Center Degenerated Walking-Primer PCR: A Novel and Universal Genome-Walking Method
Abstract
1. Introduction
2. Materials and Methods
2.1. Genomic Templates
2.2. Design of Primer
2.3. Procedure of CDWP-PCR
2.4. Effects of Degeneracy of cdWP
2.5. DNA Sequencing and Analysis
3. Results
3.1. Overview of CDWP-PCR
3.2. Effects of Degeneracy of cdWP
3.3. Validation of CDWP-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yik, M.H.Y.; Lo, Y.T.; Lin, X.; Sun, W.; Chan, T.F.; Shaw, P.C. Authentication of Hedyotis products by adaptor ligation-mediated PCR and metabarcoding. J. Pharm. Biomed. Anal. 2021, 196, 113920. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, A.; Makarenko, M.; Yunusova, A. Transgene mapping in animals: What to choose? Int. J. Mol. Sci. 2025, 26, 4705. [Google Scholar] [CrossRef]
- Christy, S.M.E.; Arun, V. Isolation of actin regulatory region from medicinal plants by thermal asymmetric interlaced PCR (TAIL PCR) and its bioinformatic analysis. Braz. J. Bot. 2024, 47, 67–78. [Google Scholar] [CrossRef]
- Han, H.J.; Kim, D.H.; Baik, J.Y. A splinkerette PCR-based genome walking technique for the identification of transgene integration sites in CHO cells. J. Biotechnol. 2023, 371, 1–9. [Google Scholar] [CrossRef]
- Li, H.X.; Wei, C.; Pan, Z.K.; Liu, X.H. Arbitrarily suffixed sequence-specific primer PCR for reliable genome walking: Taking genome walkings of Levilactobacillus brevis and rice as examples. Iran. J. Biotechnol. 2024, 22, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Alquezar-Planas, D.E.; Löber, U.; Cui, P.; Quedenau, C.; Chen, W.; Greenwood, A.D.; Johnston, S. DNA sonication inverse PCR for genome scale analysis of uncharacterized flanking sequences. Methods Ecol. Evol. 2020, 12, 182–195. [Google Scholar] [CrossRef]
- Yu, D.; Zhou, T.S.; Sun, X.W.; Sun, Z.Z.; Sheng, X.B.; Tan, Y.; Liu, L.; Ouyang, N.; Xu, K.; Shi, K.; et al. Cyclic digestion and ligation-mediated PCR used for flanking sequence walking. Sci. Rep. 2020, 10, 3434. [Google Scholar] [CrossRef]
- Stefano, B.; Patrizia, B.; Matteo, C.; Massimo, G. Inverse PCR and quantitative PCR as alternative methods to southern blotting analysis to assess transgene copy number and characterize the integration site in transgenic woody plants. Biochem. Genet. 2016, 54, 291–305. [Google Scholar] [CrossRef]
- Trinh, Q.; Zhu, P.Y.; Shi, H.; Xu, W.T.; Hao, J.R.; Luo, Y.B.; Huang, K. A-T linker adapter polymerase chain reaction for determining flanking sequences by rescuing inverse PCR or thermal asymmetric interlaced PCR products. Anal. Biochem. 2014, 466, 24–26. [Google Scholar] [CrossRef]
- Okulova, E.S.; Burlakovskiy, M.S.; Lutova, L.A. PCR-based genome walking methods (review). Ecol. Genet. 2024, 22, 75–104. [Google Scholar] [CrossRef]
- Noguchi, A.; Takekawa, N.; Einarsdottir, T.; Koura, M.; Noguchi, Y.; Takano, K.; Yamamoto, Y.; Matsuda, J.; Suzuki, O. Chromosomal mapping and zygosity check of transgenes based on flanking genome sequences determined by genomic walking. Exp. Anim. 2004, 53, 103–111. [Google Scholar] [CrossRef][Green Version]
- Zhao, S.; Wang, Y.; Zhu, Z.H.; Chen, P.; Liu, W.G.; Wang, C.R.; Lu, H.; Xiang, Y.; Liu, Y.; Qian, Q.; et al. Streamlined whole-genome genotyping through NGS-enhanced thermal asymmetric interlaced (TAIL)-PCR. Plant Commun. 2024, 5, 100983. [Google Scholar] [CrossRef] [PubMed]
- Lyu, M.J.; Liu, H.F.; Waititu, J.K.; Sun, Y.; Wang, H.; Fu, J.J.; Chen, Y.; Liu, J.; Ku, L.; Cheng, X. TEAseq-based identification of 35,696 Dissociation insertional mutations facilitates functional genomic studies in maize. J. Genet. Genom. 2021, 48, 961–971. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.Y.; Feng, S.L.; Li, X.J.; Li, Y.J.; Wang, W.; Gilliham, M.; Chen, Z.H.; Xu, S.C. TTLOC: A Tn5 transposase-based approach to localize T-DNA integration sites. Plant Physiol. 2025, 197, kiaf102. [Google Scholar] [CrossRef]
- Pillai, M.M.; Venkataraman, G.M.; Kosak, S.; Torok-Storb, B. Integration site analysis in transgenic mice by thermal asymmetric interlaced (TAIL)-PCR: Segregating multiple-integrant founder lines and determining zygosity. Transgenic Res. 2008, 17, 749–754. [Google Scholar] [CrossRef]
- O’Malley, R.C.; Alonso, J.M.; Kim, C.J.; Leisse, T.J.; Ecker, J.R. An adapter ligation-mediated PCR method for high-throughput mapping of T-DNA inserts in the Arabidopsis genome. Nat. Protoc. 2007, 2, 2910–2917. [Google Scholar] [CrossRef] [PubMed]
- Potter, C.J.; Luo, L.Q. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS ONE 2010, 5, e10168. [Google Scholar] [CrossRef]
- Lung, J.; Hung, M.S.; Chen, C.Y.; Yang, T.M.; Lin, C.K.; Fang, Y.H.; Jiang, Y.Y.; Liao, H.F.; Lin, Y.C. An optimized ligation-mediated PCR method for chromosome walking and fusion gene chromosomal breakpoints identification. Biol. Methods Protoc. 2024, 9, bpae037. [Google Scholar] [CrossRef]
- Tian, B.; Wu, H.; Wang, R.; Chen, H.; Li, H. N7-ended walker PCR: An efficient genome-walking tool. Biochem. Genet. 2024, 63, 3758–3772. [Google Scholar] [CrossRef]
- Chen, H.; Tian, B.; Wang, R.; Pan, Z.; Gao, D.; Li, H. Uracil walking primer PCR: An accurate and efficient genome-walking tool. J. Genet. Eng. Biotechnol. 2025, 23, 100478. [Google Scholar] [CrossRef]
- Kotik, M. Novel genes retrieved from environmental DNA by polymerase chain reaction: Current genome-walking techniques for future metagenome applications. J. Biotechnol. 2009, 144, 75–82. [Google Scholar] [CrossRef]
- Jones, D.H.; Winistorfer, S.C. Sequence specific generation of a DNA panhandle permits PCR amplification of unknown flanking DNA. Nucleic Acids Res. 1992, 20, 595–600. [Google Scholar] [CrossRef]
- Robinson, B.W.; Slater, D.J.; Felix, C.A. BglII-based panhandle and reverse panhandle PCR approaches increase capability for cloning der(II) and der(other) genomic breakpoint junctions of translocations. Genes Chromosom. Cancer 2006, 45, 740–753. [Google Scholar] [CrossRef]
- Raffini, L.D.; Slater, D.J.; Rappaport, E.F.; Lo Nigro, L.; Cheung, N.K.V.; Felix, C.A. Reverse panhandle PCR for rapid identification of der(other) genomic breakpoint junctions of gene translocations. Blood 2000, 96, 691a. [Google Scholar]
- Yan, Y.X.; An, C.C.; Li, L.; Gu, J.Y.; Tan, G.H.; Chen, Z.L. T-linker-specific ligation PCR (T-linker PCR): An advanced PCR technique for chromosome walking or for isolation of tagged DNA ends. Nucleic Acids Res. 2003, 31, e68. [Google Scholar] [CrossRef]
- Tsaftaris, A.; Pasentzis, K.; Argiriou, A. Rolling circle amplification of genomic templates for inverse PCR (RCA-GIP): A method for 5′- and 3′-genome walking without anchoring. Biotechnol. Lett. 2010, 32, 157–161. [Google Scholar] [CrossRef]
- Gutiérrez, L.G.; Abelleyro, M.M.; Ruiz, M.S.; Anchordoqui, M.S.; Freitas, J.; Bianchini, M.; De Brasi, C.D.; Larripa, I.B. Development of an inverse-PCR approach for characterization of the major breakpoint sequences on genomic DNA: Proof of concept. Clin. Chem. Lab. Med. 2021, 59, E449–E453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.C.; Xu, W.W.; Feng, Z.Y.; Hong, Z. A low degenerate primer pool improved the efficiency of high-efficiency thermal asymmetric interlaced PCR to amplify T-DNA flanking sequences in Arabidopsis thaliana. 3 Biotech 2017, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jia, M.; Li, Z.; Liu, X.; Sun, T.; Pei, J.; Wei, C.; Lin, Z.; Li, H. Protocol to access unknown flanking DNA sequences using Wristwatch-PCR for genome-walking. STAR Protoc. 2023, 4, 102037. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Di, D.W.; Zhang, D.; Song, B.; Luo, P.; Guo, G.Q. Frequent problems and their resolutions by using thermal asymmetric interlaced PCR (TAIL-PCR) to clone genes in T-DNA tagged mutants. Biotechnol. Biotechnol. Equip. 2015, 29, 260–267. [Google Scholar] [CrossRef]
- Chenchik, A.; Diachenko, L.; Moqadam, F.; Tarabykin, V.; Lukyanov, S.; Siebert, P.D. Full-length cDNA cloning and determination of mRNA 5′ and 3′ ends by amplification of adaptor-ligated cDNA. Biotechniques 1996, 21, 526–534. [Google Scholar] [CrossRef]
- Siebert, P.D.; Chenchik, A.; Kellogg, D.E.; Lukyanov, K.A.; Lukyanov, S.A. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995, 23, 1087–1088. [Google Scholar] [CrossRef]
- Gao, D.D.; Chang, K.P.; Ding, G.T.; Wu, H.J.; Chen, Y.H.; Jia, M.Y.; Liu, X.; Wang, S.; Jin, Y.; Pan, H.; et al. Genomic insights into a robust gamma-aminobutyric acid-producer Lactobacillus brevis CD0817. AMB Express 2019, 9, 72. [Google Scholar] [CrossRef]
- Sun, T.Y.; Jia, M.Y.; Wang, L.Q.; Li, Z.Q.; Lin, Z.Y.; Wei, C.; Pei, J.; Li, H. DAR-PCR: A new tool for efficient retrieval of unknown flanking genomic DNA. AMB Express 2022, 12, 131. [Google Scholar] [CrossRef]
- Tan, J.T.; Gong, Q.; Yu, S.Z.; Hou, Y.K.; Zeng, D.C.; Zhu, Q.L.; Liu, Y.G. A modified high-efficiency thermal asymmetric interlaced PCR method for amplifying long unknown flanking sequences. J. Genet. Genom. 2019, 46, 363–366. [Google Scholar] [CrossRef]
- Dai, S.F.; Xue, X.F.; Wang, Y.F.; Xie, Y.L.; Song, Z.P.; Xu, D.Y.; Wen, Z.; Yan, Z. Novel D-hordein-like HMW glutenin sequences isolated from Psathyrostachys juncea by thermal asymmetric interlaced PCR. Cereal Res. Commun. 2018, 46, 135–145. [Google Scholar] [CrossRef]
- Li, H.X.; Lin, Z.Y.; Guo, X.Y.; Pan, Z.K.; Pan, H.; Wang, D.Y. Primer extension refractory PCR: An efficient and reliable genome walking method. Mol. Genet. Genom. 2024, 299, 27. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.Y.; Zhu, Y.S.; Pan, Z.K.; Pan, H.; Li, H.X. Single primer site-specific nested PCR for accurate and rapid genome-walking. J. Microbiol. Methods 2024, 220, 106926. [Google Scholar] [CrossRef] [PubMed]
- Tonooka, Y.; Fujishima, M. Comparison and critical evaluation of PCR-mediated methods to walk along the sequence of genomic DNA. Appl. Microbiol. Biotechnol. 2009, 85, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Grin, I.R.; Mechetin, G.V.; Kasymov, R.D.; Diatlova, E.A.; Yudkina, A.V.; Shchelkunov, S.N.; Gileva, I.P.; Denisova, A.A.; Stepanov, G.A.; Chilov, G.G.; et al. A new class of uracil–DNA glycosylase inhibitors active against human and vaccinia virus enzyme. Molecules 2021, 26, 6668. [Google Scholar] [CrossRef]
- Li, J.; Chen, R.; Yang, Y.; Zhang, Z.M.; Fang, G.C.; Xie, W.; Cao, W. An unconventional family 1 uracil DNA glycosylase in Nitratifractor salsuginis. FEBS J. 2017, 284, 4017–4034. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.R.; Liu, Z.H. Integration site analysis in transgenic pigs by thermal asymmetric interlaced (TAIL)-PCR and junction PCR. Reprod. Fertil. Dev. 2010, 22, 371. [Google Scholar] [CrossRef]
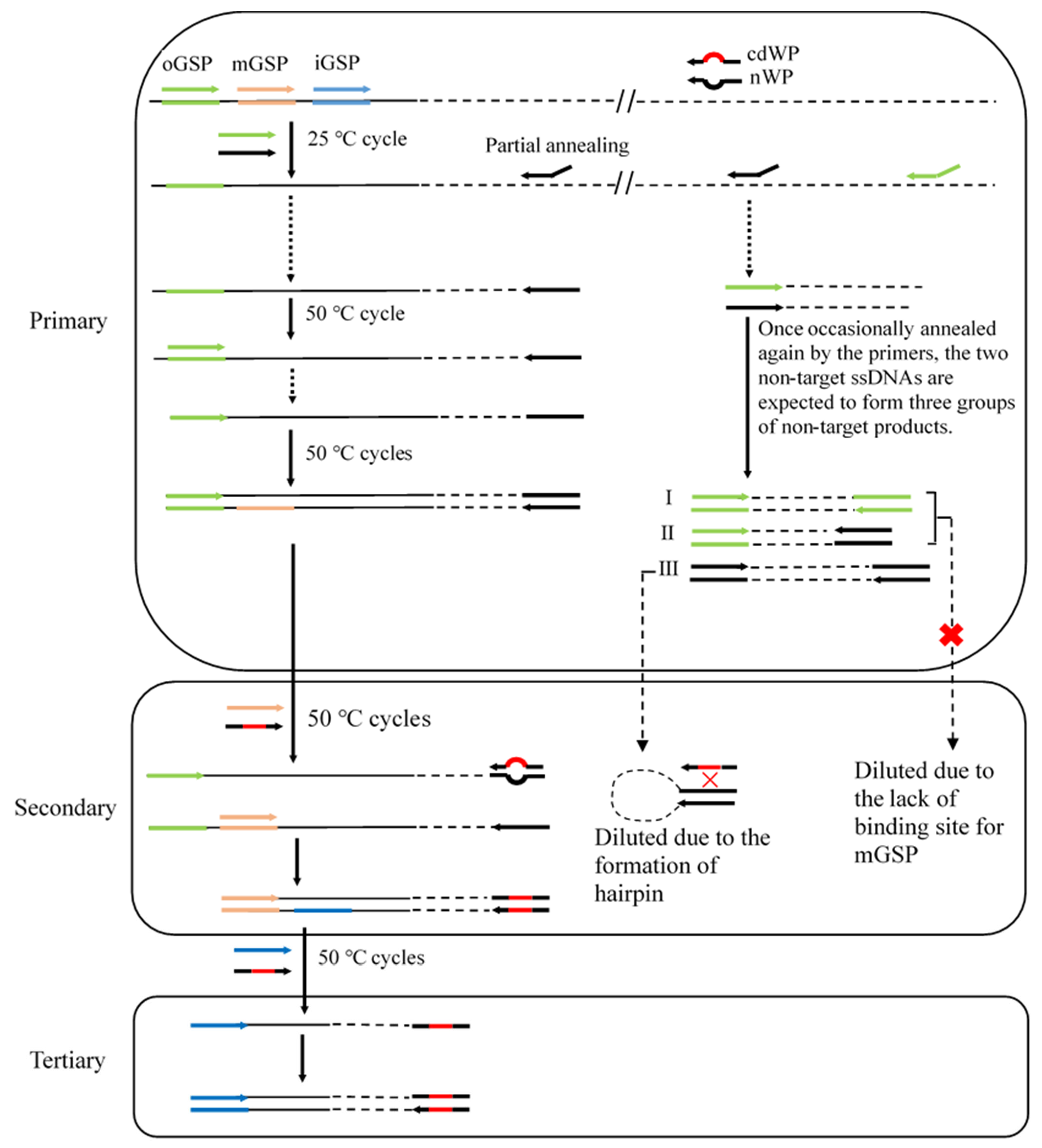
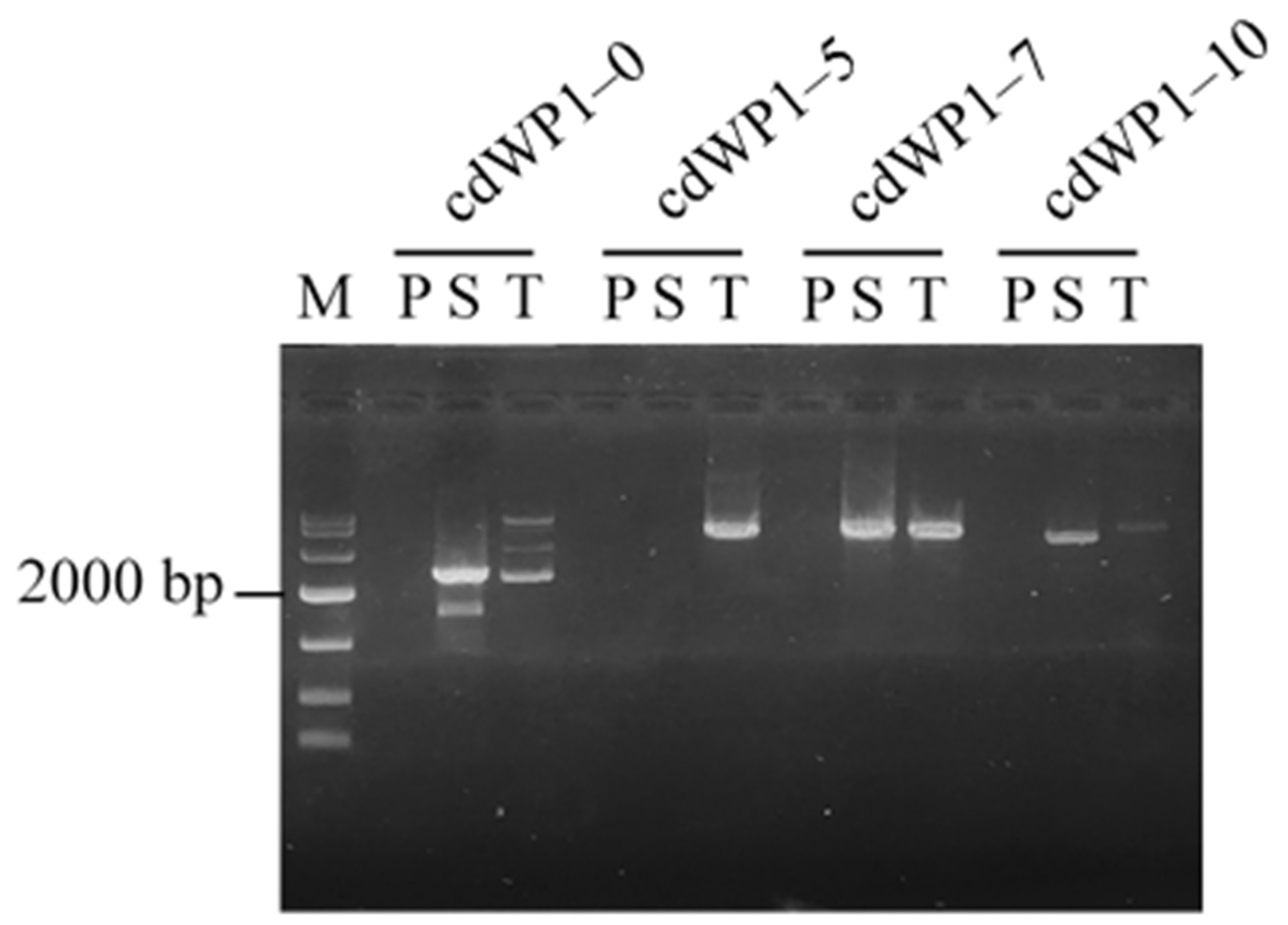
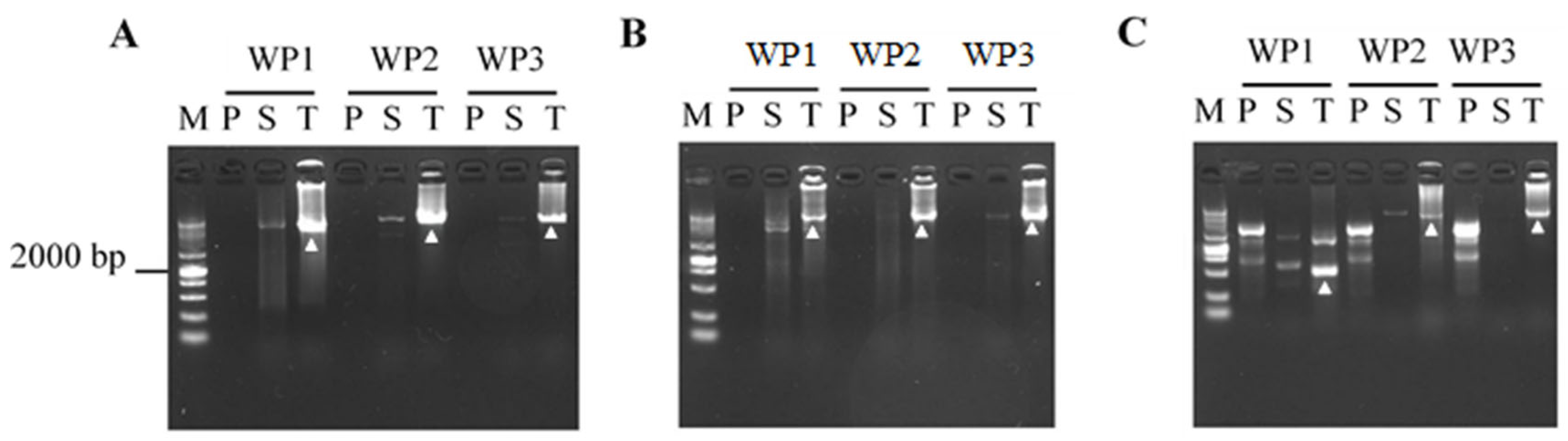
| Primer Set a | Primary CDWP-PCR | Secondary CDWP-PCR | Tertiary CDWP-PCR |
|---|---|---|---|
| hyp | CCAAGGACTTAGGCTTTGACATTGA | GGATGCTGCCTTCGGTGGGTTATTT | TGGTCACAAGTACGGCATGGTTTAC |
| gluT | TCCTTCGTTCTTGATTCCATACCCT | CCATTTCCATAGGTTGCTCCAAGG | GGATACTGGCTAAAATGAATTAACTCGGATAA |
| hyg | TTTCAGCTTCGATGTAGGAGGGCGT | GTAAATAGCTGCGCCGATGGTTTCT | TTGACATTGGGGAGTTTAGCGAGAG |
| WP1 | GGCTATGAAAACAGAACTCGACCTC | GGCTATGAAAACANNNNNNNACCTC | GGCTATGAAAACANNNNNNNACCTC |
| WP2 | CCTGACCGCCTTCTACACCT | CCTGACCGNNNNNNNCACCT | CCTGACCGNNNNNNNCACCT |
| WP3 | ATGCTGCTCGTGGATGACTCT | ATGCTGCTCNNNNNNNACTCT | ATGCTGCTCNNNNNNNACTCT |
| Round of PCR | Thermal Parameters | Cycle Number |
|---|---|---|
| Primary | 94 °C, 2 min | 1 |
| 94 °C, 30 s; 25 °C, 30 s; 72 °C, 2 min | 1 | |
| 94 °C, 30 s; 50 °C, 30 s; 72 °C, 2 min | 40 | |
| 72 °C, 5 min | 1 | |
| Secondary | 94 °C, 2 min | 1 |
| 94 °C, 30 s; 50 °C, 30 s; 72 °C, 2 min | 30 | |
| 72 °C, 5 min | 1 | |
| Tertiary | 94 °C, 2 min | 1 |
| 94 °C, 30 s; 50 °C, 30 s; 72 °C, 2 min | 30 | |
| 72 °C, 5 min | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, D.; Pan, Z.; Pan, H.; Gu, Y.; Li, H. Center Degenerated Walking-Primer PCR: A Novel and Universal Genome-Walking Method. Curr. Issues Mol. Biol. 2025, 47, 602. https://doi.org/10.3390/cimb47080602
Gao D, Pan Z, Pan H, Gu Y, Li H. Center Degenerated Walking-Primer PCR: A Novel and Universal Genome-Walking Method. Current Issues in Molecular Biology. 2025; 47(8):602. https://doi.org/10.3390/cimb47080602
Chicago/Turabian StyleGao, Dandan, Zhenkang Pan, Hao Pan, Yinwei Gu, and Haixing Li. 2025. "Center Degenerated Walking-Primer PCR: A Novel and Universal Genome-Walking Method" Current Issues in Molecular Biology 47, no. 8: 602. https://doi.org/10.3390/cimb47080602
APA StyleGao, D., Pan, Z., Pan, H., Gu, Y., & Li, H. (2025). Center Degenerated Walking-Primer PCR: A Novel and Universal Genome-Walking Method. Current Issues in Molecular Biology, 47(8), 602. https://doi.org/10.3390/cimb47080602








