The Complete Chloroplast Genomes of Three Manglietia Species and Phylogenetic Insight into the Genus Manglietia Blume
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, DNA Extraction, and Sequencing
2.2. Chloroplast Genome Assembly and Annotation
2.3. Repeat Sequence Analysis
2.4. Analysis of Codon Usageis
2.5. Genomic Comparative Analysis
2.6. Ka/Ks Ratio Analysis
2.7. Nucleotide Diversity Analysis
2.8. Phylogenetic Trees Construction and Analysis
3. Results
3.1. Genome Size and Basic Structural Features
3.2. Analysis of Repeats and SSRs
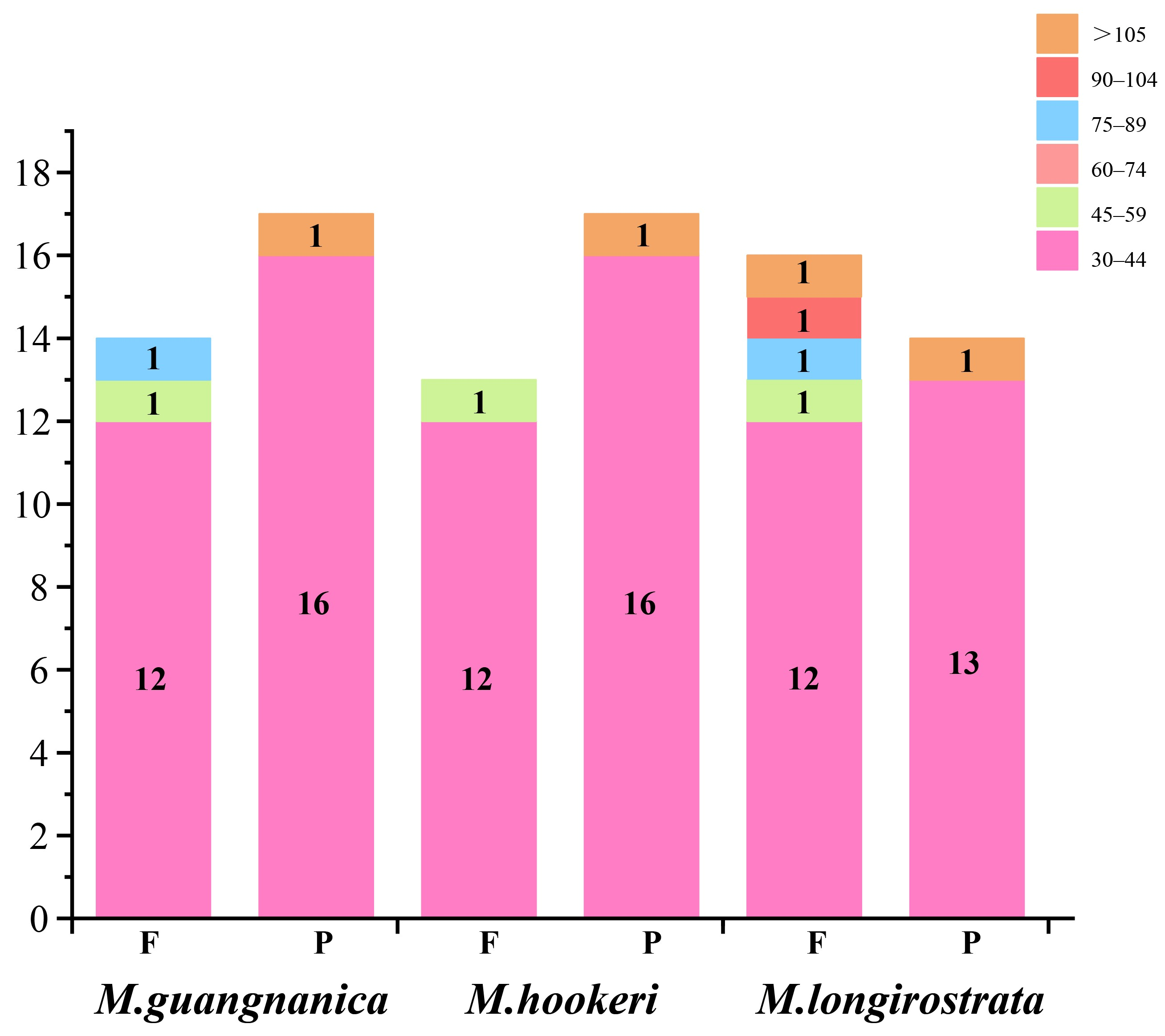
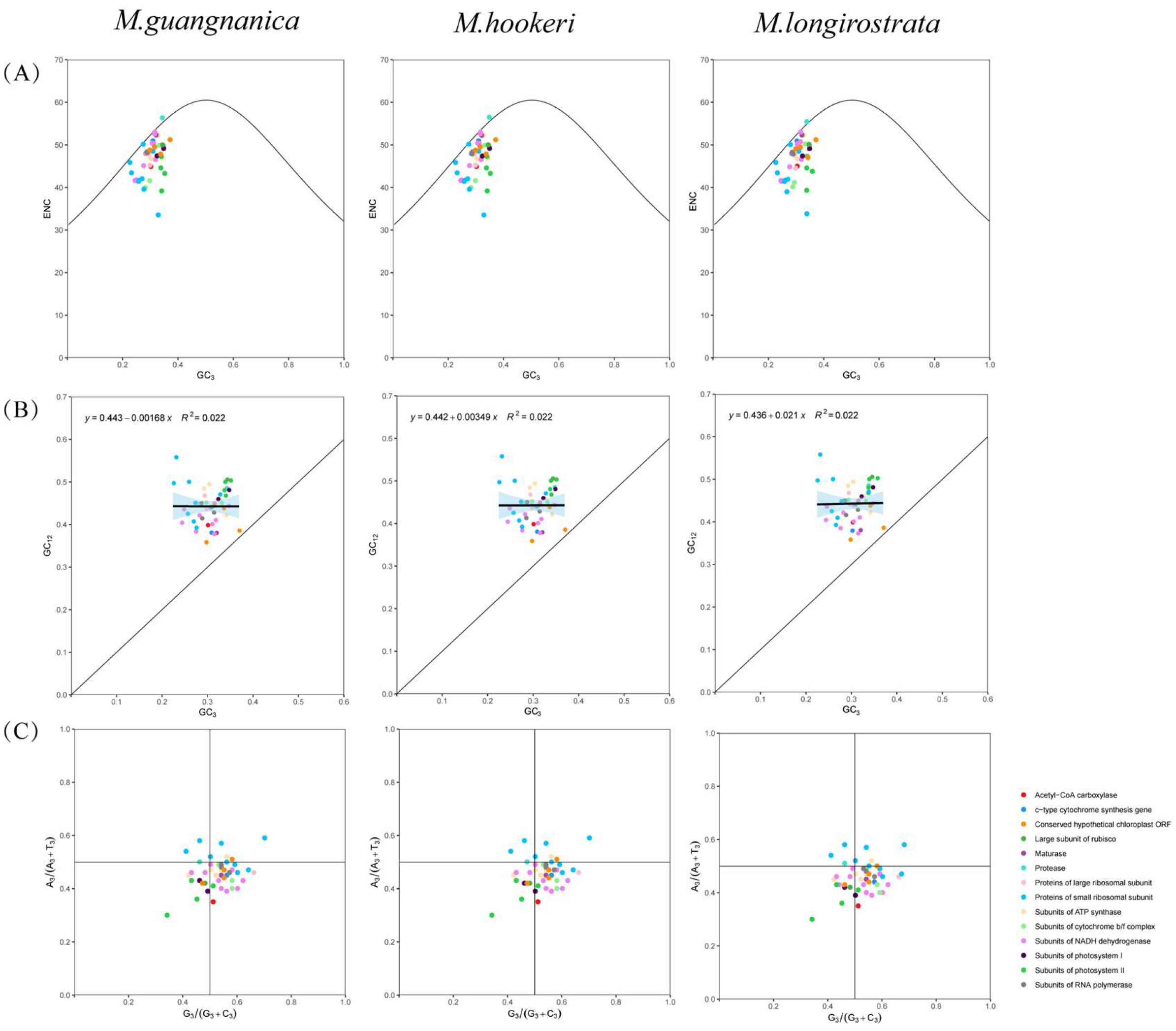
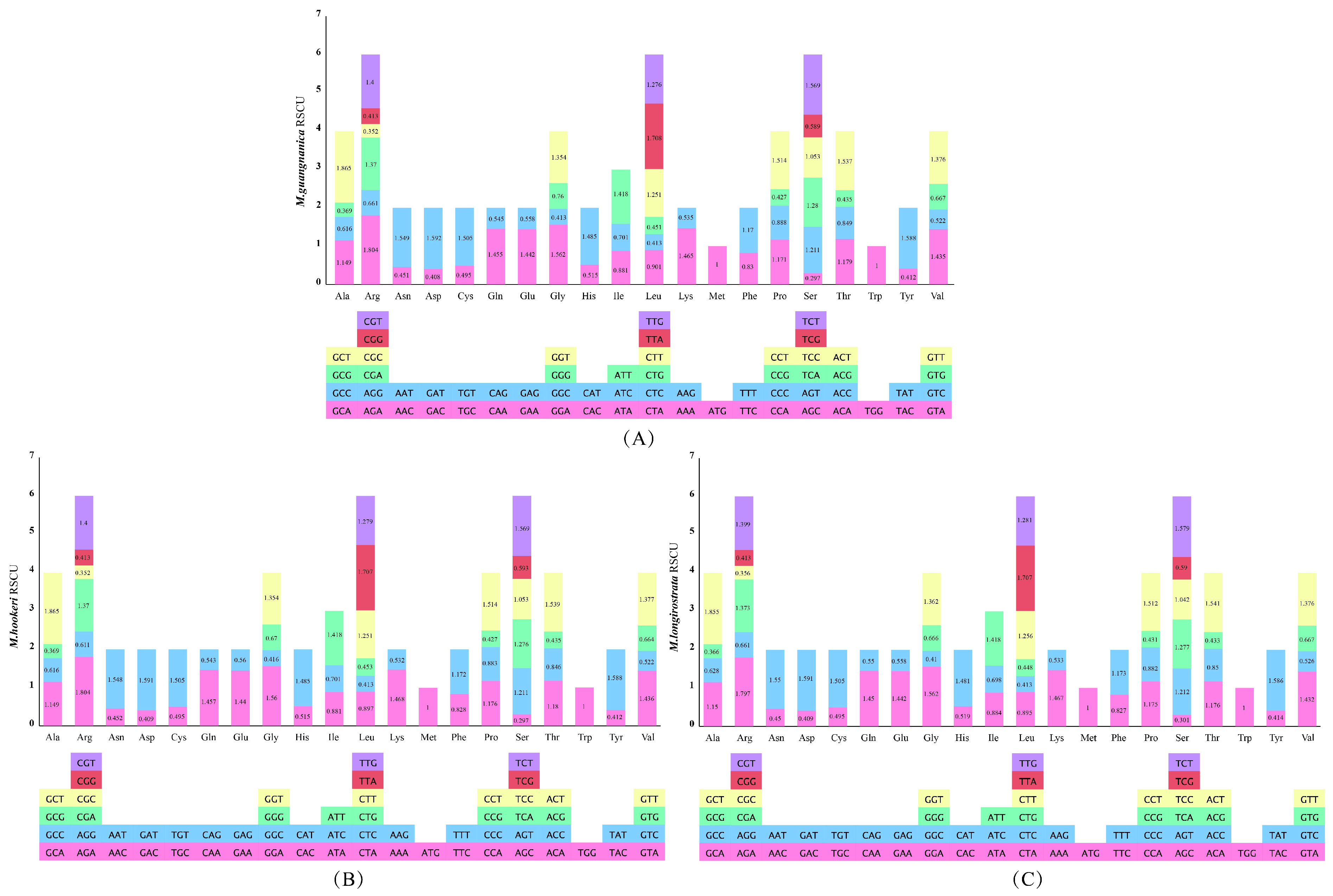
3.3. Codon Usage Bias Analysis
3.4. Comparative Genomic Analysis
3.5. Selection Pressure Analysis
3.6. Nucleotide Diversity and Highly Variable Regions
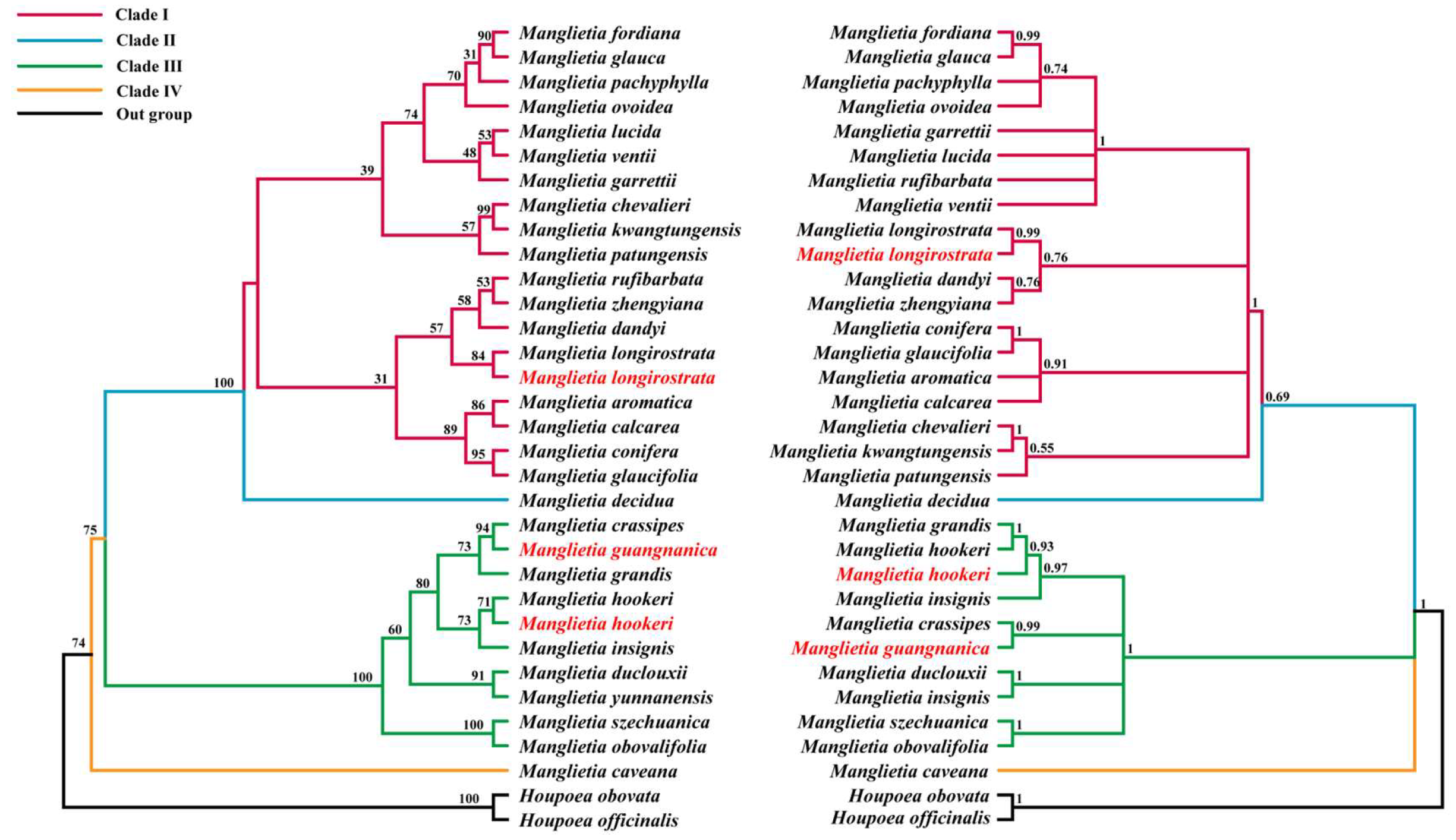
3.7. Phylogenetic Analysis
4. Discussion
4.1. Chloroplast Genome Characteristics of Three Manglietia Species
4.2. Interspecific Divergence of Chloroplast Genomes
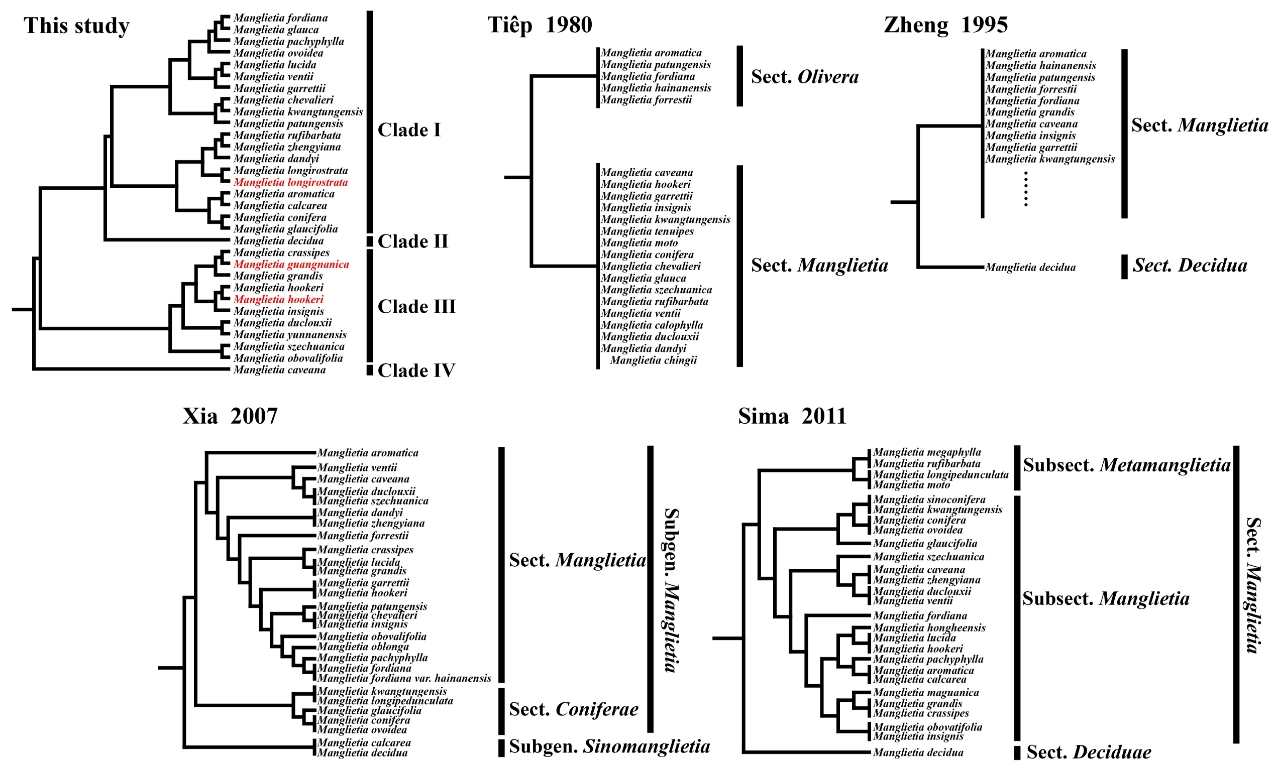
4.3. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hu, Y.; Zhang, Q.; Rao, G. Sodmergen Occurrence of plastids in the sperm cells of Caprifoliaceae: Biparental plastid inheritance in angiosperms is unilaterally derived from maternal inheritance. Plant Cell Physiol. 2008, 49, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Li, Z.; Wu, N.; Yang, J.; Yuan, L.; Zhao, T.; Sima, Y.; Xu, T. Comparitive Analysis of the Chloroplast Genomes of Three Houpoea Plants. Genes 2023, 14, 51–61. [Google Scholar] [CrossRef]
- Kim, S.; Park, C.; Kim, Y.; Suh, Y. Phylogenetic relationships in family Magnoliaceae inferred from ndhF sequences. Am. J. Bot. 2001, 88, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kan, S.; Liao, X.; Zhou, J.; Tembrock, L.R.; Daniell, H.; Jin, S.; Wu, Z.Q. Plant organellar genomes: Much done, much more to do. Trends Plant Sci. 2024, 29, 754–769. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, B.; Nie, Z.; Chen, H.; Chen, F.; Figlar, R.B.; Wen, J. Major clades and a revised classification of Magnolia and Magnoliaceae based on whole plastid genome sequences via genome skimming. J. Syst. Evol. 2020, 58, 673–695. [Google Scholar] [CrossRef]
- Wu, T.; Sima, Y.; Chen, S.; Fu, Y.; Ma, H.; Hao, J.; Zhu, Y. Comparative Analysis of the Chloroplast Genomes of Eight Species of the Genus Lirianthe Spach with Its Generic Delimitation Implications. Int. J. Mol. Sci. 2024, 25, 3506. [Google Scholar] [CrossRef]
- Blume, C.L. Beschrijving van Eenige Gewassen, Waargenomen op Eenen Togtnaar den Salak in den Jaare 1822; Het Bataviaasch Genoot. V. Kunsten En Wet: Batavia, Indonesia, 1823; pp. 149–150. [Google Scholar]
- Xia, N.H.; Liu, Y.H.; Nooteboom, H.P. Wu, Z.Y., Raven, P.H., Eds.; Magnoliaceae. In Flora of China; Science Press: Beijing, China, 2008; Volume 7, pp. 48–91. [Google Scholar]
- Sima, Y. A Taxonomic Revision of the Magnoliaceae from China. Ph.D. Thesis, Yunnan University, Kunming, China, 2011. [Google Scholar]
- Zhang, D. Exploring the versatile world of magnolias: Urban landscape enhancement and metabolite applications in drug discovery and cosmetics. Discov. Plants 2025, 2, 139. [Google Scholar] [CrossRef]
- Tiêṕ, N.V. Beiträge zur Sippenstruktur der Gattung Manglietia BL. (Magnoliaceae). Feddes Repert. 1980, 91, 497–576. [Google Scholar] [CrossRef]
- Zheng, Q. A revised name of Huamulian. J. Nanjing For. Univ. 1995, 19, 46. [Google Scholar]
- Xia, N. Taxonomic revision on the Famliy Magnoliaceae from China. Ph.D. Thesis, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China, 2007. [Google Scholar]
- Li, T.; Zhang, S.; Deng, Y.; Li, Y. Comparative Analysis of Chloroplast Genomes for the Genus Manglietia Blume (Magnoliaceae): Molecular Structure and Phylogenetic Evolution. Genes 2024, 15, 406. [Google Scholar] [CrossRef]
- Hu, X.; Zeng, Q.; Fu, L. Magnolia hookeri var. Longirostrata (Magnoliaceae), a New Taxon from Yunnan, China. Ann. Bot. Fenn. 2012, 49, 417–421. [Google Scholar] [CrossRef]
- Luo, W.; Li, Z.; Sima, Y.; Xu, T. The complete chloroplast genome sequence of the Manglietia longirostrata Sima, a rare and endemic species to china. Mitochondrial DNA B Resour. 2020, 5, 2872–2873. [Google Scholar] [CrossRef]
- Li, M.; Shen, J.; Zhang, Y.; Yang, C. Study on structure and plant diversity of the Manglietia guangnanica community in Guizhou Province. J. Cent. South Univ. For. Technol. 2022, 42, 147–158. [Google Scholar] [CrossRef]
- Hu, X.; Zeng, Q.; Fu, L. Manglietia guangnanica (Magnoliaceae), a New Species from Yunnan, China. Novon 2014, 23, 171–175. [Google Scholar] [CrossRef]
- Govaerts, R.; Nic Lughadha, E.; Black, N.; Turner, R.; Paton, A. The World Checklist of Vascular Plants, a continuously updated resource for exploring global plant diversity. Sci. Data 2021, 8, 215. [Google Scholar] [CrossRef]
- Jin, J.; Yu, W.; Yang, J.; Song, Y.; de Pamphilis, C.; Yi, T.; Li, D. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Qu, X.; Moore, M.; Li, D.; Yi, T. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 2019, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.; Wang, G. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Li, H.; Guo, Q.; Xu, L.; Gao, H.; Liu, L.; Zhou, X. CPJSdraw: Analysis and visualization of junction sites of chloroplast genomes. PeerJ 2023, 11, e15326. [Google Scholar] [CrossRef]
- Darling, A.; Mau, B.; Blattner, F.; Perna, N. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Frazer, K.; Pachter, L.; Poliakov, A.; Rubin, E.; Dubchak, I. mVISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhao, X.; Wang, J.; Wong, G.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinf. 2006, 4, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.; Wong, T.; von Haeseler, A.; Jermiin, L. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.; Huelsenbeck, J. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, B.; Li, B.; Zhou, Q.; Wang, G.; Jiang, X.; Wang, C.; Xu, Z. Comparative analysis of codon usage patterns in chloroplast genomes of six Euphorbiaceae species. PeerJ 2020, 8, e8251. [Google Scholar] [CrossRef]
- Wu, D.; Fu, W.; Fan, G.; Huang, D.; Wu, K.; Zhan, Y.; Tu, X.; He, J. Characteristics and Comparative Analysis of the Special-Structure (Non-Single-Circle) Mitochondrial Genome of Capsicum pubescens Ruiz & Pav. Genes 2024, 15, 152. [Google Scholar] [CrossRef]
- Zhu, W.; Ma, J.; Tan, Y.; Song, Y.; Xin, P. Phylogenetic analysis of Asiatic species in the tropical genus Beilschmiedia (Lauraceae). BMC Genom. 2025, 26, 226. [Google Scholar] [CrossRef]
- Shi, C.; Hu, N.; Huang, H.; Gao, J.; Zhao, Y.; Gao, L. An Improved Chloroplast DNA Extraction Procedure for Whole Plastid Genome Sequencing. PLoS ONE 2012, 7, e31468. [Google Scholar] [CrossRef] [PubMed]
- Leng, Z.; Pang, Z.; He, Z.; Gao, Q. Comparative Analysis of Chloroplast Genomes of 19 Saxifraga Species, Mostly from the European Alps. Int. J. Mol. Sci. 2025, 26, 6015. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Liu, Z.; Lan, W.; Yang, M.; Mo, Q.; Huang, X.; Wu, P.; Huang, H.; Huang, M. Complete Chloroplast Genomes of 9 Impatiens Species: Genome Structure, Comparative Analysis, and Phylogenetic Relationships. Int. J. Mol. Sci. 2025, 26, 536. [Google Scholar] [CrossRef]
- Rudenko, V.; Korotkov, E. Study of Dispersed Repeats in the Cyanidioschyzon merolae Genome. Int. J. Mol. Sci. 2024, 25, 4441. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, L.; Yang, T.; Tang, J.; Miao, Y.; Ren, T. Genome-wide simple sequence repeat analysis and specific molecular marker development of rye. BMC Genom. 2024, 25, 780. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, J.; Lv, Y.; Zhang, X.; Xia, C.; Zhao, H.; Wen, C. Genetic diversity analysis and variety identification using SSR and SNP markers in melon. BMC Plant Biol. 2023, 23, 39. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Zhou, L.; Wei, X.; Shang, C.; Zhang, Z. Comparative Chloroplast Genomics Reveals Intrageneric Divergence in Salix. Int. J. Mol. Sci. 2025, 26, 2248. [Google Scholar] [CrossRef]
- Lin, Y.; Chen, Y.; Zhao, Y.; Wu, W.; Yang, C.; Zheng, Y.; Huang, M. Comparative Analysis of Complete Chloroplast Genomes and Phylogenetic Relationships in Medicinally Important Pantropical Genus Bauhinia s.s. (Leguminosae) from Southern Africa and Eastern Asia. Int. J. Mol. Sci. 2025, 26, 397. [Google Scholar] [CrossRef]
- Yang, M.; Liu, J.; Yang, W.; Li, Z.; Hai, Y.; Duan, B.; Zhang, H.; Yang, X.; Xia, C. Analysis of codon usage patterns in 48 Aconitum species. BMC Genom. 2023, 24, 703. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Hu, Y.; Hu, Z. Comparative analysis of codon usage patterns in 16 chloroplast genomes of suborder Halimedineae. BMC Genom. 2024, 25, 945. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Li, T.; Ma, Z.; Wang, X.; Xu, L.; Zhang, Y.; Cai, Y.; Tang, Z. Codon usage pattern of the ancestor of green plants revealed through Rhodophyta. BMC Genom. 2023, 24, 538. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L. A DNA barcode for land plants. Proc. Natl. Acad. Sci. 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.; Schilling, E.; Small, R. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef]
- Yu, W.; Li, X.; Lv, Z.; Yang, L.; Peng, D. The complete chloroplast genome sequences of monotypic genus Pseudogalium, and comparative analyses with its relative genera. BMC Genom. 2025, 26, 93. [Google Scholar] [CrossRef]
- Raman, G.; Nam, G.H.; Park, S. Extensive reorganization of the chloroplast genome of Corydalis platycarpa: A comparative analysis of their organization and evolution with other Corydalis plastomes. Front. Plant Sci. 2022, 13, 1043740. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Tian, J.; Yang, J.; Dong, X.; Zhong, Z.; Mwachala, G.; Zhang, C.; Hu, G.; Wang, Q. Comparative and phylogenetic analyses of six Kenya Polystachya (Orchidaceae) species based on the complete chloroplast genome sequences. BMC Plant Biol. 2022, 22, 177. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bai, S.; Zhang, Z.; Zheng, F.; Song, L.; Wen, L.; Guo, M.; Cheng, G.; Yao, W.; Gao, Y.; et al. Comparative analysis of chloroplast genomes of 29 tomato germplasms: Genome structures, phylogenetic relationships, and adaptive evolution. Front. Plant Sci. 2023, 14, 1179009. [Google Scholar] [CrossRef]
- Schöttler, M.; Albus, C.; Bock, R. Photosystem I: Its biogenesis and function in higher plants. J. Plant Physiol. 2011, 168, 1452–1461. [Google Scholar] [CrossRef] [PubMed]
- Amar, M. ycf1-ndhF genes, the most promising plastid genomic barcode, sheds light on phylogeny at low taxonomic levels in Prunus persica. J. Genet. Eng. Biotechnol. 2020, 18, 42. [Google Scholar] [CrossRef]
- Börner, T.; Aleynikova, A.; Zubo, Y.; Kusnetsov, V. Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim. Biophys. Acta 2015, 1847, 761–769. [Google Scholar] [CrossRef]
- Balaji, R.; Easwaran, S.; Devanathan, K.; Sharma, S.; Alqahtani, T.; Uti, D.; Malik, T. Unfolding the Complete Chloroplast Genome of Myrica esculenta Buch.-Ham. ex D.Don (1825): Advancing Phylogenetic Insights Within Fagales and Pioneering DNA Barcodes for Precise Species Identification. Ecol. Evol. 2025, 15, e71566. [Google Scholar] [CrossRef]
- Li, H.; Xiao, W.; Tong, T.; Li, Y.; Zhang, M.; Lin, X.; Zou, X.; Wu, Q.; Guo, X. The specific DNA barcodes based on chloroplast genes for species identification of Orchidaceae plants. Sci. Rep. 2021, 11, 1424. [Google Scholar] [CrossRef]
- Song, Z.-K.; Zhu, A.-H.; Liu, Z.-D.; Qu, Z.; Li, Y.; Ma, H.-X. Three New Species of Hypoxylon (Xylariales, Ascomycota) on a Multigene Phylogeny from Medog in Southwest China. Journal of Fungi. 2022, 8, 500. [Google Scholar] [CrossRef]
- Wang, G.; Hou, N.; Zhang, S.; Luo, Y. Characterization of the complete chloroplast genomes of seven Manglietia and one Michelia species (Magnoliales: Magnoliaceae). Conserv. Genet. Resour. 2018, 10, 705–708. [Google Scholar] [CrossRef]
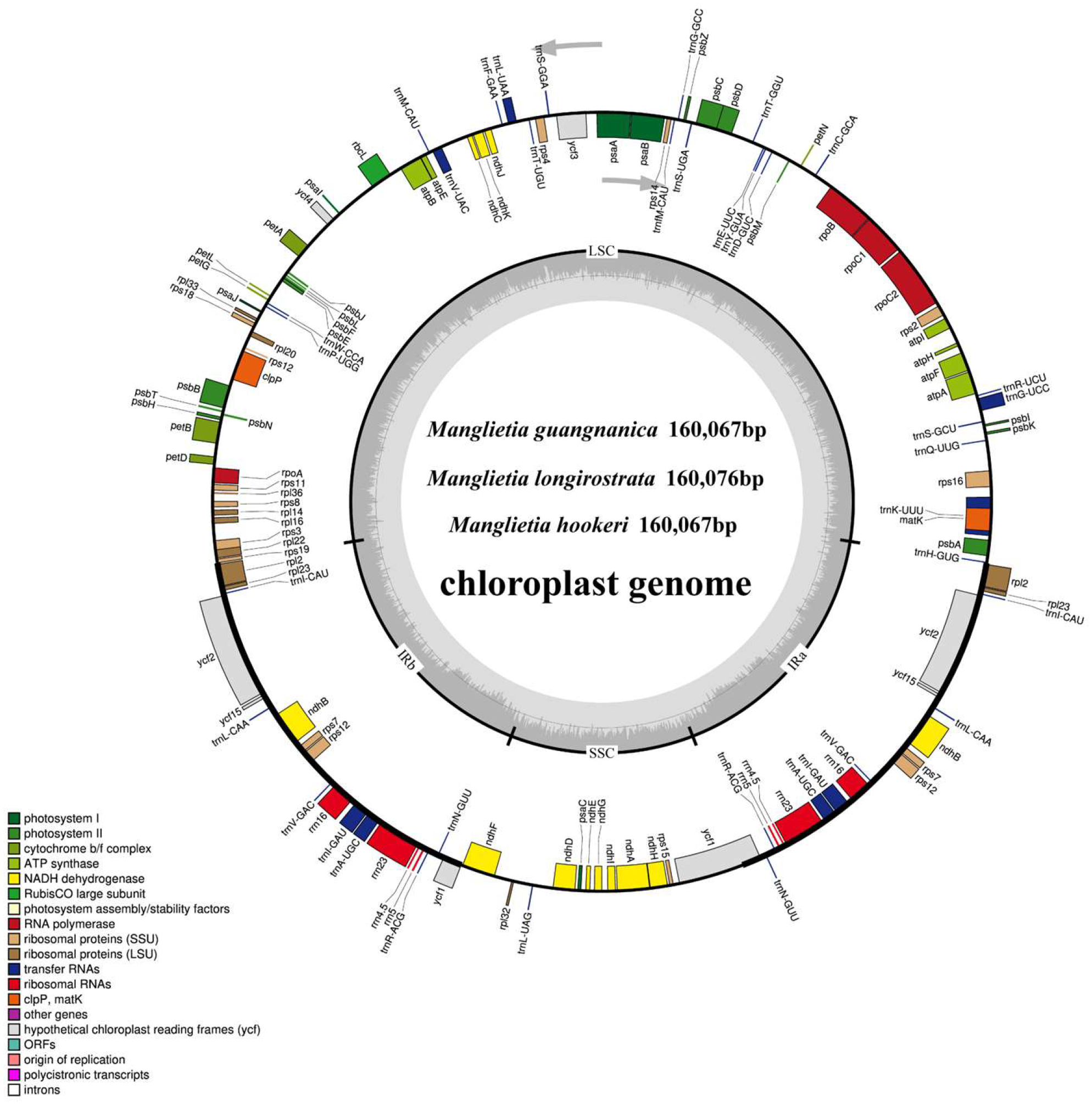

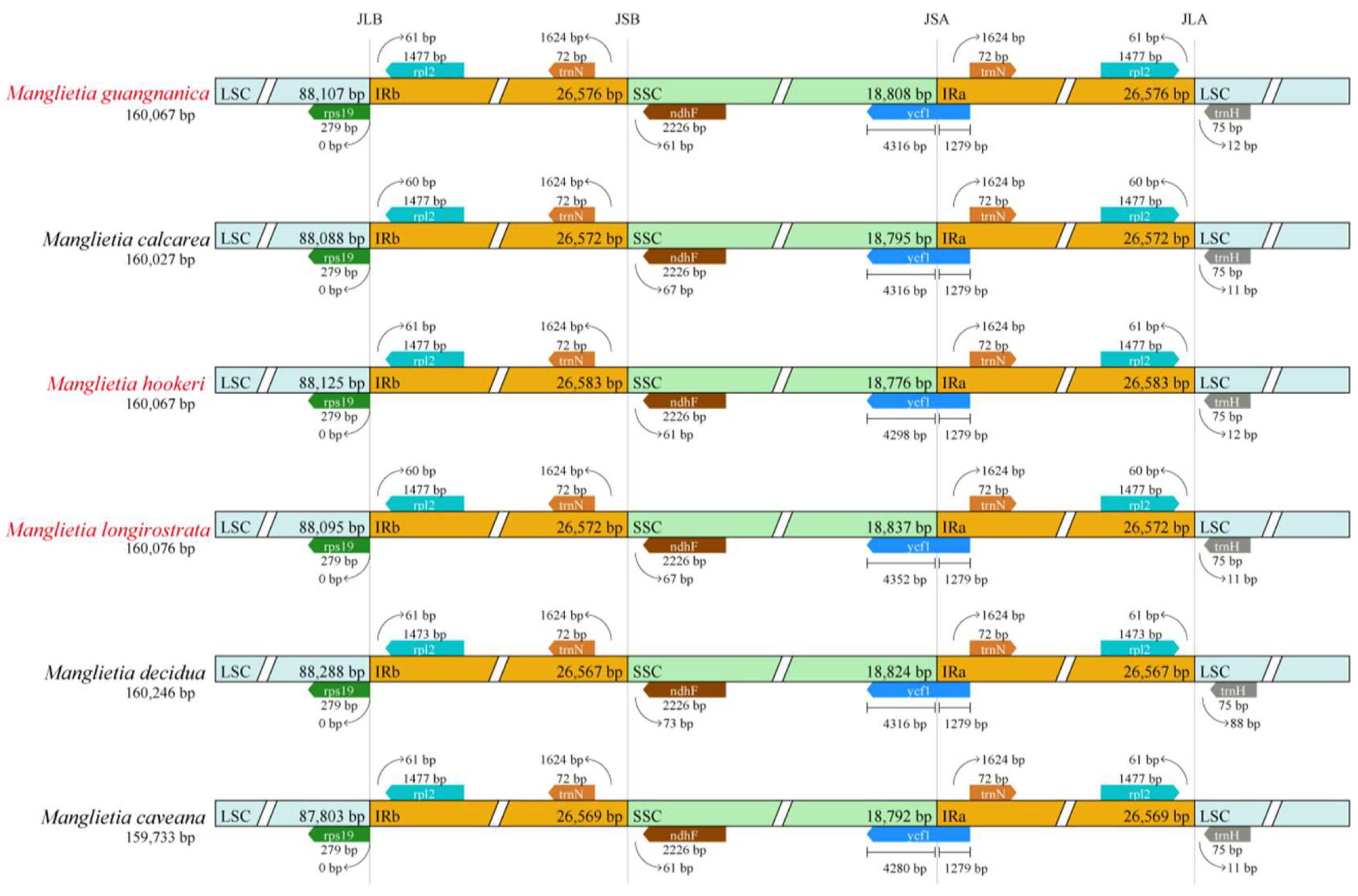
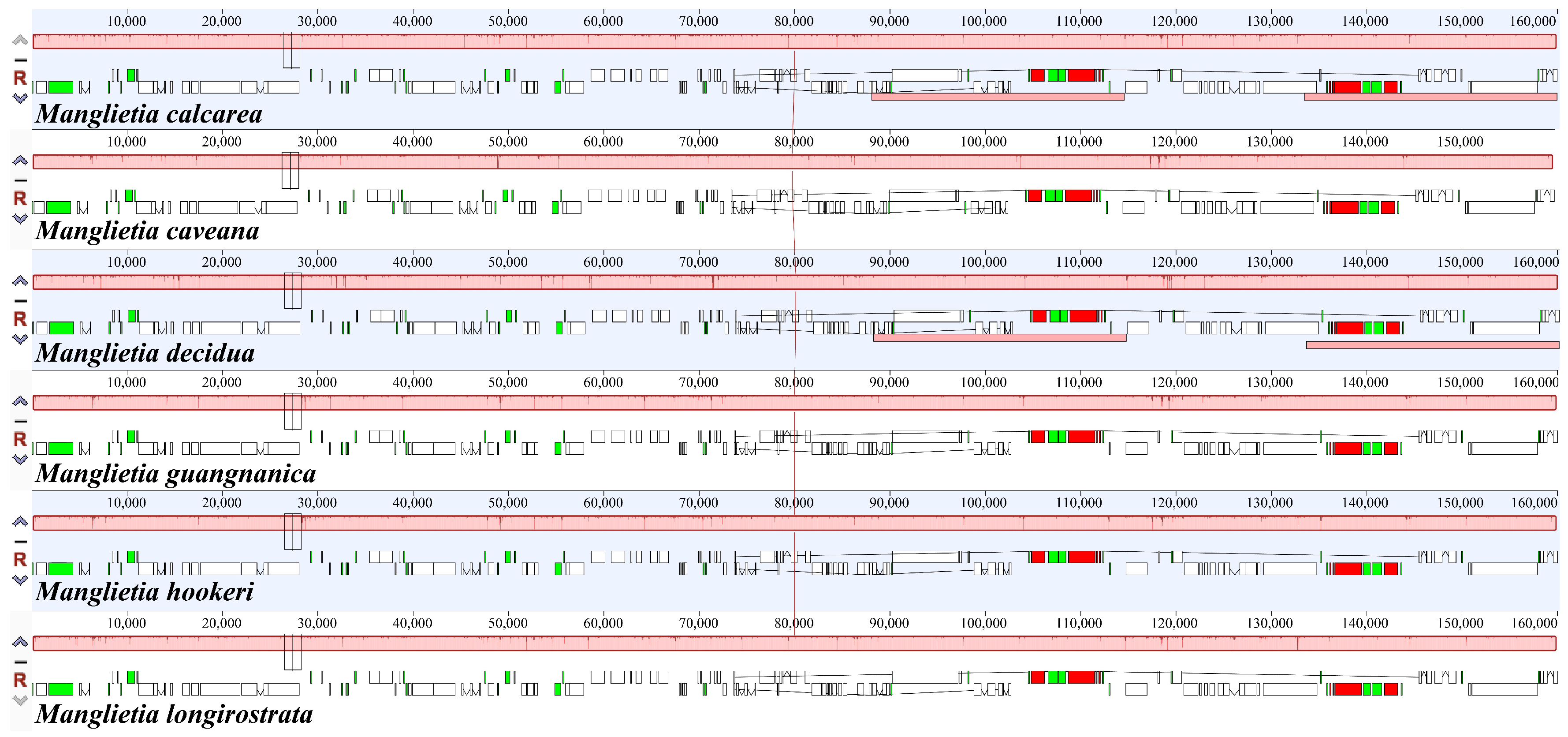

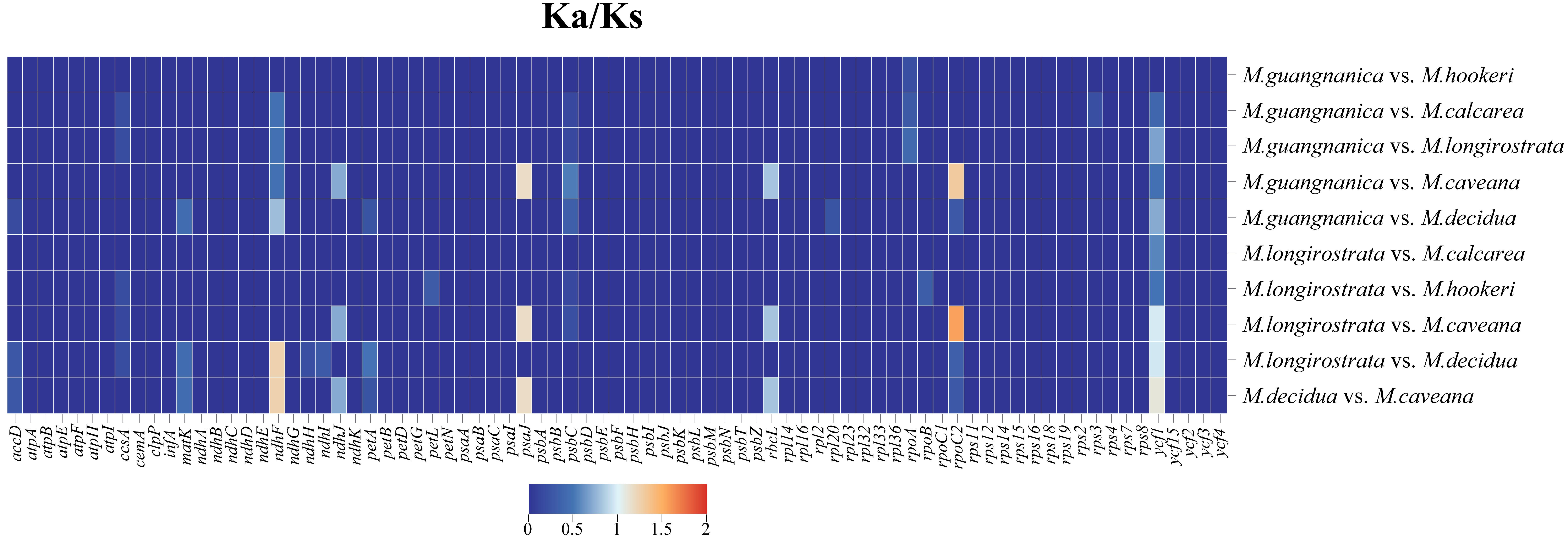
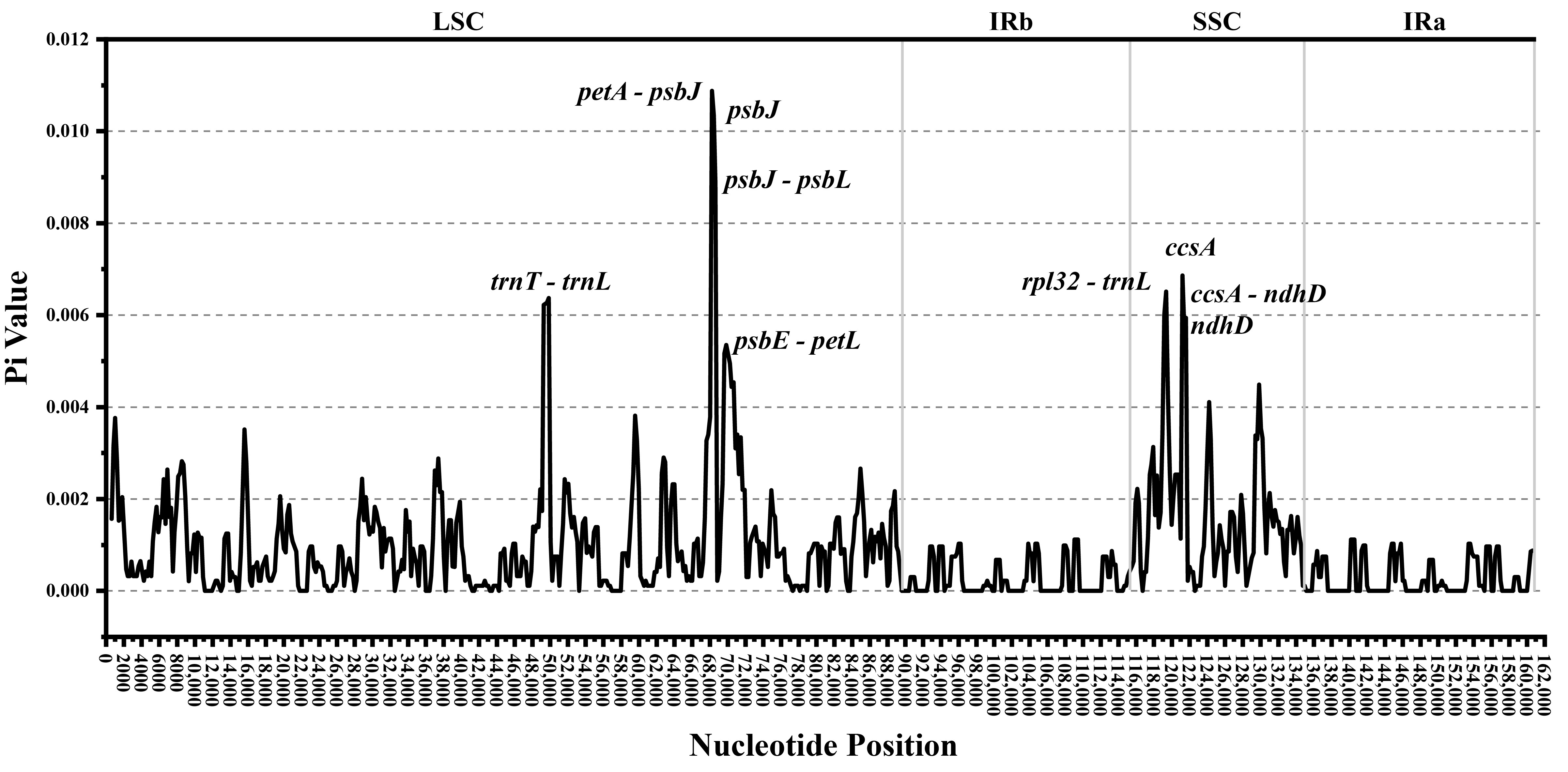
| Species | Total Length (bp) | Total GC (%) | LSC Length (bp) | LCS GC (%) | SSC Length (bp) | SSC GC (%) | IR Length (bp) | IR GC (%) | Genes | CDS | tRNAs | rRNAs | Number of Accession |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M.guangnanica | 160,067 | 39.3 | 88,107 | 38 | 18,808 | 34.2 | 26,576 | 43.2 | 133 | 88 | 37 | 8 | PQ246290 |
| M.longirostrata | 160,076 | 39.3 | 88,095 | 38 | 18,837 | 34.2 | 26,572 | 43.2 | 133 | 88 | 37 | 8 | PV239601 |
| M.hookeri | 160,067 | 39.3 | 88,125 | 38 | 18,776 | 34.2 | 26,583 | 43.2 | 133 | 88 | 37 | 8 | PV239602 |
| Category | Gene |
|---|---|
| Large ribosomal subunits | rpl2(2) *, rpl14, rpl16, rpl20, rpl22, rpl23(2), rpl32, rpl33, rpl36 |
| Small ribosomal subunits | rps2, rps3, rps4, rps7(2), rps8, rps11, rps12(2) **, rps14, rps15, rps16 *, rps18, rps19 |
| DNA-dependent RNA Polymerase protease | rpoA, rpoB, rpoC1 *, rpoC2 |
| Translation initiation factor | infA |
| Ribosomal RNA genes | rrn4.5(2), rrn5(2), rrn16(2), rrn23(2) |
| Transfer RNA genes | trnA-UGC(2) *, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC *, trnH-GUG, trnI-CAU(2), trnI-GAU(2) *, trnK-UUU *, trnL-CAA(2), trnL-UAA *, trnL-UAG, trnM-CAU, trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC *, trnW-CCA, trnY-GUA |
| Photosystem I Photosystem II Rubisco large subunit Cytochrome b6/f complex F-type ATP synthase DNA-dependent RNA Polymerase protease NAD(P)H dehydrogenase complex Maturase Inner membrane protein Subunit of acetyl-CoA-carboxylase Cytochrome C biogenesis protein Function uncertain | psaA, psaB, psaC, psaI, psaJ psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ rbcL petA, petB *, petD, petG, petL, petN atpA, atpB, atpE, atpF *, atpH, atpI clpP ** ndhA *, ndhB(2) *, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK matK cemA accD ccsA ycf1(2), ycf2(2), ycf3 **, ycf4, ycf15(2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Y.; Luo, W.; Zhao, T.; Yang, J.; Yuan, L.; Zhang, P.; Gong, Z.; Li, H.; Sima, Y.; Xu, T. The Complete Chloroplast Genomes of Three Manglietia Species and Phylogenetic Insight into the Genus Manglietia Blume. Curr. Issues Mol. Biol. 2025, 47, 737. https://doi.org/10.3390/cimb47090737
Luo Y, Luo W, Zhao T, Yang J, Yuan L, Zhang P, Gong Z, Li H, Sima Y, Xu T. The Complete Chloroplast Genomes of Three Manglietia Species and Phylogenetic Insight into the Genus Manglietia Blume. Current Issues in Molecular Biology. 2025; 47(9):737. https://doi.org/10.3390/cimb47090737
Chicago/Turabian StyleLuo, Yuan, Wei Luo, Tongxing Zhao, Jing Yang, Lang Yuan, Pinzheng Zhang, Zixin Gong, Haizhu Li, Yongkang Sima, and Tao Xu. 2025. "The Complete Chloroplast Genomes of Three Manglietia Species and Phylogenetic Insight into the Genus Manglietia Blume" Current Issues in Molecular Biology 47, no. 9: 737. https://doi.org/10.3390/cimb47090737
APA StyleLuo, Y., Luo, W., Zhao, T., Yang, J., Yuan, L., Zhang, P., Gong, Z., Li, H., Sima, Y., & Xu, T. (2025). The Complete Chloroplast Genomes of Three Manglietia Species and Phylogenetic Insight into the Genus Manglietia Blume. Current Issues in Molecular Biology, 47(9), 737. https://doi.org/10.3390/cimb47090737






