The Construction of ceRNA Regulatory Network Unraveled Prognostic Biomarkers and Repositioned Drug Candidates for the Management of Pancreatic Ductal Adenocarcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Microarray Datasets
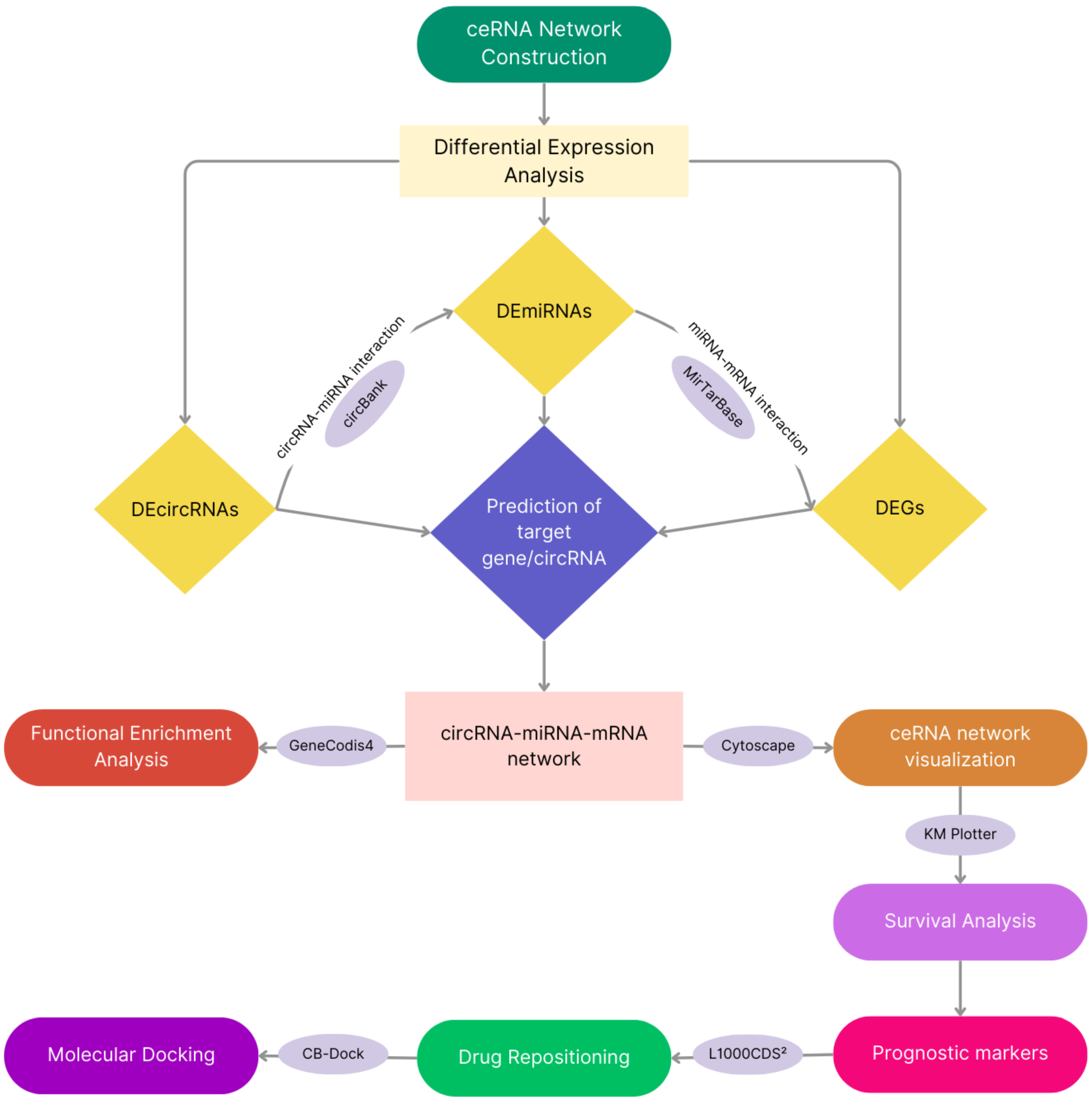
2.2. Identification of DEGs, DEmiRNAs, and DEcircRNAs
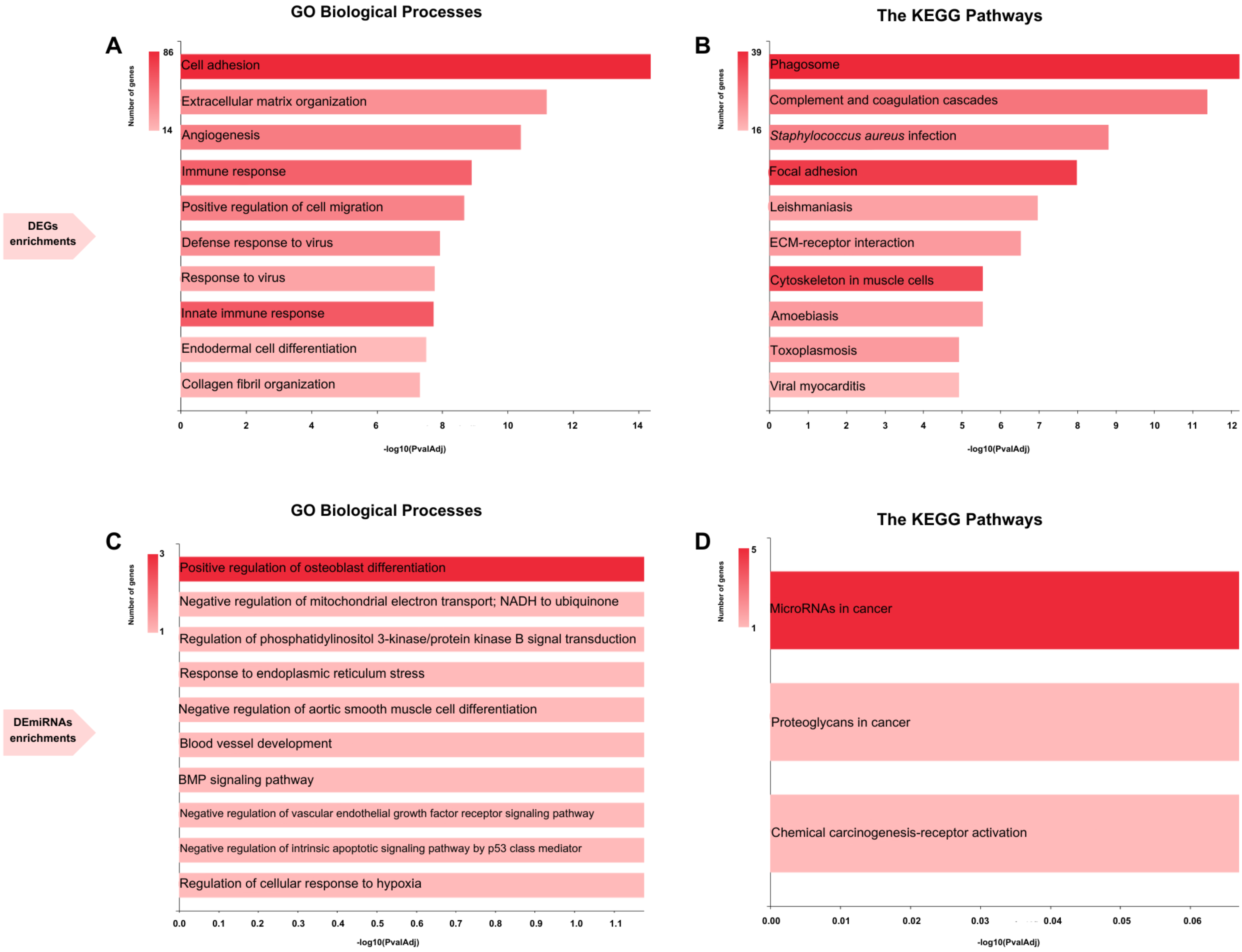
2.3. Functional Enrichment Analysis
2.4. Construction of ceRNA Network
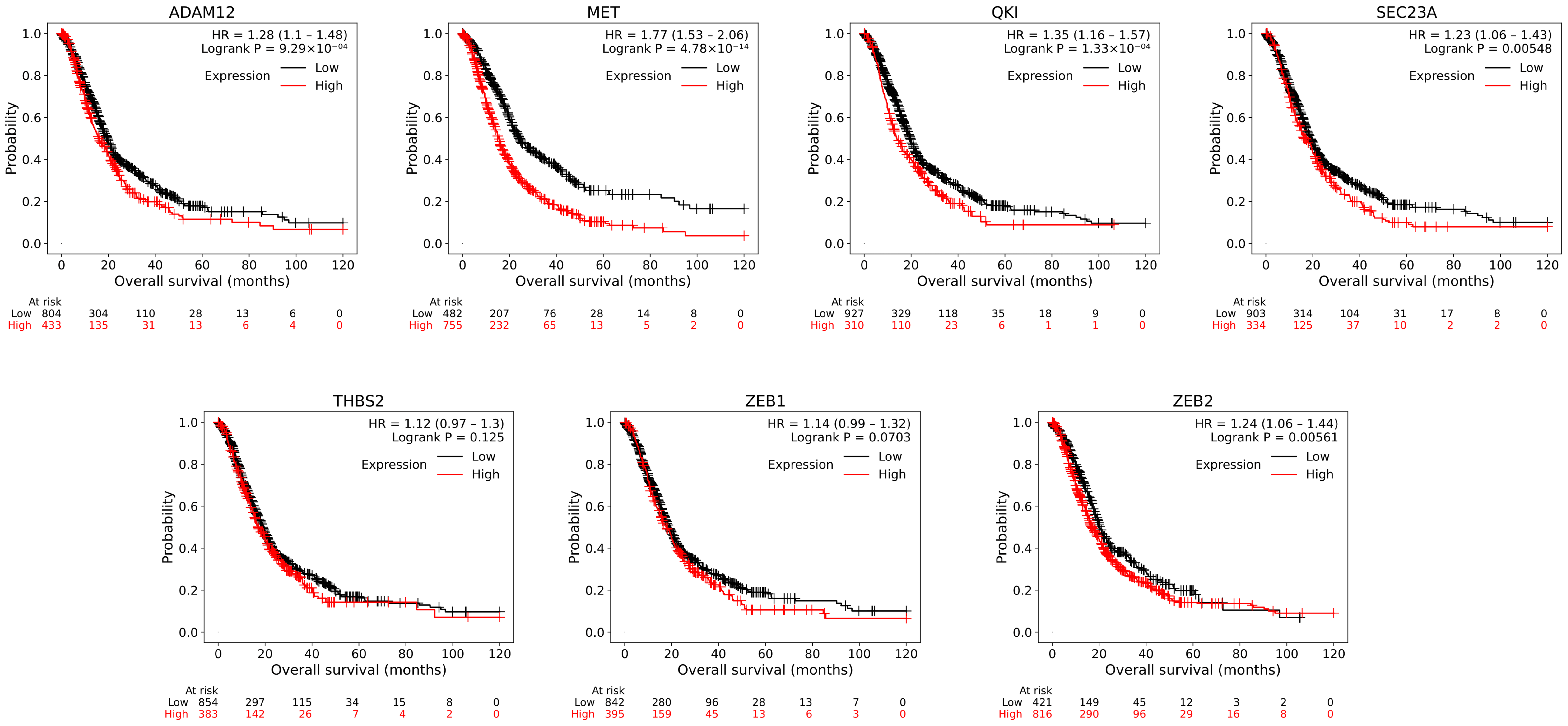
2.5. Survival Analysis
2.6. Determination of Secretion Levels of Hub Genes-Associated Proteins in Different Tissues
2.7. Drug Repositioning
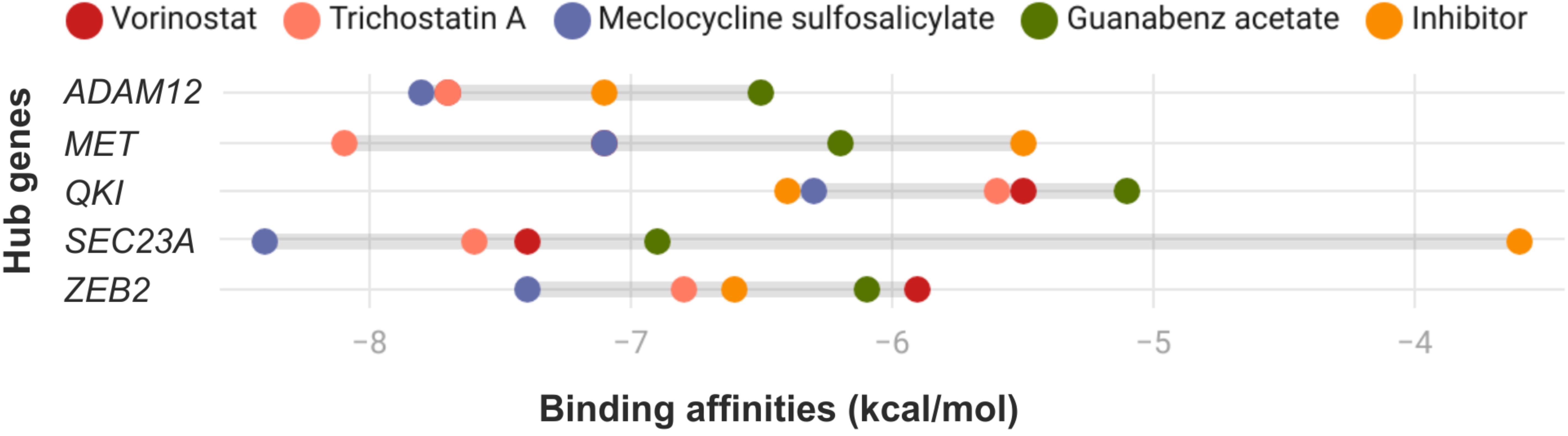
2.8. Molecular Docking
3. Results
3.1. Identification of DEGs, DEcircRNAs, DEmiRNAs, and Enrichments of DEGs
3.2. Visualization of ceRNA Network
3.3. Survival Analysis and Subnetwork Construction of ceRNA
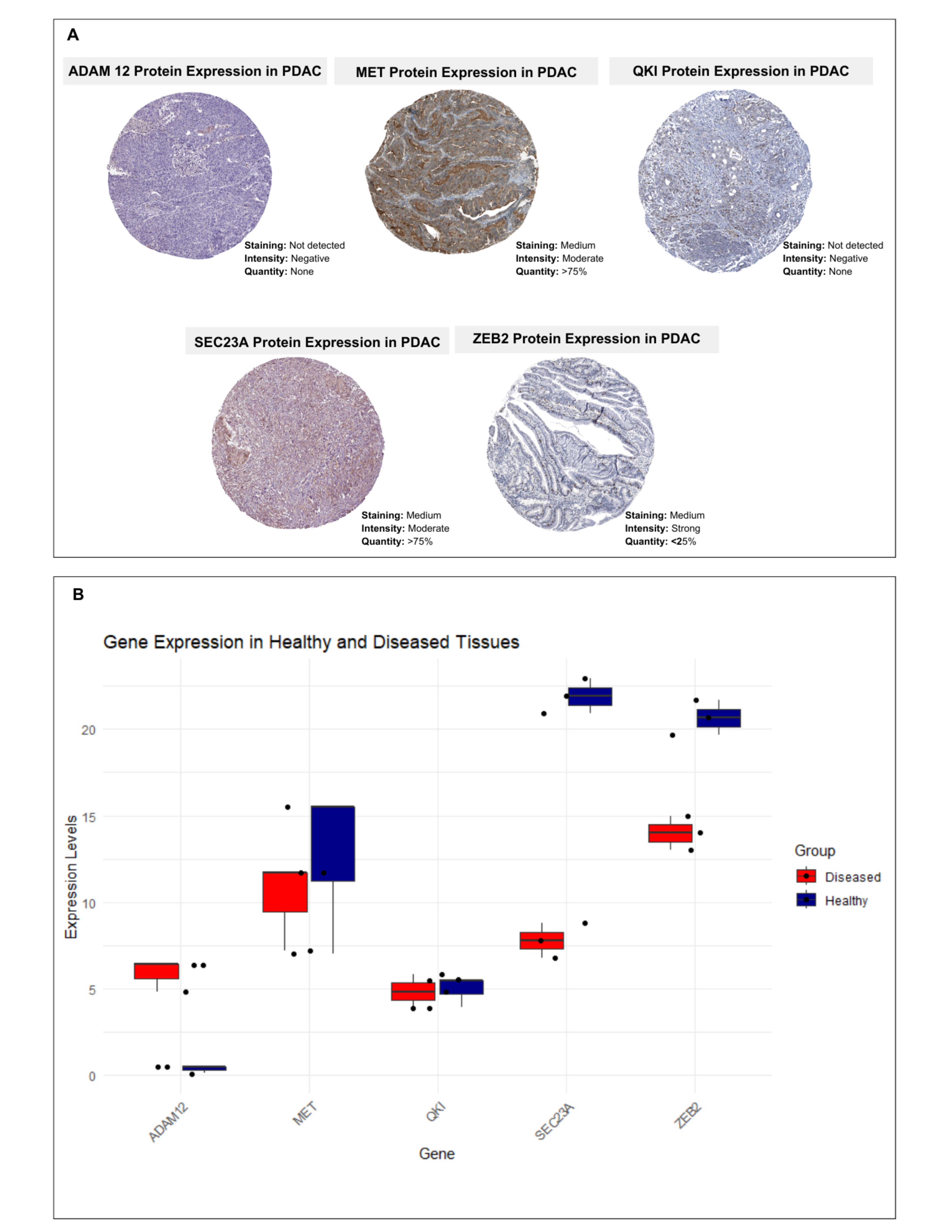
3.4. Secretion Levels of Hub Genes-Associated Proteins in Different Tissues
3.5. Drug Repositioning with ceRNA Subnetwork
3.6. Molecular Docking Analysis with Candidate Repositioned Drugs
3.7. Cross-Validation of Protein and Gene Expressions of Proposed Diseased Signatures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Grossberg, A.J.; Chu, L.C.; Deig, C.R.; Fishman, E.K.; Hwang, W.L.; Maitra, A.; Marks, D.L.; Mehta, A.; Nabavizadeh, N.; Simeone, D.M.; et al. Multidisciplinary Standards of Care and Recent Progress in Pancreatic Ductal Adenocarcinoma. CA Cancer J. Clin. 2020, 70, 375–403. [Google Scholar] [CrossRef]
- Klöppel, G.; Adsay, N.V. Chronic Pancreatitis and the Differential Diagnosis versus Pancreatic Cancer. Arch. Pathol. Lab. Med. 2009, 133, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Nagtegaal, I.D.; Odze, R.D.; Klimstra, D.; Paradis, V.; Rugge, M.; Schirmacher, P.; Washington, K.M.; Carneiro, F.; Cree, I.A. The 2019 WHO Classification of Tumours of the Digestive System. Histopathology 2020, 76, 182–188. [Google Scholar] [CrossRef]
- de Wilde, R.F.; Hruban, R.H.; Maitra, A.; Offerhaus, G.J.A. Reporting Precursors to Invasive Pancreatic Cancer: Pancreatic Intraepithelial Neoplasia, Intraductal Neoplasms and Mucinous Cystic Neoplasm. Diagn. Histopathol. 2012, 18, 17–30. [Google Scholar] [CrossRef]
- Samuel, N.; Hudson, T.J. The Molecular and Cellular Heterogeneity of Pancreatic Ductal Adenocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 2011, 9, 77–87. [Google Scholar] [CrossRef]
- Chen, L.-L.; Yang, L. Regulation of CircRNA Biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef]
- Adelman, K.; Egan, E. Non-Coding RNA: More Uses for Genomic Junk. Nature 2017, 543, 183–185. [Google Scholar] [CrossRef]
- Hashemi, M.; Khosroshahi, E.M.; Daneii, P.; Hassanpoor, A.; Eslami, M.; Koohpar, Z.K.; Asadi, S.; Zabihi, A.; Jamali, B.; Ghorbani, A.; et al. Emerging Roles of CircRNA-MiRNA Networks in Cancer Development and Therapeutic Response. Non-Coding RNA Res. 2025, 10, 98–115. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The Biogenesis, Biology and Characterization of Circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.S.; Kim, S.E.; Oh, S.Y.; Ahn, Y.-H. Clinical Implications of Circulating Circular RNAs in Lung Cancer. Biomedicines 2022, 10, 871. [Google Scholar] [CrossRef]
- Xu, J.-Z.; Shao, C.-C.; Wang, X.-J.; Zhao, X.; Chen, J.-Q.; Ouyang, Y.-X.; Feng, J.; Zhang, F.; Huang, W.-H.; Ying, Q.; et al. circTADA2As Suppress Breast Cancer Progression and Metastasis via Targeting MiR-203a-3p/SOCS3 Axis. Cell Death Dis. 2019, 10, 175. [Google Scholar] [CrossRef]
- Ji, H.; Hu, N. Circular RNA 0001823 Aggravates the Growth and Metastasis of the Cervical Cancer Cells through Modulating the MicroRNA-613/RAB8A Axis. Bioengineered 2022, 13, 10335–10349. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A CeRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Wooten, D.J.; Sinha, I.; Sinha, R. Selenium Induces Pancreatic Cancer Cell Death Alone and in Combination with Gemcitabine. Biomedicines 2022, 10, 149. [Google Scholar] [CrossRef]
- Carbone, D.; Parrino, B.; Cascioferro, S.; Pecoraro, C.; Giovannetti, E.; Di Sarno, V.; Musella, S.; Auriemma, G.; Cirrincione, G.; Diana, P. 1,2,4-Oxadiazole Topsentin Analogs with Antiproliferative Activity against Pancreatic Cancer Cells, Targeting GSK3β Kinase. Chem. Med. Chem. 2021, 16, 537–554. [Google Scholar] [CrossRef]
- Carbone, D.; Pecoraro, C.; Panzeca, G.; Xu, G.; Roeten, M.S.F.; Cascioferro, S.; Giovannetti, E.; Diana, P.; Parrino, B. 1,3,4-Oxadiazole and 1,3,4-Thiadiazole Nortopsentin Derivatives against Pancreatic Ductal Adenocarcinoma: Synthesis, Cytotoxic Activity, and Inhibition of CDK1. Mar. Drugs 2023, 21, 412. [Google Scholar] [CrossRef]
- Clough, E.; Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; et al. NCBI GEO: Archive for Gene Expression and Epigenomics Data Sets: 23-Year Update. Nucleic Acids Res. 2024, 52, D138–D144. [Google Scholar] [CrossRef]
- Badea, L.; Herlea, V.; Dima, S.O.; Dumitrascu, T.; Popescu, I. Combined Gene Expression Analysis of Whole-Tissue and Microdissected Pancreatic Ductal Adenocarcinoma Identifies Genes Specifically Overexpressed in Tumor Epithelia. Hepatogastroenterology 2008, 55, 2016–2027. [Google Scholar]
- Idichi, T.; Seki, N.; Kurahara, H.; Yonemori, K.; Osako, Y.; Arai, T.; Okato, A.; Kita, Y.; Arigami, T.; Mataki, Y.; et al. Regulation of Actin-Binding Protein ANLN by Antitumor MiR-217 Inhibits Cancer Cell Aggressiveness in Pancreatic Ductal Adenocarcinoma. Oncotarget 2017, 8, 53180–53193. [Google Scholar] [CrossRef]
- Frampton, A.E.; Castellano, L.; Colombo, T.; Giovannetti, E.; Krell, J.; Jacob, J.; Pellegrino, L.; Roca-Alonso, L.; Funel, N.; Gall, T.M.H.; et al. MicroRNAs Cooperatively Inhibit a Network of Tumor Suppressor Genes to Promote Pancreatic Tumor Growth and Progression. Gastroenterology 2014, 146, 268–277.e18. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Azevedo-Pouly, A.C.P.; Redis, R.S.; Lee, E.J.; Gusev, Y.; Allard, D.; Sutaria, D.S.; Badawi, M.; Elgamal, O.A.; Lerner, M.R.; et al. Globally Increased Ultraconserved Noncoding RNA Expression in Pancreatic Adenocarcinoma. Oncotarget 2016, 7, 53165–53177. [Google Scholar] [CrossRef] [PubMed]
- Janky, R.; Binda, M.M.; Allemeersch, J.; Van den Broeck, A.; Govaere, O.; Swinnen, J.V.; Roskams, T.; Aerts, S.; Topal, B. Prognostic Relevance of Molecular Subtypes and Master Regulators in Pancreatic Ductal Adenocarcinoma. BMC Cancer 2016, 16, 632. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, V.; Bowitz Lothe, I.M.; Labori, K.J.; Lingjærde, O.C.; Buanes, T.; Dalsgaard, A.M.; Skrede, M.L.; Hamfjord, J.; Haaland, T.; Eide, T.J.; et al. Molecular Signatures of MRNAs and MiRNAs as Prognostic Biomarkers in Pancreatobiliary and Intestinal Types of Periampullary Adenocarcinomas. Mol. Oncol. 2015, 9, 758–771. [Google Scholar] [CrossRef]
- Donahue, T.R.; Tran, L.M.; Hill, R.; Li, Y.; Kovochich, A.; Calvopina, J.H.; Patel, S.G.; Wu, N.; Hindoyan, A.; Farrell, J.J.; et al. Integrative Survival-Based Molecular Profiling of Human Pancreatic Cancer. Clin. Cancer Res. 2012, 18, 1352–1363. [Google Scholar] [CrossRef]
- Toste, P.A.; Li, L.; Kadera, B.E.; Nguyen, A.H.; Tran, L.M.; Wu, N.; Madnick, D.L.; Patel, S.G.; Dawson, D.W.; Donahue, T.R. P85α Is a MicroRNA Target and Affects Chemosensitivity in Pancreatic Cancer. J. Surg. Res. 2015, 196, 285–293. [Google Scholar] [CrossRef]
- Li, H.; Hao, X.; Wang, H.; Liu, Z.; He, Y.; Pu, M.; Zhang, H.; Yu, H.; Duan, J.; Qu, S. Circular RNA Expression Profile of Pancreatic Ductal Adenocarcinoma Revealed by Microarray. Cell. Physiol. Biochem. 2016, 40, 1334–1344. [Google Scholar] [CrossRef]
- Qu, S.; Song, W.; Yang, X.; Wang, J.; Zhang, R.; Zhang, Z.; Zhang, H.; Li, H. Microarray Expression Profile of Circular RNAs in Human Pancreatic Ductal Adenocarcinoma. Genom. Data 2015, 5, 385–387. [Google Scholar] [CrossRef]
- Guo, S.; Xu, X.; Ouyang, Y.; Wang, Y.; Yang, J.; Yin, L.; Ge, J.; Wang, H. Microarray Expression Profile Analysis of Circular RNAs in Pancreatic Cancer. Mol. Med. Rep. 2018, 17, 7661–7671. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An Interactive Venn Diagram Viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Moreno, A.; López-Domínguez, R.; Villatoro-García, J.A.; Ramirez-Mena, A.; Aparicio-Puerta, E.; Hackenberg, M.; Pascual-Montano, A.; Carmona-Saez, P. Functional Enrichment Analysis of Regulatory Elements. Biomedicines 2022, 10, 590. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Q.; Shen, J.; Yang, B.B.; Ding, X. Circbank: A Comprehensive Database for CircRNA with Standard Nomenclature. RNA Biol. 2019, 16, 899–905. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Lin, Y.-C.-D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. MiRTarBase Update 2022: An Informative Resource for Experimentally Validated MiRNA-Target Interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Posta, M.; Győrffy, B. Analysis of a Large Cohort of Pancreatic Cancer Transcriptomic Profiles to Reveal the Strongest Prognostic Factors. Clin. Transl. Sci. 2023, 16, 1479–1491. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Samaras, P.; Schmidt, T.; Frejno, M.; Gessulat, S.; Reinecke, M.; Jarzab, A.; Zecha, J.; Mergner, J.; Giansanti, P.; Ehrlich, H.-C.; et al. ProteomicsDB: A Multi-Omics and Multi-Organism Resource for Life Science Research. Nucleic Acids Res. 2020, 48, D1153–D1163. [Google Scholar] [CrossRef]
- Schaab, C.; Geiger, T.; Stoehr, G.; Cox, J.; Mann, M. Analysis of High Accuracy, Quantitative Proteomics Data in the MaxQB Database. Mol. Cell. Proteom. 2012, 11, M111.014068. [Google Scholar] [CrossRef] [PubMed]
- Montague, E.; Stanberry, L.; Higdon, R.; Janko, I.; Lee, E.; Anderson, N.; Choiniere, J.; Stewart, E.; Yandl, G.; Broomall, W.; et al. MOPED 2.5—An Integrated Multi-Omics Resource: Multi-Omics Profiling Expression Database Now Includes Transcriptomics Data. OMICS J. Integr. Biol. 2014, 18, 335–343. [Google Scholar] [CrossRef]
- Duan, Q.; Reid, S.P.; Clark, N.R.; Wang, Z.; Fernandez, N.F.; Rouillard, A.D.; Readhead, B.; Tritsch, S.R.; Hodos, R.; Hafner, M.; et al. L1000CDS2: LINCS L1000 Characteristic Direction Signatures Search Engine. npj Syst. Biol. Appl. 2016, 2, 16015. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 Update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Wiegers, T.C.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2023. Nucleic Acids Res. 2023, 51, D1257–D1262. [Google Scholar] [CrossRef]
- Knox, C.; Wilson, M.; Klinger, C.M.; Franklin, M.; Oler, E.; Wilson, A.; Pon, A.; Cox, J.; Chin, N.E.L.; Strawbridge, S.A.; et al. DrugBank 6.0: The DrugBank Knowledgebase for 2024. Nucleic Acids Res. 2024, 52, D1265–D1275. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Liu, Y.; Grimm, M.; Dai, W.-T.; Hou, M.-C.; Xiao, Z.-X.; Cao, Y. CB-Dock: A Web Server for Cavity Detection-Guided Protein-Ligand Blind Docking. Acta Pharmacol. Sin. 2020, 41, 138–144. [Google Scholar] [CrossRef]
- Wewer, U.M. Barrett, A.J., Rawlings, N.D., Woessner, J.F., Eds.; 212—ADAM12. In Handbook of Proteolytic Enzymes; Academic Press: London, UK, 2004; pp. 724–726. [Google Scholar] [CrossRef]
- Mendaza, S.; Ulazia-Garmendia, A.; Monreal-Santesteban, I.; Córdoba, A.; de Azúa, Y.R.; Aguiar, B.; Beloqui, R.; Armendáriz, P.; Arriola, M.; Martín-Sánchez, E.; et al. ADAM12 Is A Potential Therapeutic Target Regulated by Hypomethylation in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2020, 21, 903. [Google Scholar] [CrossRef]
- Christiansen, M.; Spencer, K.; Laigaard, J.; Cowans, N.J.; Larsen, S.O.; Wewer, U.M. ADAM 12 as a Second-Trimester Maternal Serum Marker in Screening for Down Syndrome. Prenat. Diagn. 2007, 27, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Nyren-Erickson, E.K.; Jones, J.M.; Srivastava, D.K.; Mallik, S. A Disintegrin and Metalloproteinase-12 (ADAM12): Function, Roles in Disease Progression, and Clinical Implications. Biochim. Biophys. Acta 2013, 1830, 4445–4455. [Google Scholar] [CrossRef] [PubMed]
- Malik, R.; Mambetsariev, I.; Fricke, J.; Chawla, N.; Nam, A.; Pharaon, R.; Salgia, R. Kumar, R., Fisher, P.B., Eds.; Chapter Seven—MET Receptor in Oncology: From Biomarker to Therapeutic Target. In Advances in Cancer Research; Academic Press: London, UK, 2020; Volume 147, pp. 259–301. [Google Scholar] [CrossRef]
- Furlan, A.; Kherrouche, Z.; Montagne, R.; Copin, M.-C.; Tulasne, D. Thirty Years of Research on Met Receptor to Move a Biomarker from Bench to Bedside. Cancer Res. 2014, 74, 6737–6744. [Google Scholar] [CrossRef]
- Benvenuti, S.; Comoglio, P.M. The MET Receptor Tyrosine Kinase in Invasion and Metastasis. J. Cell. Physiol. 2007, 213, 316–325. [Google Scholar] [CrossRef]
- Neumann, D.P.; Goodall, G.J.; Gregory, P.A. The Quaking RNA-Binding Proteins as Regulators of Cell Differentiation. WIREs RNA 2022, 13, e1724. [Google Scholar] [CrossRef]
- Darbelli, L.; Richard, S. Emerging Functions of the Quaking RNA-Binding Proteins and Link to Human Diseases. WIREs RNA 2016, 7, 399–412. [Google Scholar] [CrossRef]
- Lang, M.R.; Lapierre, L.A.; Frotscher, M.; Goldenring, J.R.; Knapik, E.W. Secretory COPII Coat Component Sec23a Is Essential for Craniofacial Chondrocyte Maturation. Nat. Genet. 2006, 38, 1198–1203. [Google Scholar] [CrossRef]
- Boyadjiev, S.A.; Fromme, J.C.; Ben, J.; Chong, S.S.; Nauta, C.; Hur, D.J.; Zhang, G.; Hamamoto, S.; Schekman, R.; Ravazzola, M.; et al. Cranio-Lenticulo-Sutural Dysplasia Is Caused by a SEC23A Mutation Leading to Abnormal Endoplasmic-Reticulum-to-Golgi Trafficking. Nat. Genet. 2006, 38, 1192–1197. [Google Scholar] [CrossRef]
- Grabitz, A.L.; Duncan, M.K. Focus on Molecules: Smad Interacting Protein 1 (Sip1, ZEB2, ZFHX1B). Exp. Eye Res. 2012, 101, 105–106. [Google Scholar] [CrossRef]
- Hegarty, S.V.; Sullivan, A.M.; O’Keeffe, G.W. Zeb2: A Multifunctional Regulator of Nervous System Development. Prog. Neurobiol. 2015, 132, 81–95. [Google Scholar] [CrossRef]
- Cui, J.; Pan, G.; He, Q.; Yin, L.; Guo, R.; Bi, H. MicroRNA-545 Targets ZEB2 to Inhibit the Development of Non-small Cell Lung Cancer by Inactivating Wnt/β-Catenin Pathway. Oncol. Lett. 2019, 18, 2931–2938. [Google Scholar] [CrossRef] [PubMed]
- Mann, B.S.; Johnson, J.R.; Cohen, M.H.; Justice, R.; Pazdur, R. FDA Approval Summary: Vorinostat for Treatment of Advanced Primary Cutaneous T-Cell Lymphoma. Oncologist 2007, 12, 1247–1252. [Google Scholar] [CrossRef] [PubMed]
- Kozeretska, I.A.; Serga, S.V.; Koliada, A.K.; Vaiserman, A.M. Verlinden, H., Ed.; Chapter Four—Epigenetic Regulation of Longevity in Insects. In Advances in Insect Physiology; Academic Press: London, UK, 2017; Volume 53, pp. 87–114. [Google Scholar] [CrossRef]
- Cacabelos, R.; Torrellas, C. Tollefsbol, T.O., Ed.; Chapter 32—Pharmacoepigenomics. In Medical Epigenetics; Academic Press: Boston, MA, USA, 2016; pp. 585–617. [Google Scholar] [CrossRef]
- Gelmetti, C. Local Antibiotics in Dermatology. Dermatol. Ther. 2008, 21, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Holmes, B.; Brogden, R.N.; Heel, R.C.; Speight, T.M.; Avery, G.S. Guanabenz: A Review of its Pharmacodynamic Properties and Therapeutic Efficacy in Hypertension. Drugs 1983, 26, 212–229. [Google Scholar] [CrossRef]
- Rishi, A.; Goggins, M.; Wood, L.D.; Hruban, R.H. Pathological and Molecular Evaluation of Pancreatic Neoplasms. Semin. Oncol. 2015, 42, 28–39. [Google Scholar] [CrossRef]
- Ferlay, J.; Partensky, C.; Bray, F. More Deaths from Pancreatic Cancer than Breast Cancer in the EU by 2017. Acta Oncol. 2016, 55, 1158–1160. [Google Scholar] [CrossRef]
- Rebelo, R.; Polónia, B.; Santos, L.L.; Vasconcelos, M.H.; Xavier, C.P.R. Drug Repurposing Opportunities in Pancreatic Ductal Adenocarcinoma. Pharmaceuticals 2021, 14, 280. [Google Scholar] [CrossRef]
- Aydin, B.; Arga, K.Y.; Karadag, A.S. Omics-Driven Biomarkers of Psoriasis: Recent Insights, Current Challenges, and Future Prospects. Clin. Cosmet. Investig. Dermatol. 2020, 13, 611–625. [Google Scholar] [CrossRef]
- Beklen, H.; Gulfidan, G.; Arga, K.Y.; Mardinoglu, A.; Turanli, B. Drug Repositioning for P-Glycoprotein Mediated Co-Expression Networks in Colorectal Cancer. Front. Oncol. 2020, 10, 1273. [Google Scholar] [CrossRef]
- Turanli, B.; Yildirim, E.; Gulfidan, G.; Arga, K.Y.; Sinha, R. Current State of “Omics” Biomarkers in Pancreatic Cancer. J. Pers. Med. 2021, 11, 127. [Google Scholar] [CrossRef]
- Jiang, F.; Shen, X. Current Prevalence Status of Gastric Cancer and Recent Studies on the Roles of Circular RNAs and Methods Used to Investigate Circular RNAs. Cell. Mol. Biol. Lett. 2019, 24, 53. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, Z.; Hu, G.; Jiang, Y. Roles of Circular RNA in Breast Cancer: Present and Future. Am. J. Transl. Res. 2019, 11, 3945–3954. [Google Scholar]
- Chen, J.; Chen, T.; Zhu, Y.; Li, Y.; Zhang, Y.; Wang, Y.; Li, X.; Xie, X.; Wang, J.; Huang, M.; et al. CircPTN Sponges MiR-145-5p/MiR-330-5p to Promote Proliferation and Stemness in Glioma. J. Exp. Clin. Cancer Res. 2019, 38, 398. [Google Scholar] [CrossRef]
- Ren, S.; Liu, J.; Feng, Y.; Li, Z.; He, L.; Li, L.; Cao, X.; Wang, Z.; Zhang, Y. Knockdown of CircDENND4C Inhibits Glycolysis, Migration and Invasion by up-Regulating MiR-200b/c in Breast Cancer under Hypoxia. J. Exp. Clin. Cancer Res. 2019, 38, 388. [Google Scholar] [CrossRef]
- Gong, Y.; Jiao, Y.; Qi, X.; Fu, J.; Qian, J.; Zhu, J.; Yang, H.; Tang, L. Construction of a CircRNA-MiRNA-MRNA Network Based on Differentially Co-Expressed Circular RNA in Gastric Cancer Tissue and Plasma by Bioinformatics Analysis. World J. Surg. Oncol. 2022, 20, 34. [Google Scholar] [CrossRef]
- Jin, J.; Xu, B.; Hu, Z.; He, X. Circular RNA CircTADA2A Promotes the Proliferation, Invasion, and Migration of Non-Small Cell Lung Cancer Cells via the MiR-450b-3p/HMGN5 Signaling Pathway. Transl. Cancer Res. 2022, 11, 242–251. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, L.; Hou, J.; Zhong, S.; Zhou, S.; Zhu, L.; Li, J.; Wang, D.; Sun, D.; Ji, Z.; et al. Circular RNA Hsa_circ_0052112 Promotes Cell Migration and Invasion by Acting as Sponge for MiR-125a-5p in Breast Cancer. Biomed. Pharmacother. 2018, 107, 1342–1353. [Google Scholar] [CrossRef]
- Jing, X.F.; Ren, M.Y.; Fan, Y.S.; Fu, Y.H.; Wang, C.F. Circular RNA_0001073 (Circ_0001073) Suppresses The Progression of Non-Small Cell Lung Cancer via MiR-582-3p/RGMB Axis. Cell J. 2021, 23, 684–691. [Google Scholar] [CrossRef] [PubMed]
- Kurahara, H.; Takao, S.; Maemura, K.; Mataki, Y.; Kuwahata, T.; Maeda, K.; Sakoda, M.; Iino, S.; Ueno, S.; Natsugoe, S. Epithelial–Mesenchymal Transition and Mesenchymal–Epithelial Transition via Regulation of ZEB-1 and ZEB-2 Expression in Pancreatic Cancer. J. Surg. Oncol. 2012, 105, 655–661. [Google Scholar] [CrossRef]
- Krebs, A.M.; Mitschke, J.; Lasierra Losada, M.; Schmalhofer, O.; Boerries, M.; Busch, H.; Boettcher, M.; Mougiakakos, D.; Reichardt, W.; Bronsert, P.; et al. The EMT-Activator Zeb1 Is a Key Factor for Cell Plasticity and Promotes Metastasis in Pancreatic Cancer. Nat. Cell Biol. 2017, 19, 518–529. [Google Scholar] [CrossRef]
- Yang, Y.; Ishak Gabra, M.B.; Hanse, E.A.; Lowman, X.H.; Tran, T.Q.; Li, H.; Milman, N.; Liu, J.; Reid, M.A.; Locasale, J.W.; et al. MiR-135 Suppresses Glycolysis and Promotes Pancreatic Cancer Cell Adaptation to Metabolic Stress by Targeting Phosphofructokinase-1. Nat. Commun. 2019, 10, 809. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Jiang, T.; Yong, J.; Peng, J.; Dong, S.; Gu, Y.; Ji, X.; Luo, L.; Chang, W.-L. MiR-135b-5p Targets ADAM12 to Suppress Invasion and Accelerate Trophoblast Apoptosis in Preeclampsia. Placenta 2023, 143, 69–79. [Google Scholar] [CrossRef]
- Lulli, V.; Buccarelli, M.; Martini, M.; Signore, M.; Biffoni, M.; Giannetti, S.; Morgante, L.; Marziali, G.; Ilari, R.; Pagliuca, A.; et al. MiR-135b Suppresses Tumorigenesis in Glioblastoma Stem-like Cells Impairing Proliferation, Migration and Self-Renewal. Oncotarget 2015, 6, 37241–37256. [Google Scholar] [CrossRef]
- Kapplingattu, S.V.; Bhattacharya, S.; Adlakha, Y.K. MiRNAs as Major Players in Brain Health and Disease: Current Knowledge and Future Perspectives. Cell Death Discov. 2025, 11, 7. [Google Scholar] [CrossRef]
- Katoch, A.; Jamwal, V.L.; Faheem, M.M.; Kumar, S.; Senapati, S.; Yadav, G.; Gandhi, S.G.; Goswami, A. Overlapping Targets Exist between the Par-4 and MiR-200c Axis Which Regulate EMT and Proliferation of Pancreatic Cancer Cells. Transl. Oncol. 2021, 14, 100879. [Google Scholar] [CrossRef]
- Yu, J.; Ohuchida, K.; Mizumoto, K.; Sato, N.; Kayashima, T.; Fujita, H.; Nakata, K.; Tanaka, M. MicroRNA, Hsa-MiR-200c, Is an Independent Prognostic Factor in Pancreatic Cancer and Its Upregulation Inhibits Pancreatic Cancer Invasion but Increases Cell Proliferation. Mol. Cancer 2010, 9, 169. [Google Scholar] [CrossRef]
- Ahmad, A.; Aboukameel, A.; Kong, D.; Wang, Z.; Sethi, S.; Chen, W.; Sarkar, F.H.; Raz, A. Phosphoglucose Isomerase/Autocrine Motility Factor Mediates Epithelial-Mesenchymal Transition Regulated by MiR-200 in Breast Cancer Cells. Cancer Res. 2011, 71, 3400–3409. [Google Scholar] [CrossRef]
- Hurteau, G.J.; Spivack, S.D.; Brock, G.J. Potential MRNA Degradation Targets of Hsa-MiR-200c, Identified Using Informatics and QRT-PCR. Cell Cycle 2006, 5, 1951–1956. [Google Scholar] [CrossRef]
- Korpal, M.; Lee, E.S.; Hu, G.; Kang, Y. The MiR-200 Family Inhibits Epithelial-Mesenchymal Transition and Cancer Cell Migration by Direct Targeting of E-Cadherin Transcriptional Repressors ZEB1 and ZEB2. J. Biol. Chem. 2008, 283, 14910–14914. [Google Scholar] [CrossRef]
- Sundararajan, V.; Gengenbacher, N.; Stemmler, M.P.; Kleemann, J.A.; Brabletz, T.; Brabletz, S. The ZEB1/MiR-200c Feedback Loop Regulates Invasion via Actin Interacting Proteins MYLK and TKS5. Oncotarget 2015, 6, 27083–27096. [Google Scholar] [CrossRef]
- Title, A.C.; Hong, S.-J.; Pires, N.D.; Hasenöhrl, L.; Godbersen, S.; Stokar-Regenscheit, N.; Bartel, D.P.; Stoffel, M. Genetic Dissection of the MiR-200–Zeb1 Axis Reveals Its Importance in Tumor Differentiation and Invasion. Nat. Commun. 2018, 9, 4671. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, U.J.; Kim, M.N.; Lee, E.-J.; Kim, J.Y.; Lee, M.Y.; Choung, S.; Kim, Y.J.; Choi, Y.-C. MicroRNA MiR-199a* Regulates the MET Proto-Oncogene and the Downstream Extracellular Signal-Regulated Kinase 2 (ERK2). J. Biol. Chem. 2008, 283, 18158–18166. [Google Scholar] [CrossRef] [PubMed]
- Würdinger, T.; Tannous, B.A.; Saydam, O.; Skog, J.; Grau, S.; Soutschek, J.; Weissleder, R.; Breakefield, X.O.; Krichevsky, A.M. MiR-296 Regulates Growth Factor Receptor Overexpression in Angiogenic Endothelial Cells. Cancer Cell. 2008, 14, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ma, J.; Si, Y.; Cheng, S.; Hu, M.; Zhi, X.; Li, B.; Yu, H.; Jiang, W.G. Differential Expression and Functions of Ehm2 Transcript Variants in Lung Adenocarcinoma. Int. J. Oncol. 2019, 54, 1747–1758. [Google Scholar] [CrossRef]
- Shao, Y.; Zhang, S.; Pan, Y.; Peng, Z.; Dong, Y. miR-135b: A Key Role in Cancer Biology and Therapeutic Targets. Non-Coding RNA Res. 2025, 12, 67–80. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, J.; Tao, J.; Zhu, J.; Zheng, R.; Wang, H. Emerging Roles of Circular RNAs in Cancer: A Narrative Review. J. Pancreatol. 2022, 5, 41–48. [Google Scholar] [CrossRef]
- Hill, K.S.; Lorinczi, M.; Elferink, L.A. Altered down Regulation of the Receptor Tyrosine Kinase Met in Pancreatic Adenocarcinoma Cells. J. Exp. Ther. Oncol. 2010, 8, 297–312. [Google Scholar]
- Ruta, V.; Naro, C.; Pieraccioli, M.; Leccese, A.; Archibugi, L.; Cesari, E.; Panzeri, V.; Allgöwer, C.; Arcidiacono, P.G.; Falconi, M.; et al. An Alternative Splicing Signature Defines the Basal-like Phenotype and Predicts Worse Clinical Outcome in Pancreatic Cancer. Cell Rep. Med. 2024, 5, 101411. [Google Scholar] [CrossRef]
- Zeng, B.; Zhao, Q.; Sun, Z.; Liu, D.; Chen, H.; Li, X.; Wang, J.; Xing, H.R. SEC23A Is an Independent Prognostic Biomarker in Bladder Cancer Correlated With MAPK Signaling. Front. Genet. 2021, 12, 672832. [Google Scholar] [CrossRef]
- Veenstra, V.L.; Damhofer, H.; Waasdorp, C.; van Rijssen, L.B.; van de Vijver, M.J.; Dijk, F.; Wilmink, H.W.; Besselink, M.G.; Busch, O.R.; Chang, D.K.; et al. ADAM12 Is a Circulating Marker for Stromal Activation in Pancreatic Cancer and Predicts Response to Chemotherapy. Oncogenesis 2018, 7, 87. [Google Scholar] [CrossRef]
- Gimotty, P.A.; Till, J.E.; Udgata, S.; Takenaka, N.; Yee, S.S.; LaRiviere, M.J.; O’Hara, M.H.; Reiss, K.A.; O’Dwyer, P.; Katona, B.W.; et al. THSB2 as a Prognostic Biomarker for Patients Diagnosed with Metastatic Panreatic Ductal Adenocarcinoma. Oncotarget 2021, 12, 2266–2272. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Yoshida, M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef] [PubMed]
- Bubna, A.K. Vorinostat-An Overview. Indian J. Dermatol. 2015, 60, 419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Adachi, M.; Kawamura, R.; Imai, K. Bmf Is a Possible Mediator in Histone Deacetylase Inhibitors FK228 and CBHA-Induced Apoptosis. Cell Death Differ. 2006, 13, 129–140. [Google Scholar] [CrossRef]
- Zhao, C.; Dong, H.; Xu, Q.; Zhang, Y. Histone Deacetylase (HDAC) Inhibitors in Cancer: A Patent Review (2017–Present). Expert Opin. Ther. Pat. 2020, 30, 263–274. [Google Scholar] [CrossRef]
- Bali, P.; Pranpat, M.; Bradner, J.; Balasis, M.; Fiskus, W.; Guo, F.; Rocha, K.; Kumaraswamy, S.; Boyapalle, S.; Atadja, P.; et al. Inhibition of Histone Deacetylase 6 Acetylates and Disrupts the Chaperone Function of Heat Shock Protein 90: A Novel Basis for Antileukemia Activity of Histone Deacetylase Inhibitors. J. Biol. Chem. 2005, 280, 26729–26734. [Google Scholar] [CrossRef]
- Richon, V.M.; Sandhoff, T.W.; Rifkind, R.A.; Marks, P.A. Histone Deacetylase Inhibitor Selectively Induces P21WAF1 Expression and Gene-Associated Histone Acetylation. Proc. Natl. Acad. Sci. USA 2000, 97, 10014–10019. [Google Scholar] [CrossRef]
- Rikiishi, H. Autophagic and Apoptotic Effects of HDAC Inhibitors on Cancer Cells. J. Biomed. Biotechnol. 2011, 2011, 830260. [Google Scholar] [CrossRef]
- Herrera-Martínez, M.; Orozco-Samperio, E.; Montaño, S.; Ariza-Ortega, J.A.; Flores-García, Y.; López-Contreras, L. Vorinostat as Potential Antiparasitic Drug. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7412–7419. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.; Raslan, S.; Tombuloglu, H.; Shamseddin, A.; Cevik, E.; Said, O.A.; Madyan, E.F.; Senel, M.; Bozkurt, A.; Rehman, S.; et al. Vorinostat-Loaded Titanium Oxide Nanoparticles (Anatase) Induce G2/M Cell Cycle Arrest in Breast Cancer Cells via PALB2 Upregulation. 3 Biotech 2020, 10, 407. [Google Scholar] [CrossRef]
- Modesitt, S.C.; Sill, M.; Hoffman, J.S.; Bender, D.P. A Phase II Study of Vorinostat in the Treatment of Persistent or Recurrent Epithelial Ovarian or Primary Peritoneal Carcinoma: A Gynecologic Oncology Group Study. Gynecol. Oncol. 2008, 109, 182–186. [Google Scholar] [CrossRef] [PubMed]
- You, B.R.; Park, W.H. Suberoylanilide Hydroxamic Acid Induces Thioredoxin1-Mediated Apoptosis in Lung Cancer Cells via up-Regulation of MiR-129-5p. Mol. Carcinog. 2017, 56, 2566–2577. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Ida, H.; Ito, K.; Zhang, H.; Ito, Y. Contribution of Reactivated RUNX3 to Inhibition of Gastric Cancer Cell Growth Following Suberoylanilide Hydroxamic Acid (Vorinostat) Treatment. Biochem. Pharmacol. 2007, 73, 990–1000. [Google Scholar] [CrossRef] [PubMed]
- Beljanski, V. Enna, S.J., Bylund, D.B., Eds.; Trichostatin A. In xPharm: The Comprehensive Pharmacology Reference; Enna, S.J., Bylund, D.B., Eds.; Elsevier: New York, NY, USA. In xPharm: The Comprehensive Pharmacology Reference; Elsevier: New York, NY, USA, 2009; pp. 1–4. [Google Scholar] [CrossRef]
- Vigushin, D.M.; Ali, S.; Pace, P.E.; Mirsaidi, N.; Ito, K.; Adcock, I.; Coombes, R.C. Trichostatin A Is a Histone Deacetylase Inhibitor with Potent Antitumor Activity against Breast Cancer in Vivo1. Clin. Cancer Res. 2001, 7, 971–976. [Google Scholar]
- Platta, C.S.; Greenblatt, D.Y.; Kunnimalaiyaan, M.; Chen, H. The HDAC Inhibitor Trichostatin A Inhibits Growth of Small Cell Lung Cancer Cells. J. Surg. Res. 2007, 142, 219–226. [Google Scholar] [CrossRef]
- Donadelli, M.; Costanzo, C.; Faggioli, L.; Scupoli, M.T.; Moore, P.S.; Bassi, C.; Scarpa, A.; Palmieri, M. Trichostatin A, an Inhibitor of Histone Deacetylases, Strongly Suppresses Growth of Pancreatic Adenocarcinoma Cells. Mol. Carcinog. 2003, 38, 59–69. [Google Scholar] [CrossRef]
- Knutson, D.D.; Swinyer, L.J.; Smoot, W.H. Meclocycline Sulfosalicylate. Topical Antibiotic Agent for the Treatment of Acne Vulgaris. Cutis 1981, 27, 203–204. [Google Scholar]
- Shutter, M.C.; Akhondi, H. Tetracycline. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2024. [Google Scholar]
- Tribouillard-Tanvier, D.; Béringue, V.; Desban, N.; Gug, F.; Bach, S.; Voisset, C.; Galons, H.; Laude, H.; Vilette, D.; Blondel, M. Antihypertensive Drug Guanabenz Is Active in Vivo against Both Yeast and Mammalian Prions. PLoS ONE 2008, 3, e1981. [Google Scholar] [CrossRef]
- Kang, H.J.; Seol, H.S.; Lee, S.E.; Suh, Y.-A.; Kim, J.; Jang, S.J.; Yu, E. Guanabenz Acetate Induces Endoplasmic Reticulum Stress–Related Cell Death in Hepatocellular Carcinoma Cells. J. Pathol. Transl. Med. 2019, 53, 94–103. [Google Scholar] [CrossRef]
- Hamamura, K.; Minami, K.; Tanjung, N.; Wan, Q.; Koizumi, M.; Matsuura, N.; Na, S.; Yokota, H. Attenuation of Malignant Phenotypes of Breast Cancer Cells through EIF2α-Mediated Downregulation of Rac1 Signaling. Int. J. Oncol. 2014, 44, 1980–1988. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Zhao, Y.; Tian, W.; Zhai, L.; Pang, H.; Kang, J.; Hou, H.; Chen, Y.; Li, D. Rhein Derivative 4F Inhibits the Malignant Phenotype of Breast Cancer by Downregulating Rac1 Protein. Front. Pharmacol. 2020, 11, 754. [Google Scholar] [CrossRef] [PubMed]
- Birchmeier, C.; Birchmeier, W.; Gherardi, E.; Vande Woude, G.F. Met, Metastasis, Motility and More. Nat. Rev. Mol. Cell Biol. 2003, 4, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Lamszus, K.; Laterra, J.; Westphal, M.; Rosen, E.M. Scatter Factor/Hepatocyte Growth Factor (SF/HGF) Content and Function in Human Gliomas. Int. J. Dev. Neurosci. 1999, 17, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Trusolino, L.; Bertotti, A.; Comoglio, P.M. MET Signalling: Principles and Functions in Development, Organ Regeneration and Cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 834–848. [Google Scholar] [CrossRef]
- Sato, K.; Nakano, A. Mechanisms of COPII Vesicle Formation and Protein Sorting. FEBS Lett. 2007, 581, 2076–2082. [Google Scholar] [CrossRef]
- Boyadjiev, S.A.; Kim, S.-D.; Hata, A.; Haldeman-Englert, C.; Zackai, E.H.; Naydenov, C.; Hamamoto, S.; Schekman, R.W.; Kim, J. Cranio-Lenticulo-Sutural Dysplasia Associated with Defects in Collagen Secretion. Clin. Genet. 2011, 80, 169–176. [Google Scholar] [CrossRef]
- Zhu, M.; Tao, J.; Vasievich, M.P.; Wei, W.; Zhu, G.; Khoriaty, R.N.; Zhang, B. Neural Tube Opening and Abnormal Extraembryonic Membrane Development in SEC23A Deficient Mice. Sci. Rep. 2015, 5, 15471. [Google Scholar] [CrossRef]


| Detected RNA Types | GEO Reference Series | Platform | Samples | Study Design | Ref. |
|---|---|---|---|---|---|
| Messenger RNA | GSE15471 | Affymetrix Human Genome U133 Plus 2.0 Array | 78 | 39 N/ 39 PDAC | [20,21] |
| GSE41368 | Affymetrix Human Gene 1.0 ST Array | 12 | 6 N/ 6 PDAC | [22] | |
| GSE71989 | Affymetrix Human Genome U133 Plus 2.0 Array | 22 | 8 N/ 14 PDAC | [23] | |
| GSE62165 | Affymetrix Human Genome U219 Array | 131 | 13 N/ 118 PDAC | [24] | |
| MicroRNA | GSE60978 | Agilent-031181 Unrestricted Human miRNA V16.0 Microarray 030840 | 57 | 6 N/ 51 PDAC | [25] |
| GSE32678 | miRCURY LNA microRNA Array, v.11.0—hsa, mmu, and rno | 32 | 7 N/ 25 PDAC | [26,27] | |
| GSE41369 | NanoString nCounter Human miRNA assay V1 | 18 | 9 N/ 9 PDAC | [22] | |
| Circular RNA | GSE69362 | Agilent-069978 Arraystar Human CircRNA microarray V1 | 12 | 6 N/ 6 PDAC | [28,29] |
| GSE79634 | Agilent-069978 Arraystar Human CircRNA microarray V1 | 40 | 20 N/ 20 PDAC | [30] |
| Hub Gene | Name | Function | Uniprot ID | Inhibitor | PubChem ID | Ref. |
|---|---|---|---|---|---|---|
| ADAM12 | Disintegrin and metalloproteinase domain-containing protein 12 | Functions in the development of preimplantation embryos | O43184 | Abrine | 160511 | [50,51,52,53] |
| MET | MET Proto-Oncogene, Receptor Tyrosine Kinase | Affects angiogenesis, wound healing, morphogenesis, cell proliferation, cancer invasion, and survival | P08581 | Acetaminophen | 1983 | [54,55,56] |
| QKI | Quaking Homolog, KH Domain Containing RNA Binding | Controls mRNA stability, translation, export of mRNAs from the nucleus, and pre-mRNA splicing | Q96PU8 | Aristolochic acid I | 2236 | [57,58] |
| SEC23A | SEC23 Homolog A, COPII Coat Complex Component | Serving as a GTPase-activating protein | Q15436 | 1,2-Dimethylhydrazine | 1322 | [59,60] |
| ZEB2 | Zinc Finger E-Box-Binding Homeobox 2 | Activating Wnt/β-catenin signaling | O60315 | Antimycin A | 14957 | [61,62,63] |
| Drug | Drug Class | Mechanism of Action | Approval Status | Trial Number/Trial Status | Ref. |
|---|---|---|---|---|---|
| Vorinostat | Histone deacetylase (HDAC) inhibitor | Inhibits the enzymatic activity of HDAC | Approved | PAAD/NCT00948688/Phase I/II | [64] |
| PA/NCT00983268/Phase I/II (Terminated) | |||||
| PAAD/NCT02349867/Phase I | |||||
| Trichostatin A | Antifungal agent | Induces terminal differentiation, cell cycle arrest, and apoptosis in various cancer cell lines | Investigational | HM/NCT03838926/Phase I | [65,66] |
| Meclocycline sulfosalicylate | Antimicrobial agent | Inhibits protein synthesis in bacteria by binding to the 30S ribosomal subunit | Approved but discontinued | OM/NCT00385515/Phase II | [67] |
| Guanabenz acetate | Antihypertensive agent | Stimulates central alpha-2 adrenergic receptors, which reduces the release of norepinephrine | Investigational | MS/NCT02423083 /Phase I | [68] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aydin, B.; Okutan, K.; Aydogan, O.O.; Sinha, R.; Turanli, B. The Construction of ceRNA Regulatory Network Unraveled Prognostic Biomarkers and Repositioned Drug Candidates for the Management of Pancreatic Ductal Adenocarcinoma. Curr. Issues Mol. Biol. 2025, 47, 496. https://doi.org/10.3390/cimb47070496
Aydin B, Okutan K, Aydogan OO, Sinha R, Turanli B. The Construction of ceRNA Regulatory Network Unraveled Prognostic Biomarkers and Repositioned Drug Candidates for the Management of Pancreatic Ductal Adenocarcinoma. Current Issues in Molecular Biology. 2025; 47(7):496. https://doi.org/10.3390/cimb47070496
Chicago/Turabian StyleAydin, Busra, Keziban Okutan, Ozge Onluturk Aydogan, Raghu Sinha, and Beste Turanli. 2025. "The Construction of ceRNA Regulatory Network Unraveled Prognostic Biomarkers and Repositioned Drug Candidates for the Management of Pancreatic Ductal Adenocarcinoma" Current Issues in Molecular Biology 47, no. 7: 496. https://doi.org/10.3390/cimb47070496
APA StyleAydin, B., Okutan, K., Aydogan, O. O., Sinha, R., & Turanli, B. (2025). The Construction of ceRNA Regulatory Network Unraveled Prognostic Biomarkers and Repositioned Drug Candidates for the Management of Pancreatic Ductal Adenocarcinoma. Current Issues in Molecular Biology, 47(7), 496. https://doi.org/10.3390/cimb47070496







