The SMIM25-COX-2 Axis Modulates the Immunosuppressive Tumor Microenvironment and Predicts Immunotherapy Response in Hepatocellular Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. RNA Extraction and Reverse Transcription–Polymerase Chain Reaction (RT-PCR)
2.3. RNA In Situ Hybridization (ISH) Assay
2.4. Immunohistochemistry
2.5. Cell Culture
2.6. Cell Transfection and Drug Treatment
2.7. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Assay
2.8. Scratch Migration Assay
2.9. Animal Experiments
2.10. Whole—Transcriptome Sequencing
2.11. Differentially Expressed Genes and Enrichment Analysis
2.12. Single-Sample GSEA (ssGSEA)
2.13. Cytometry Based on Time of Flight (CyTOF)
2.13.1. Preparation of Single-Cell Suspensions
2.13.2. Antibody Labeling and Cell Staining
2.13.3. CyTOF Data Acquisition and Analysis
2.13.4. Imaging Mass Cytometry(IMC)
2.14. Quantitative Analysis Using ImageJ
2.15. CMap Connectivity Analysis for Drug Prediction
2.16. Statistical Analysisy
3. Results
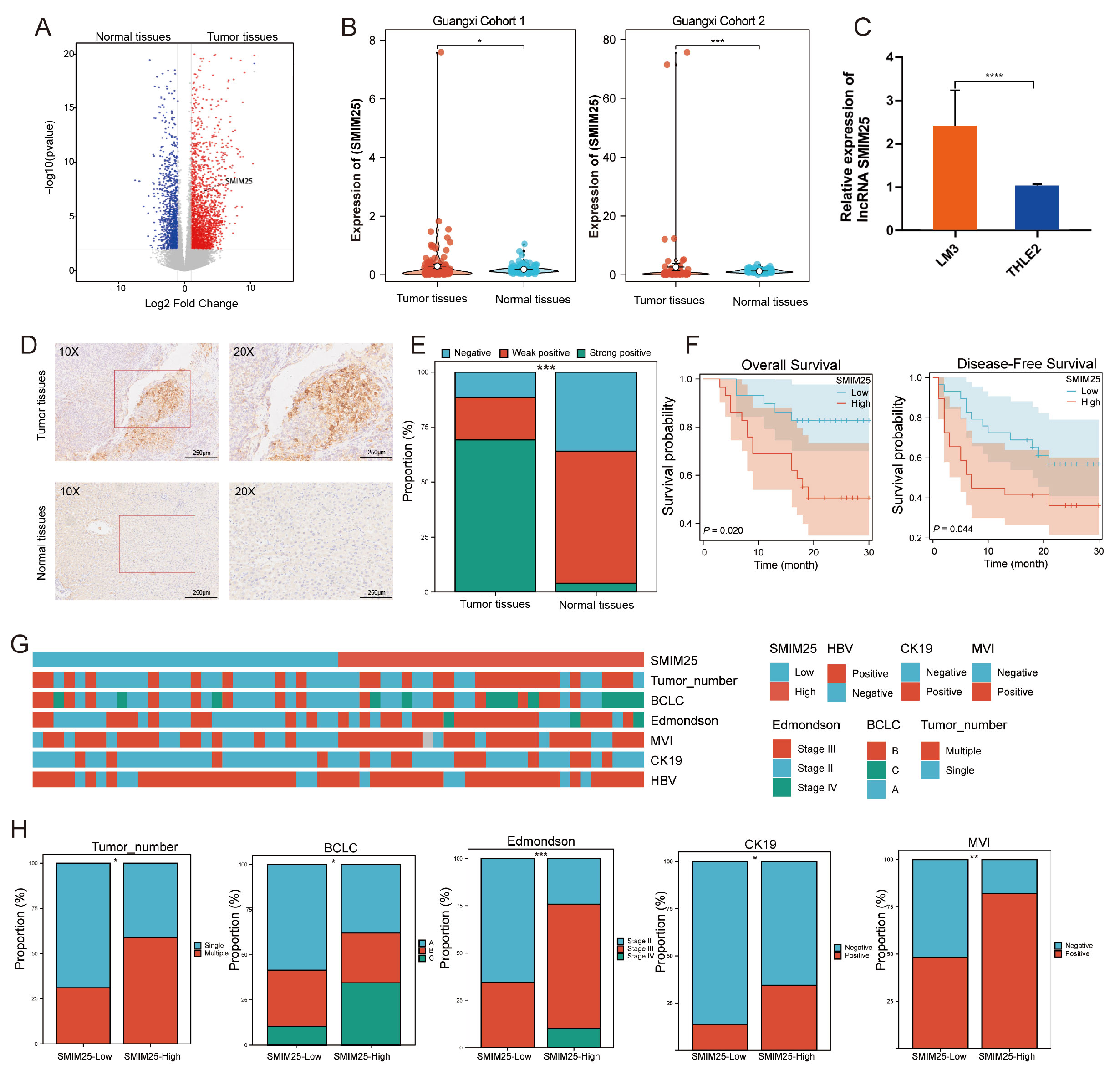
3.1. SMIM25 as a Potential Prognostic Biomarker for HCC
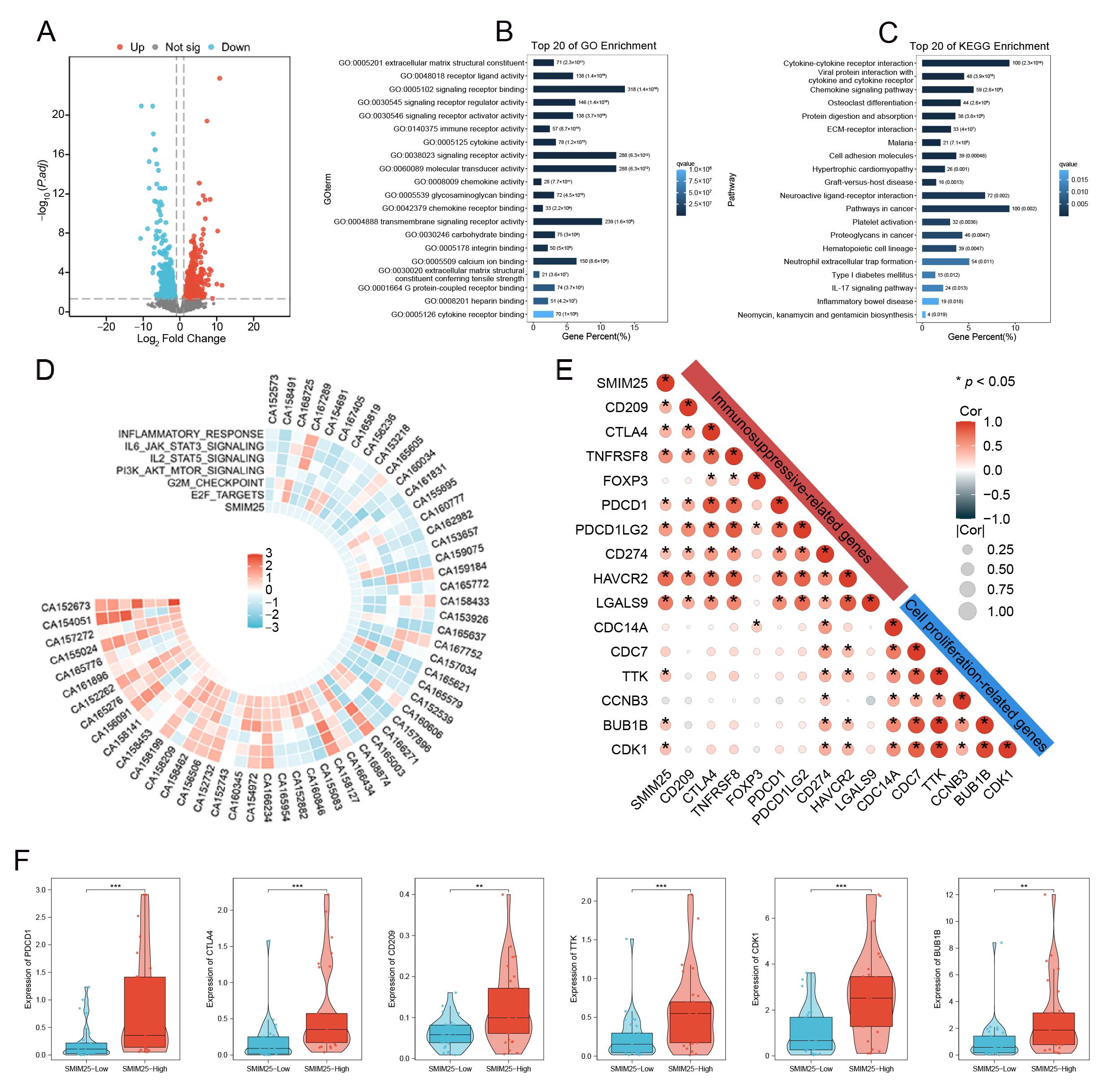
3.2. Transcriptome Analysis of SMIM25 Expression and Immunosuppression Markers in HCC
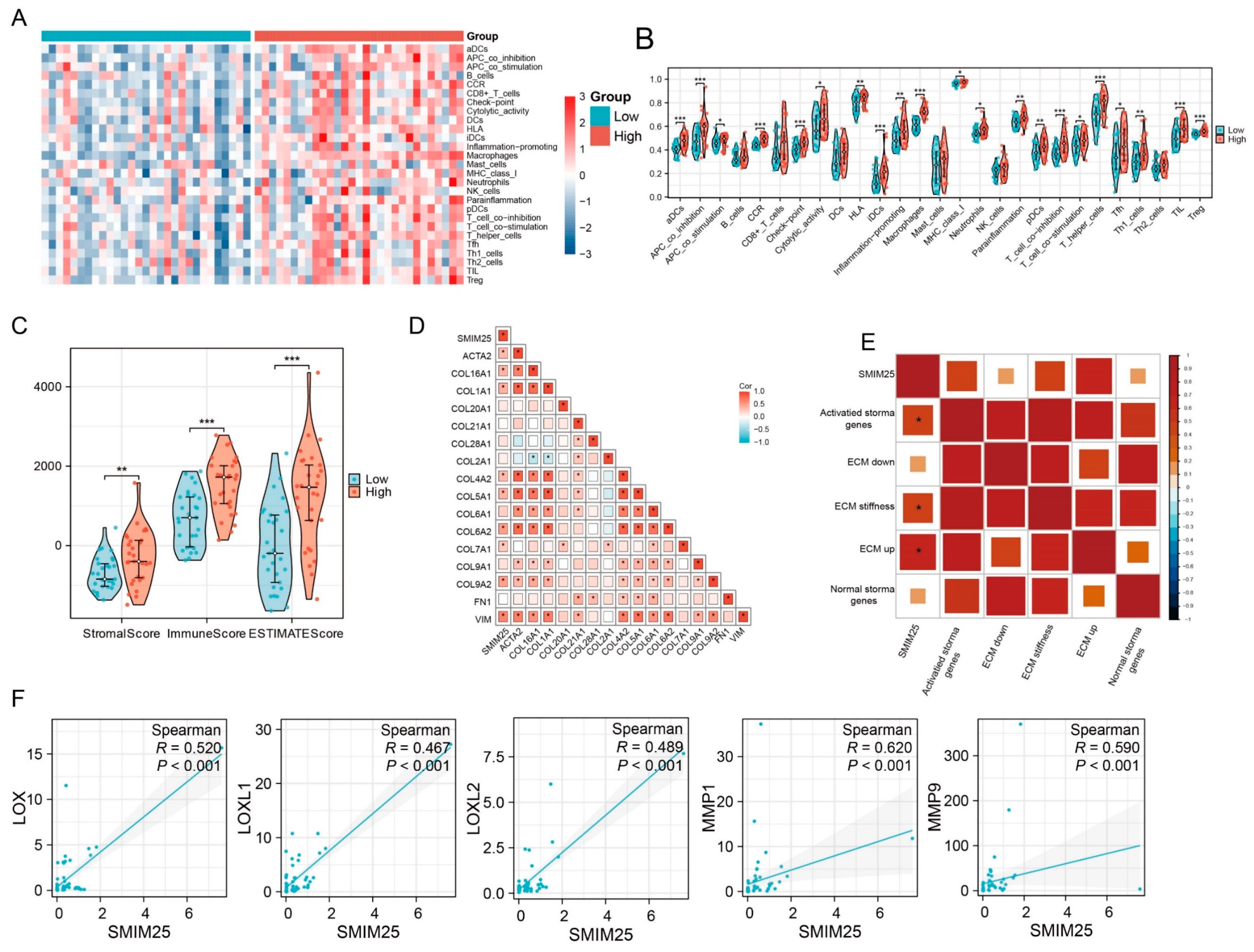
3.3. SMIM25 Plays a Central Role in Remodeling the Tumor Microenvironment in HCC
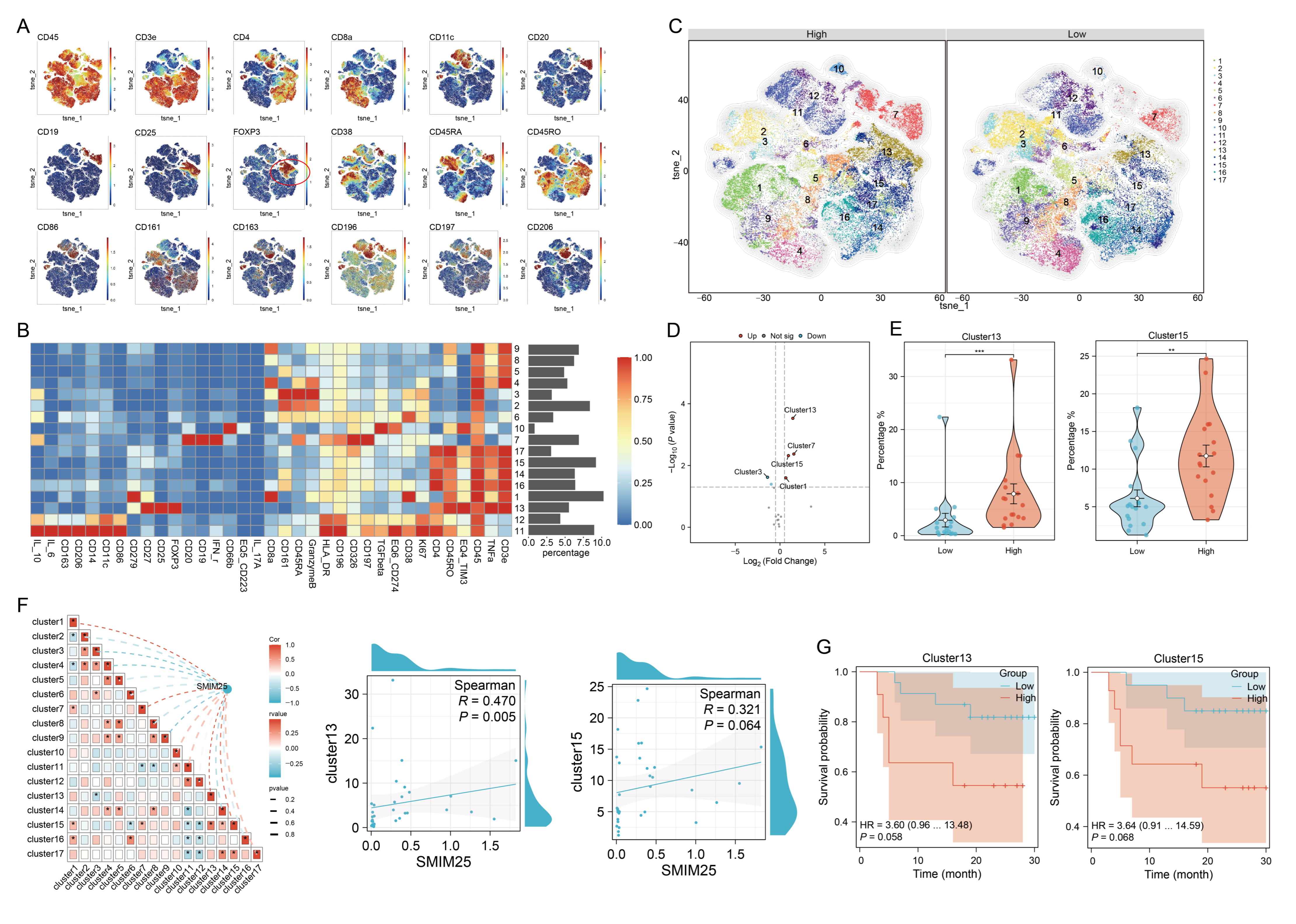
3.4. An Immunosuppression-Related Cell Cluster That Has High SMIM25 Expression Is Recruited to the TME in HCC
3.5. SMIM25 and COX-2 Appear to Have Synergistic Roles in Promoting HCC Development
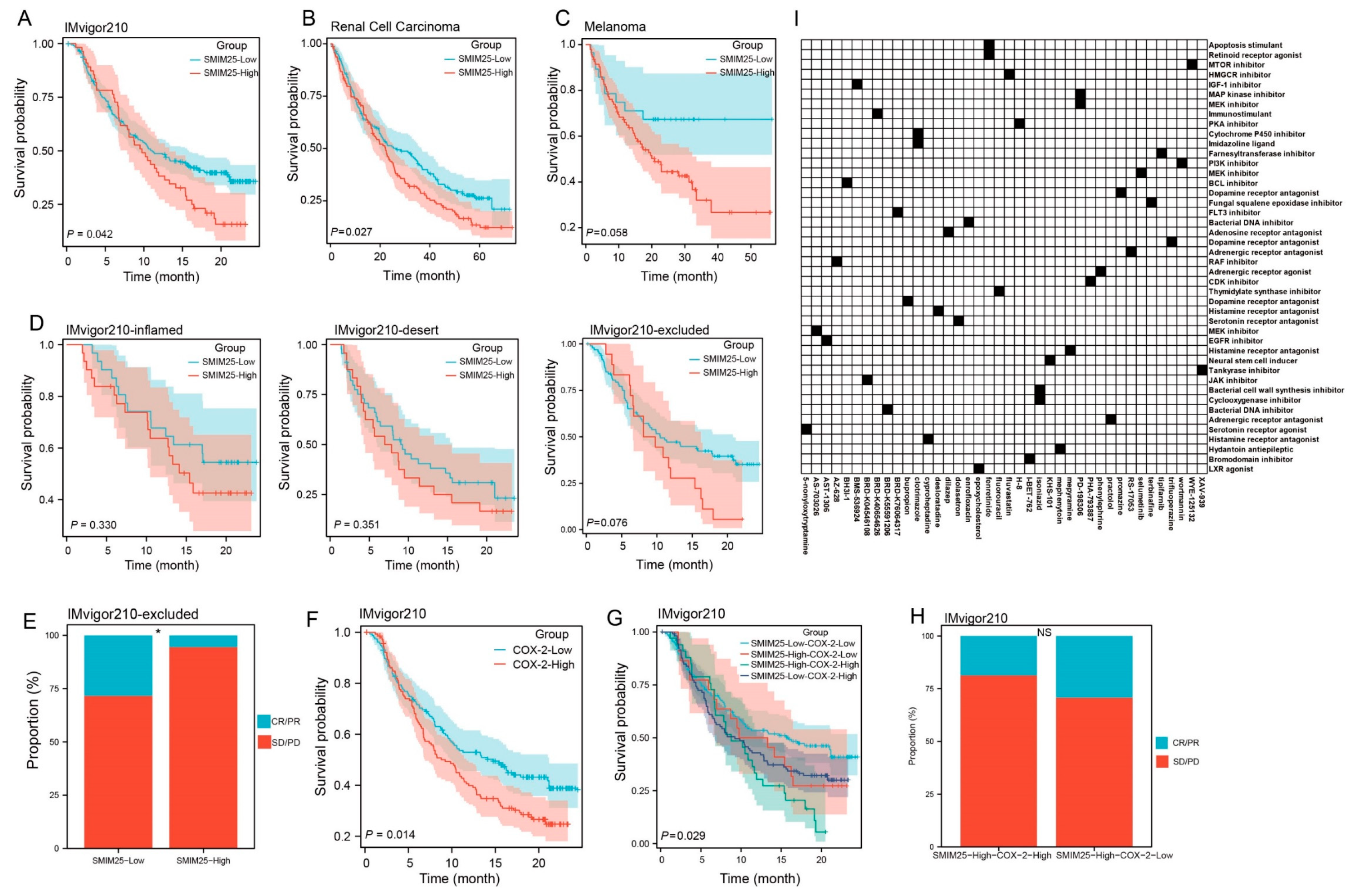
3.6. Predictive Value of the Synergistic SMIM25-COX-2 Axis in Determining Immunotherapy Response in Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HCC | Hepatocellular Carcinoma |
| TME | The Tumor Microenvironment |
| LncRNA | Long Non-Coding RNA |
| SMIM25 | Small Integral Membrane Protein 25 |
| ECM | The Extracellular Matrix |
| GSEA | Gene Set Enrichment Analysis |
| ssGSEA | Single-Sample GSEA |
| CyTOF | Cytometry Based on Time of Flight |
| ROIs | Regions of Interest |
| DEGs | Differentially Expressed Genes |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Wang, W.; Wei, C. Advances in the Early Diagnosis of Hepatocellular Carcinoma. Genes Dis. 2020, 7, 308–319. [Google Scholar] [CrossRef]
- Guo, D.Z.; Zhang, X.; Zhang, S.Q.; Zhang, S.Y.; Zhang, X.Y.; Yan, J.Y.; Dong, S.Y.; Zhu, K.; Yang, X.R.; Fan, J.; et al. Single-Cell Tumor Heterogeneity Landscape of Hepatocellular Carcinoma: Unraveling the Pro-Metastatic Subtype and Its Interaction Loop with Fibroblasts. Mol. Cancer 2024, 23, 157. [Google Scholar] [CrossRef]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 711–723, Correction in N. Engl. J. Med. 2010, 363, 1290. [Google Scholar] [CrossRef]
- Versluis, J.M.; Menzies, A.M.; Sikorska, K.; Rozeman, E.A.; Saw, R.P.M.; van Houdt, W.J.; Eriksson, H.; Klop, W.M.C.; Ch’ng, S.; van Thienen, J.V.; et al. Survival Update of Neoadjuvant Ipilimumab Plus Nivolumab in Macroscopic Stage III Melanoma in the Opacin and Opacin-Neo Trials. Ann. Oncol. 2023, 34, 420–430. [Google Scholar] [CrossRef]
- Shi, J.; Liu, J.; Tu, X.; Li, B.; Tong, Z.; Wang, T.; Zheng, Y.; Shi, H.; Zeng, X.; Chen, W.; et al. Single-Cell Immune Signature for Detecting Early-Stage Hcc and Early Assessing Anti-Pd-1 Immunotherapy Efficacy. J. Immunother. Cancer 2022, 10, e003133. [Google Scholar] [CrossRef]
- Finn, R.S.; Ryoo, B.Y.; Merle, P.; Kudo, M.; Bouattour, M.; Lim, H.Y.; Breder, V.; Edeline, J.; Chao, Y.; Ogasawara, S.; et al. Pembrolizumab as Second-Line Therapy in Patients with Advanced Hepatocellular Carcinoma in Keynote-240: A Randomized, Double-Blind, Phase III Trial. J. Clin. Oncol. 2020, 38, 193–202. [Google Scholar] [CrossRef]
- Qin, S.; Ren, Z.; Meng, Z.; Chen, Z.; Chai, X.; Xiong, J.; Bai, Y.; Yang, L.; Zhu, H.; Fang, W.; et al. Camrelizumab in Patients with Previously Treated Advanced Hepatocellular Carcinoma: A Multicentre, Open-Label, Parallel-Group, Randomised, Phase 2 Trial. Lancet Oncol. 2020, 21, 571–580. [Google Scholar] [CrossRef]
- El-Khoueiry, A.B.; Sangro, B.; Yau, T.; Crocenzi, T.S.; Kudo, M.; Hsu, C.; Kim, T.Y.; Choo, S.P.; Trojan, J.; Welling, T.H.R.; et al. Nivolumab in Patients with Advanced Hepatocellular Carcinoma (CheckMate 040): An Open-Label, Non-Comparative, Phase 1/2 Dose Escalation and Expansion Trial. Lancet 2017, 389, 2492–2502. [Google Scholar] [CrossRef]
- Qiu, X.; Zhou, T.; Li, S.; Wu, J.; Tang, J.; Ma, G.; Yang, S.; Hu, J.; Wang, K.; Shen, S.; et al. Spatial Single-Cell Protein Landscape Reveals Vimentin(High) Macrophages as Immune-Suppressive in the Microenvironment of Hepatocellular Carcinoma. Nat. Cancer 2024, 5, 1557–1578. [Google Scholar] [CrossRef]
- Araste, F.; Abnous, K.; Hashemi, M.; Taghdisi, S.M.; Ramezani, M.; Alibolandi, M. Peptide-Based Targeted Therapeutics: Focus on Cancer Treatment. J. Control. Release 2018, 292, 141–162. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.K.; Stanslas, J. Peptide-Based and Small Molecule Pd-1 and Pd-L1 Pharmacological Modulators in the Treatment of Cancer. Pharmacol. Ther. 2021, 227, 107870. [Google Scholar] [CrossRef]
- Fu, X.Y.; Yin, H.; Chen, X.T.; Yao, J.F.; Ma, Y.N.; Song, M.; Xu, H.; Yu, Q.Y.; Du, S.S.; Qi, Y.K.; et al. Three Rounds of Stability-Guided Optimization and Systematical Evaluation of Oncolytic Peptide LTX-315. J. Med. Chem. 2024, 67, 3885–3908. [Google Scholar] [CrossRef]
- Yin, H.; Chen, X.T.; Chi, Q.N.; Ma, Y.N.; Fu, X.Y.; Du, S.S.; Qi, Y.K.; Wang, K.W. The Hybrid Oncolytic Peptide Ntp-385 Potently Inhibits Adherent Cancer Cells by Targeting the Nucleus. Acta Pharmacol. Sin. 2023, 44, 201–210. [Google Scholar] [CrossRef]
- Yin, H.; Fu, X.Y.; Gao, H.Y.; Ma, Y.N.; Yao, J.F.; Du, S.S.; Qi, Y.K.; Wang, K.W. Design, Synthesis and Anticancer Evaluation of Novel Oncolytic Peptide-Chlorambucil Conjugates. Bioorg Chem. 2023, 138, 106674. [Google Scholar] [CrossRef]
- Llovet, J.M.; Brú, C.; Bruix, J. Prognosis of hepatocellular carcinoma: The BCLC staging classification. Semin. Liver Dis. 1999, 19, 329–338. [Google Scholar] [CrossRef]
- Reig, M.; Cabibbo, G. Antiviral Therapy in the Palliative Setting of Hcc (Bclc-B and -C). J. Hepatol. 2021, 74, 1225–1233. [Google Scholar] [CrossRef]
- Zhou, L.; Rui, J.A.; Zhou, W.X.; Wang, S.B.; Chen, S.G.; Qu, Q. Edmondson-Steiner Grade: A Crucial Predictor of Recurrence and Survival in Hepatocellular Carcinoma Without Microvascular Invasio. Pathol. Res. Pract. 2017, 213, 824–830. [Google Scholar] [CrossRef]
- Zhou, Z.; Cao, S.; Chen, C.; Chen, J.; Xu, X.; Liu, Y.; Liu, Q.; Wang, K.; Han, B.; Yin, Y. A Novel Nomogram for the Preoperative Prediction of Edmondson-Steiner Grade III-IV in Hepatocellular Carcinoma Patients. J. Hepatocell. Carcinoma 2023, 10, 1399–1409. [Google Scholar] [CrossRef]
- Allred, D.C.; Harvey, J.M.; Berardo, M.; Clark, G.M. Prognostic and Predictive Factors in Breast Cancer by Immunohistochemical Analysis. Mod. Pathol. 1998, 11, 155–168. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with Hisat2 and Hisat-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from Rna-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Jiang, Z.; Chen, C.; Wang, X. Classification of Triple-Negative Breast Cancers Based on Immunogenomic Profiling. J. Exp. Clin. Cancer Res. 2018, 37, 327. [Google Scholar] [CrossRef]
- Sun, X.; Wu, B.; Chiang, H.C.; Deng, H.; Zhang, X.; Xiong, W.; Liu, J.; Rozeboom, A.M.; Harris, B.T.; Blommaert, E.; et al. Tumour Ddr1 Promotes Collagen Fibre Alignment to Instigate Immune Exclusion. Nature 2021, 599, 673–678. [Google Scholar] [CrossRef]
- Dewidar, B.; Meyer, C.; Dooley, S.; Meindl-Beinker, A.N. TGF-β in Hepatic Stellate Cell Activation and Liver Fibrogenesis-Updated 2019. Cells 2019, 8, 1419. [Google Scholar] [CrossRef]
- Davis, F.M.; Melvin, W.J.; Mangum, K.; Tsoi, L.C.; Joshi, A.D.; Cai, Q.; Henke, P.K.; Gudjonsson, J.E.; Gallagher, K.A. The Histone Methyltransferase SETDB2 Modulates Tissue Inhibitors of Metalloproteinase-Matrix Metalloproteinase Activity During Abdominal Aortic Aneurysm Development. Ann. Surg. 2023, 278, 426–440. [Google Scholar] [CrossRef]
- Narvaez-Montoya, C.; Mahlknecht, J.; Torres-Martínez, J.A.; Mora, A.; Pino-Vargas, E. FlowSOM Clustering—A Novel Pattern Recognition Approach for Water Research: Application to a Hyper-Arid Coastal Aquifer System. Sci. Total Environ. 2024, 915, 169988. [Google Scholar] [CrossRef]
- Liu, X.; Song, W.; Wong, B.Y.; Zhang, T.; Yu, S.; Lin, G.N.; Ding, X. A Comparison Framework and Guideline of Clustering Methods for Mass Cytometry Data. Genome Biol. 2019, 20, 297. [Google Scholar] [CrossRef]
- Chen, L.; Huang, Y. High Expression of Lncrna Pelaton Serves as a Risk Factor for the Incidence and Prognosis of Acute Coronary Syndrome. Sci. Rep. 2022, 12, 8030. [Google Scholar] [CrossRef]
- Subhash, S.; Kalmbach, N.; Wegner, F.; Petri, S.; Glomb, T.; Dittrich-Breiholz, O.; Huang, C.; Bali, K.K.; Kunz, W.S.; Samii, A.; et al. Transcriptome-Wide Profiling of Cerebral Cavernous Malformations Patients Reveal Important Long Noncoding Rna Molecular Signatures. Sci. Rep. 2019, 9, 18203. [Google Scholar] [CrossRef]
- Fu, H.; Zhang, Z.; Li, D.; Lv, Q.; Chen, S.; Zhang, Z.; Wu, M. Lncrna Pelaton, a Ferroptosis Suppressor and Prognositic Signature for Gbm. Front. Oncol. 2022, 12, 817737. [Google Scholar] [CrossRef]
- Leng, X.; Liu, G.; Wang, S.; Song, J.; Zhang, W.; Zhang, X.; Rong, L.; Ma, Y.; Song, F. Linc01272 Promotes Migration and Invasion of Gastric Cancer Cells Via Emt. Onco Targets Ther. 2020, 13, 3401–3410. [Google Scholar] [CrossRef]
- Pardoll, D.M. The Blockade of Immune Checkpoints in Cancer Immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Li, B.; Chan, H.L.; Chen, P. Immune Checkpoint Inhibitors: Basics and Challenges. Curr. Med. Chem. 2019, 26, 3009–3025. [Google Scholar] [CrossRef]
- Maharaj, K.; Uriepero, A.; Sahakian, E.; Pinilla-Ibarz, J. Regulatory T Cells (Tregs) in Lymphoid Malignancies and the Impact of Novel Therapies. Front. Immunol. 2022, 13, 943354. [Google Scholar] [CrossRef]
- Massa, D.; Tosi, A.; Rosato, A.; Guarneri, V.; Dieci, M.V. Multiplexed in Situ Spatial Protein Profiling in the Pursuit of Precision Immuno-Oncology for Patients with Breast Cancer. Cancers 2022, 14, 4885. [Google Scholar] [CrossRef]
- Yao, H.; Yu, S.; Luo, Y.; Wang, M.; Wang, X.; Xu, S.; Chen, Y.; Xie, Z. Effects of Plasma-Derived Exosomes from the Normal and Thin Bactrian Camels on Hepatocellular Carcinoma and Their Differences at Transcriptome and Proteomics Levels. Front. Oncol. 2023, 13, 994340. [Google Scholar] [CrossRef]
- Ai, H.; Yang, H.; Li, L.; Ma, J.; Liu, K.; Li, Z. Cancer/Testis Antigens: Promising Immunotherapy Targets for Digestive Tract Cancers. Front. Immunol. 2023, 14, 1190883. [Google Scholar] [CrossRef]
- Grout, J.A.; Sirven, P.; Leader, A.M.; Maskey, S.; Hector, E.; Puisieux, I.; Steffan, F.; Cheng, E.; Tung, N.; Maurin, M.; et al. Spatial Positioning and Matrix Programs of Cancer-Associated Fibroblasts Promote T-cell Exclusion in Human Lung Tumors. Cancer Discov. 2022, 12, 2606–2625. [Google Scholar] [CrossRef]
- Liu, J.; Chiang, H.C.; Xiong, W.; Laurent, V.; Griffiths, S.C.; Dülfer, J.; Deng, H.; Sun, X.; Yin, Y.W.; Li, W.; et al. A Highly Selective Humanized Ddr1 Mab Reverses Immune Exclusion by Disrupting Collagen Fiber Alignment in Breast Cancer. J. Immunother. Cancer 2023, 11, e006720. [Google Scholar] [CrossRef] [PubMed]
- Tharp, K.M.; Kersten, K.; Maller, O.; Timblin, G.A.; Stashko, C.; Canale, F.P.; Menjivar, R.E.; Hayward, M.K.; Berestjuk, I.; Ten Hoeve, J.; et al. Tumor-Associated Macrophages Restrict Cd8+ T Cell Function Through Collagen Deposition and Metabolic Reprogramming of the Breast Cancer Microenvironment. Nat. Cancer 2024, 5, 1045–1062. [Google Scholar] [CrossRef]
- Jain, R.; Hussein, M.A.; Pierce, S.; Martens, C.; Shahagadkar, P.; Munirathinam, G. Oncopreventive and Oncotherapeutic Potential of Licorice Triterpenoid Compound Glycyrrhizin and its Derivatives: Molecular Insights. Pharmacol. Res. 2022, 178, 106138. [Google Scholar] [CrossRef]
- Hu, H.; Kang, L.; Zhang, J.; Wu, Z.; Wang, H.; Huang, M.; Lan, P.; Wu, X.; Wang, C.; Cao, W.; et al. Neoadjuvant Pd-1 Blockade with Toripalimab, with or Without Celecoxib, in Mismatch Repair-Deficient or Microsatellite Instability-High, Locally Advanced, Colorectal Cancer (Picc): A Single-Centre, Parallel-Group, Non-Comparative, Randomised, Phase 2 Trial. Lancet Gastroenterol. Hepatol. 2022, 7, 38–48. [Google Scholar] [CrossRef]
- Li, J.; Wu, C.; Hu, H.; Qin, G.; Wu, X.; Bai, F.; Zhang, J.; Cai, Y.; Huang, Y.; Wang, C.; et al. Remodeling of the Immune and Stromal Cell Compartment by Pd-1 Blockade in Mismatch Repair-Deficient Colorectal Cancer. Cancer Cell 2023, 41, 1152–1169.e1157. [Google Scholar] [CrossRef]
- Peng, K.; Liu, Y.; Liu, S.; Wang, Z.; Zhang, H.; He, W.; Jin, Y.; Wang, L.; Xia, X.; Xia, L. Targeting Mek/Cox-2 Axis Improve Immunotherapy Efficacy in Dmmr Colorectal Cancer with Pik3ca Overexpression. Cell. Oncol. 2024, 47, 1043–1058. [Google Scholar] [CrossRef]
- Zhang, A.; Zou, X.; Yang, S.; Yang, H.; Ma, Z.; Li, J. Effect of Nets/Cox-2 Pathway on Immune Microenvironment and Metastasis in Gastric Cancer. Front. Immunol. 2023, 14, 1177604. [Google Scholar] [CrossRef]
- Yu, B.; Wang, Y.; Bing, T.; Tang, Y.; Huang, J.; Xiao, H.; Liu, C.; Yu, Y. Platinum Prodrug Nanoparticles with Cox-2 Inhibition Amplify Pyroptosis for Enhanced Chemotherapy and Immune Activation of Pancreatic Cancer. Adv. Mater. 2024, 36, e2310456. [Google Scholar] [CrossRef]
- Zhang, P.; Zhong, D.; Yu, Y.; Wang, L.; Li, Y.; Liang, Y.; Shi, Y.; Duan, M.; Li, B.; Niu, H.; et al. Integration of Sting Activation and Cox-2 Inhibition Via Steric-Hindrance Effect Tuned Nanoreactors for Cancer Chemoimmunotherapy. Biomaterials 2024, 311, 122695. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Li, X.; Zhang, S.; Sun, J.; Lu, Q.; Tao, Y.; Liang, S.; Lan, X.; Zhong, J.; Wang, Q. The SMIM25-COX-2 Axis Modulates the Immunosuppressive Tumor Microenvironment and Predicts Immunotherapy Response in Hepatocellular Carcinoma. Curr. Issues Mol. Biol. 2025, 47, 693. https://doi.org/10.3390/cimb47090693
Wang Z, Li X, Zhang S, Sun J, Lu Q, Tao Y, Liang S, Lan X, Zhong J, Wang Q. The SMIM25-COX-2 Axis Modulates the Immunosuppressive Tumor Microenvironment and Predicts Immunotherapy Response in Hepatocellular Carcinoma. Current Issues in Molecular Biology. 2025; 47(9):693. https://doi.org/10.3390/cimb47090693
Chicago/Turabian StyleWang, Zhenxing, Xia Li, Shiyi Zhang, Jiamin Sun, Qinchen Lu, Yuting Tao, Shuang Liang, Xiuwan Lan, Jianhong Zhong, and Qiuyan Wang. 2025. "The SMIM25-COX-2 Axis Modulates the Immunosuppressive Tumor Microenvironment and Predicts Immunotherapy Response in Hepatocellular Carcinoma" Current Issues in Molecular Biology 47, no. 9: 693. https://doi.org/10.3390/cimb47090693
APA StyleWang, Z., Li, X., Zhang, S., Sun, J., Lu, Q., Tao, Y., Liang, S., Lan, X., Zhong, J., & Wang, Q. (2025). The SMIM25-COX-2 Axis Modulates the Immunosuppressive Tumor Microenvironment and Predicts Immunotherapy Response in Hepatocellular Carcinoma. Current Issues in Molecular Biology, 47(9), 693. https://doi.org/10.3390/cimb47090693





