular function |
| TSG | Tumor suppressor gene |
| IBC | Invasive breast cancer |
References
- Van Seijen, M.; Lips, E.H.; Thompson, A.M.; Nik-Zainal, S.; Futreal, A.; Hwang, E.S.; Verschuur, E.; Lane, J.; Jonkers, J.; Rea, D.W.; et al. Ductal carcinoma in situ: To treat or not to treat, that is the question. Br. J. Cancer 2019, 121, 285–292. [Google Scholar] [CrossRef]
- Welch, H.G.; Black, W.C. Using autopsy series to estimate the disease “reservoir” for ductal carcinoma in situ of the breast: How much more breast cancer can we find. Ann. Intern. Med. 1997, 127, 1023–1028. [Google Scholar] [CrossRef]
- Sanders, M.E.; Schuyler, P.A.; Dupont, W.D.; Page, D.L. The natural history of low-grade ductal carcinoma in situ of the breast in women treated by biopsy only revealed over 30 years of long-term follow-up. Cancer 2005, 103, 2481–2484. [Google Scholar] [CrossRef]
- Fleischer, T.; Frigessi, A.; Johnson, K.C.; Edvardsen, H.; Touleimat, N.; Klajic, J.; Riis, M.L.; Haakensen, V.D.; Wärnberg, F.; Naume, B.; et al. Genome-wide DNA methylation profiles in progression to in situ and invasive carcinoma of the breast with impact on gene transcription and prognosis. Genome Biol. 2014, 15, 435. [Google Scholar]
- Zhou, W.; Liu, G.; Hung, R.J.; Haycock, P.C.; Aldrich, M.C.; Andrew, A.S.; Arnold, S.M.; Bickeböller, H.; Bojesen, S.E.; Brennan, P.; et al. Causal relationships between body mass index, smoking and lung cancer: Univariable and multivariable Mendelian randomization. Int. J. Cancer 2021, 148, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Ma, Z.; Zhang, X.; Hang, D.; Yin, R.; Fetng, J.; Xu, L.; Shen, H. C-reactive protein and cancer risk: A pan-cancer study of prospective cohort and Mendelian randomization analysis. BMC Med. 2022, 20, 301. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sundquist, K.; Zhang, N.; Wang, X.; Sundquist, J.; Memon, A.A. Mitochondrial related genome-wide Mendelian randomization identifies putatively causal genes for multiple cancer types. eBioMedicine 2023, 88, 104432. [Google Scholar] [CrossRef]
- Ma, J.; Li, J.; Jin, C.; Yang, J.; Zheng, C.; Chen, K.; Xie, Y.; Yang, Y.; Bo, Z.; Wang, J.; et al. Association of gut microbiome and primary liver cancer: A two-sample Mendelian randomization and case-control study. Liver Int. 2023, 43, 221–233. [Google Scholar] [CrossRef]
- Constantinescu, A.E.; Bull, C.J.; Jones, N.; Mitchell, R.; Burrows, K.; Dimou, N.; Bézieau, S.; Brenner, H.; Buchanan, D.D.; D’Amato, M.; et al. Circulating white blood cell traits and colorectal cancer risk: A Mendelian randomisation study. Int. J. Cancer 2024, 154, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Rahbar, H. Imaging and Pathology of Ductal Carcinoma in Situ of the Breast: The Forest and the Trees. Radiology 2022, 303, 285–286. [Google Scholar] [CrossRef]
- Kanbayashi, C.; Iwata, H. Current approach and future perspective for ductal carcinoma in situ of the breast. Jpn. J. Clin. Oncol. 2017, 47, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Van Bockstal, M.R.; Agahozo, M.C.; Koppert, L.B.; van Deurzen, C. A retrospective alternative for active surveillance trials for ductal carcinoma in situ of the breast. Int. J. Cancer 2020, 146, 1189–1197. [Google Scholar] [CrossRef]
- Casasent, A.K.; Almekinders, M.M.; Mulder, C.; Bhattacharjee, P.; Collyar, D.; Thompson, A.M.; Jonkers, J.; Lips, E.H.; van Rheenen, J.; Hwang, E.S.; et al. Learning to distinguish progressive and non-progressive ductal carcinoma in situ. Nat. Rev. Cancer 2022, 22, 663–678, Erratum in Nat. Rev. Cancer 2023, 23, 112. [Google Scholar] [CrossRef]
- Hophan, S.L.; Odnokoz, O.; Liu, H.; Luo, Y.; Khan, S.; Gradishar, W.; Zhou, Z.; Badve, S.; Torres, M.A.; Wan, Y.; et al. Ductal Carcinoma In Situ of Breast: From Molecular Etiology to Therapeutic Management. Endocrinology 2022, 163, bqac027. [Google Scholar] [CrossRef] [PubMed]
- Rane, S.U.; Mirza, H.; Grigoriadis, A.; Pinder, S.E. Selection and evolution in the genomic landscape of copy number alterations in ductal carcinoma in situ (DCIS) and its progression to invasive carcinoma of ductal/no special type: A meta-analysis. Breast Cancer Res. Treat. 2015, 153, 101–121. [Google Scholar] [CrossRef]
- Ouattara, D.; Mathelin, C.; Özmen, T.; Lodi, M. Molecular Signatures in Ductal Carcinoma In Situ (DCIS): A Systematic Review and Meta-Analysis. J. Clin. Med. 2023, 12, 2036. [Google Scholar] [CrossRef]
- Rhee, I.; Zhong, M.C.; Reizis, B.; Cheong, C.; Veillette, A. Control of dendritic cell migration, T cell-dependent immunity, and autoimmunity by protein tyrosine phosphatase PTPN12 expressed in dendritic cells. Mol. Cell Biol. 2014, 34, 888–899. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Zhang, S.; Guo, Z.; Wang, G.; Zhang, G.; Xie, L.; Lou, J.; Chen, X.; Wu, D.; Bergmeier, W.; et al. RAS P21 Protein Activator 3 (RASA3) Specifically Promotes Pathogenic T Helper 17 Cell Generation by Repressing T-Helper-2-Cell-Biased Programs. Immunity 2018, 49, 886–898.e5. [Google Scholar] [CrossRef]
- Zhang, C.; Guo, C.; Li, Y.; Ouyang, L.; Zhao, Q.; Liu, K. The role of YTH domain containing 2 in epigenetic modification and immune infiltration of pan-cancer. J. Cell. Mol. Med. 2021, 25, 8615–8627. [Google Scholar] [CrossRef]
- Peng, T.; Liu, B.; Lin, S.; Cao, C.; Wu, P.; Zhi, W.; Wei, Y.; Chu, T.; Gui, L.; Ding, W. APOBEC3G expression correlates with unfavorable prognosis and immune infiltration in kidney renal clear cell carcinoma. Heliyon 2022, 8, e12191. [Google Scholar] [CrossRef]
- He, Q.; Chen, N.; Wang, X.; Li, P.; Liu, L.; Rong, Z.; Liu, W.; Jialng, K.; Zhao, J. Prognostic value and immunological roles of GPX3 in gastric cancer. Int. J. Med. Sci. 2023, 20, 1399–1416. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Y.; Li, H.; Xu, J. Galectin-8 alters immune microenvironment and promotes tumor progression. Am. J. Cancer Res. 2023, 13, 2517–2529. [Google Scholar]
- Zheng, Y.; Lang, Y.; Qi, B.; Li, T. TSPAN4 and migrasomes in atherosclerosis regression correlated to myocardial infarction and pan-cancer progression. Cell Adh. Migr. 2023, 17, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Chitadze, G.; Oberg, H.H.; Wesch, D.; Kabelitz, D. The Ambiguous Role of γδ T Lymphocytes in Antitumor Immunity. Trends Immunol. 2017, 38, 668–678. [Google Scholar] [CrossRef]
- Conejo-Garcia, J.R.; Innamarato, P. γδ T cells share the spotlight in cancer. Nat. Cancer 2022, 3, 657–658. [Google Scholar] [CrossRef]
- Mensurado, S.; Blanco-Domínguez, R.; Silva-Santos, B. The emerging roles of γδ T cells in cancer immunotherapy. Nat. Rev. Clin. Oncol. 2023, 20, 178–191. [Google Scholar] [CrossRef]
- Saura-Esteller, J.; de Jong, M.; King, L.A.; Ensing, E.; Winograd, B.; de Gruijl, T.D.; Parren, P.W.H.I.; van der Vliet, H.J. Gamma Delta T-Cell Based Cancer Immunotherapy: Past-Present-Future. Front. Immunol. 2022, 13, 915837. [Google Scholar] [CrossRef] [PubMed]
- Looi, C.K.; Chung, F.F.; Leong, C.O.; Wong, S.F.; Rosli, R.; Mai, C.W. Therapeutic challenges and current immunomodulatory strategies in targeting the immunosuppressive pancreatic tumor microenvironment. J. Exp. Clin. Cancer Res. 2019, 38, 162. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Khorsandi, S.; Wang, Y.; Santelli, J.; Huntoon, K.; Nguyen, N.; Yang, M.; Lee, D.; Lu, Y.; Gao, R.; et al. Cancer immunotherapy based on image-guided STING activation by nucleotide nano-complex-decorated ultrasound microbubbles. Nat. Nanotechnol. 2022, 17, 891–899. [Google Scholar] [CrossRef]
- Mangalhara, K.C.; Varanasi, S.K.; Johnson, M.A.; Burns, M.J.; Rojas, G.R.; Esparza Moltó, P.B.; Sainz, A.G.; Tadepalle, N.; Abbott, K.L.; Mendiratta, G.; et al. Manipulating mitochondrial electron flow enhances tumor immunogenicity. Science 2023, 381, 1316–1323. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.; Chung, H.C.; Sun, T.; Tyagi, S.; Dobrolecki, L.E.; Dominguez-Vidana, R.; Kurley, S.J.; Orellana, M.; Renwick, A.; Henke, D.M.; et al. Combinatorial inhibition of PTPN12-regulated receptors leads to a broadly effective therapeutic strategy in triple-negative breast cancer. Nat. Med. 2018, 24, 505–511. [Google Scholar] [CrossRef]
- Lowenfeld, L.; Mick, R.; Datta, J.; Xu, S.; Fitzpatrick, E.; Fisher, C.S.; Fox, K.R.; DeMichele, A.; Zhang, P.J.; Weinstein, S.P.; et al. Dendritic Cell Vaccination Enhances Immune Responses and Induces Regression of HER2(pos) DCIS Independent of Route: Results of Randomized Selection Design Trial. Clin. Cancer Res. 2017, 23, 2961–2971. [Google Scholar] [CrossRef]
- Lian, Y.F.; Yuan, J.; Cui, Q.; Fetng, Q.-S.; Xu, M.; Bei, J.-X.; Zeng, Y.-X.; Feng, L.; Huen, M.S.-Y. Upregulation of KLHDC4 Predicts a Poor Prognosis in Human Nasopharyngeal Carcinoma. PLoS ONE 2016, 11, e0152820. [Google Scholar] [CrossRef]
- Zhu, Z.; Shen, H.; Xu, J.; Fang, Z.; Wo, G.; Ma, Y.; Yang, K.; Wang, Y.; Yu, Q.; Tang, J.-H. GATA3 mediates doxorubicin resistance by inhibiting CYB5R2-catalyzed iron reduction in breast cancer cells. Drug Resist. Updat. 2023, 69, 100974. [Google Scholar] [CrossRef]
- Wei, Y.; An, Z.; Zou, Z.; Sumpter, R.; Su, M.; Zang, X.; Sinha, S.; Gaestel, M.; Levine, B. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. eLife 2015, 4, e05289. [Google Scholar] [CrossRef]
- Moradi-Marjaneh, R.; Hassanian, S.M.; Fiuji, H.; Soletimanpour, S.; Ferns, G.A.; Alvan, A.; Khazaei, M. Toll like receptor signaling pathway as a potential therapeutic target in colorectal cancer. J. Cell Physiol. 2018, 233, 5613–5622. [Google Scholar] [CrossRef]
- Niu, X.; Yin, L.; Yang, X.; Yang, Y.; Gu, Y.; Sun, Y.; Yalng, M.; Wang, Y.; Zhang, Q.; Ji, H. Serum amyloid A 1 induces suppressive neutrophils through the Toll-like receptor 2-mediated signaling pathway to promote progression of breast cancer. Cancer Sci. 2022, 113, 1140–1153. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, M.; Xu, F.; Jiang, S. Wnt signaling in breast cancer: Biological mechanisms, challenges and opportunities. Mol. Cancer 2020, 19, 165. [Google Scholar] [CrossRef] [PubMed]
- Lou, W.; Ding, B.; Wang, S.; Fu, P. Overexpression of GPX3, a potential biomarker for diagnosis and prognosis of breast cancer, inhibits progression of breast cancer cells in vitro. Cancer Cell Int. 2020, 20, 378. [Google Scholar] [CrossRef]
- Nirmal, A.J.; Maliga, Z.; Vallius, T.; Quattrochi, B.; Chen, A.A.; Jacobson, C.A.; Pelletier, R.J.; Yapp, C.; Arias-Camison, R.; Chen, Y.-A.; et al. The Spatial Landscape of Progression and Immunoediting in Primary Melanoma at Single-Cell Resolution. Cancer Discov. 2022, 12, 1518–1541. [Google Scholar] [CrossRef]
- Sun, Y.; Pan, Z.; Wang, Z.; Wang, H.; Weti, L.; Cui, F.; Zou, Q.; Zhalng, Z. Single-cell transcriptome analysis reveals immune microenvironment changes and insights into the transition from DCIS to IDC with associated prognostic genes. J. Transl. Med. 2024, 22, 894. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Liu, X.; Li, Z.; Zhang, T.; Yalng, B.; Su, J.; Song, Q. SpaRx: Elucidate single-cell spatial heterogeneity of drug responses for personalized treatment. Brief. Bioinform. 2023, 24, bbad338. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yi, J.; Li, T.; Wetn, J.; Huang, K.; Liu, J.; Wang, G.; Kim, P.; Song, Q.; Zhou, X. DRMref: Comprehensive reference map of drug resistance mechanisms in human cancer. Nucleic Acids Res. 2024, 52, D1253–D1264. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mo, C.-Q.; Xie, R.-W.; Li, W.-W.; Zhong, M.-J.; Li, Y.-Y.; Lin, J.-Y.; Zhang, J.-S.; Zheng, S.-K.; Lin, W.; Kong, L.-J.; et al. Integration of eQTL and GEO Datasets to Identify Genes Associated with Breast Ductal Carcinoma In Situ. Curr. Issues Mol. Biol. 2025, 47, 747. https://doi.org/10.3390/cimb47090747
Mo C-Q, Xie R-W, Li W-W, Zhong M-J, Li Y-Y, Lin J-Y, Zhang J-S, Zheng S-K, Lin W, Kong L-J, et al. Integration of eQTL and GEO Datasets to Identify Genes Associated with Breast Ductal Carcinoma In Situ. Current Issues in Molecular Biology. 2025; 47(9):747. https://doi.org/10.3390/cimb47090747
Chicago/Turabian StyleMo, Cai-Qin, Rui-Wang Xie, Wei-Wei Li, Min-Jie Zhong, Yu-Yang Li, Jun-Yu Lin, Juan-Si Zhang, Sheng-Kai Zheng, Wei Lin, Ling-Jun Kong, and et al. 2025. "Integration of eQTL and GEO Datasets to Identify Genes Associated with Breast Ductal Carcinoma In Situ" Current Issues in Molecular Biology 47, no. 9: 747. https://doi.org/10.3390/cimb47090747
APA StyleMo, C.-Q., Xie, R.-W., Li, W.-W., Zhong, M.-J., Li, Y.-Y., Lin, J.-Y., Zhang, J.-S., Zheng, S.-K., Lin, W., Kong, L.-J., Xu, S.-W., & Chen, X.-J. (2025). Integration of eQTL and GEO Datasets to Identify Genes Associated with Breast Ductal Carcinoma In Situ. Current Issues in Molecular Biology, 47(9), 747. https://doi.org/10.3390/cimb47090747




