Artificial Intelligence Approach in Machine Learning-Based Modeling and Networking of the Coronavirus Pathogenesis Pathway †
Abstract
1. Introduction
2. Materials and Methods
2.1. Coronavirus Pathogenesis Pathway and the Activation Z-Score
2.2. Network Analysis
2.3. Analysis Match
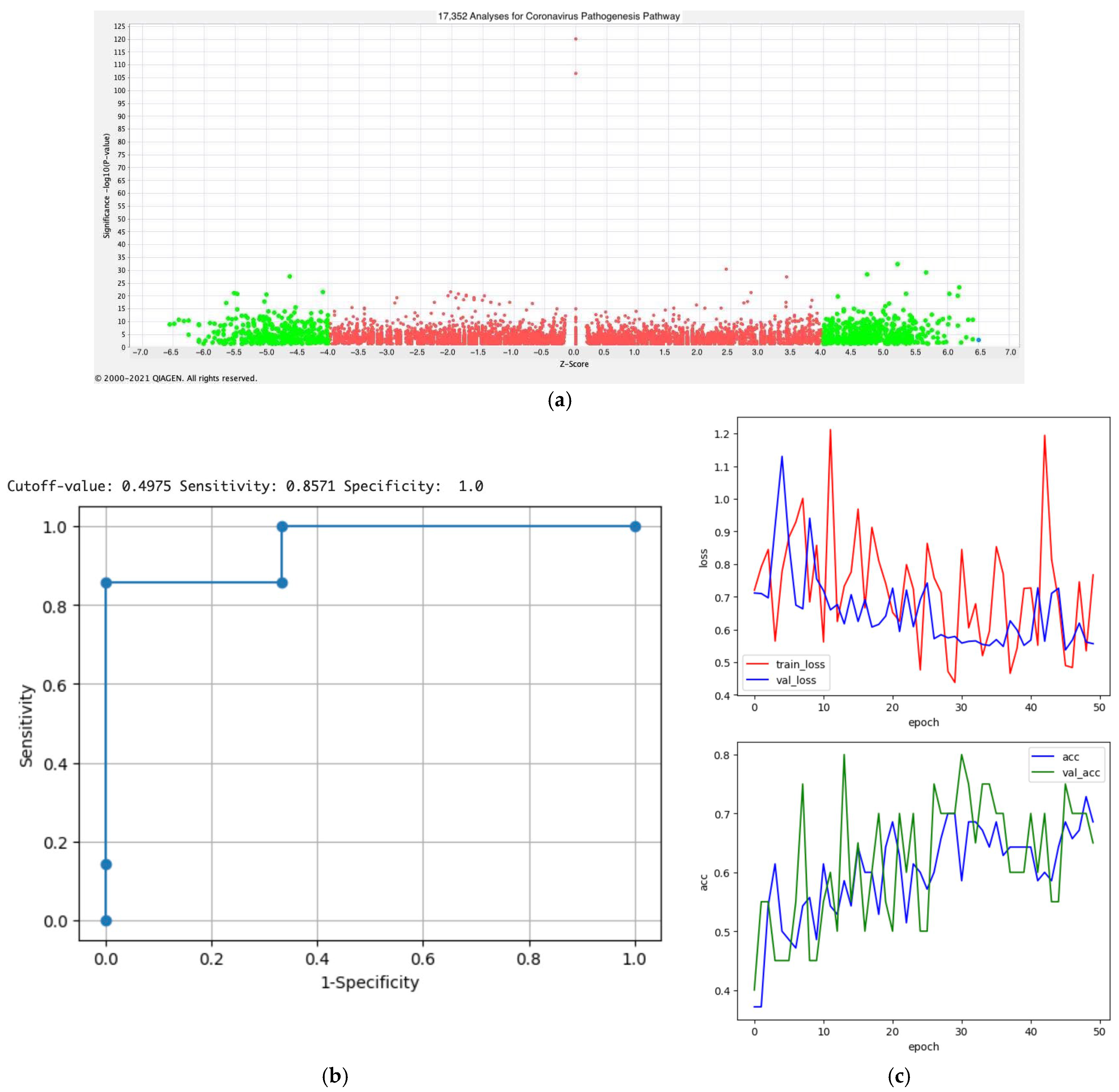
2.4. Activity Plot Analysis
2.5. Python Coding
2.6. Statistical Analysis
3. Results
3.1. Molecular Network Analysis of SARS-CoV-2
3.2. Coronavirus Pathogenesis Pathway in LUAD Samples Infected with SARS-CoV
3.3. SARS-CoV-2 Analysis Matched with Diffuse-Type Gastric Cancer
3.4. Coronavirus Pathogenesis Pathway in Stem Cells
3.5. Drugs That Interact with the Coronavirus Pathogenesis Pathway
3.6. Prediction Modeling of the Activation States of Coronavirus Pathogenesis Pathway (Python Modeling)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SARS | Severe acute respiratory syndrome |
| IPA | Ingenuity Pathway Analysis |
| SARS-CoV-2 | SARS coronavirus 2 |
| iPSC | Induced pluripotent stem cell |
| LUAD | Lung adenocarcinoma |
| ARDS | Acute respiratory distress syndrome |
| MAPK | Mitogen-activated pathway kinase |
| IFN | Interferon |
| TGF | Transforming growth factor |
| TGFβ1 | TGF beta 1 |
| JNK | c-jun N-terminal kinase |
| ERK1/2 | Extracellular signal-regulated kinase 1/2 |
| IL1B | Interleukin 1B |
| AGTR1 | Angiotensin II receptor type I |
| ACE2 | Angiotensin-converting enzyme 2 |
| COVID-19 | Coronavirus disease 2019 |
| AI | Artificial intelligence |
| EMT | Epithelial–mesenchymal transition |
| GEO | Gene Expression Omnibus |
| GSE | GEO Series |
| UR | Upstream regulator |
| CN | Master regulators in causal network |
| DE | Diseases and functions in downstream effect |
| Grad-CAM | Gradient-weighted Class Activation Mapping |
| MOI | Multiplicity of infection |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| DNMT | DNA methyltransferase |
References
- Quan, C.; Li, C.; Ma, H.; Li, Y.; Zhang, H. Immunopathogenesis of Coronavirus-Induced Acute Respiratory Distress Syndrome (ARDS): Potential Infection-Associated Hemophagocytic Lymphohistiocytosis. Clin. Microbiol. Rev. 2020, 34, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Kosyreva, A.; Dzhalilova, D.; Lokhonina, A.; Vishnyakova, P.; Fatkhudinov, T. The Role of Macrophages in the Pathogenesis of SARS-CoV-2-Associated Acute Respiratory Distress Syndrome. Front. Immunol. 2021, 12, 682871. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.S.; Liu, D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef]
- Bakadia, B.M.; He, F.; Souho, T.; Lamboni, L.; Ullah, M.W.; Boni, B.O.; Ahmed, A.A.Q.; Mukole, B.M.; Yang, G. Prevention and treatment of COVID-19: Focus on interferons, chloroquine/hydroxychloroquine, azithromycin, and vaccine. Biomed. Pharmacother. 2021, 133, 111008. [Google Scholar] [CrossRef] [PubMed]
- Hejenkowska, E.D.; Mitash, N.; Donovan, J.E.; Chandra, A.; Bertrand, C.; De Santi, C.; Greene, C.M.; Mu, F.; Swiatecka-Urban, A. TGF-β1 Inhibition of ACE2 Mediated by miRNA Uncovers Novel Mechanism of SARS-CoV-2 Pathogenesis. J. Innate Immun. 2023, 15, 629–646. [Google Scholar] [CrossRef]
- Kyriakopoulos, A.M.; Nigh, G.; McCullough, P.A.; Seneff, S. Mitogen Activated Protein Kinase (MAPK) Activation, p53, and Autophagy Inhibition Characterize the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Spike Protein Induced Neurotoxicity. Cureus 2022, 14, e32361. [Google Scholar] [CrossRef]
- Lokau, J.; Garbers, Y.; Vicente, M.M.; Dittrich, A.; Meltendorf, S.; Lingel, H.; Münster-Kühnel, A.K.; Brunner-Weinzierl, M.; Garbers, C. Long-term increase in soluble interleukin-6 receptor levels in convalescents after mild COVID-19 infection. Front. Immunol. 2024, 15, 1488745. [Google Scholar] [CrossRef] [PubMed]
- Monteonofrio, L.; Florio, M.C.; AlGhatrif, M.; Lakatta, E.G.; Capogrossi, M.C. Aging- and gender-related modulation of RAAS: Potential implications in COVID-19 disease. Vasc. Biol. 2021, 3, R1–R14. [Google Scholar] [CrossRef]
- Perez-Bermejo, J.A.; Kang, S.; Rockwood, S.J.; Simoneau, C.R.; Joy, D.A.; Silva, A.C.; Ramadoss, G.N.; Flanigan, W.R.; Fozouni, P.; Li, H.; et al. SARS-CoV-2 infection of human iPSC-derived cardiac cells reflects cytopathic features in hearts of patients with COVID-19. Sci. Transl. Med. 2021, 13, eabf7872. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Caforio, A.L.P. Receipt of mRNA Vaccine against COVID-19 and Myocarditis. N. Engl. J. Med. 2021, 385, 2189–2190. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, S. Relationship between COVID-19 vaccine and myocarditis. Adv. Clin. Med. Res. 2022, 3, 30. [Google Scholar] [CrossRef] [PubMed]
- Sessa, F.; Salerno, M.; Esposito, M.; Di Nunno, N.; Zamboni, P.; Pomara, C. Autopsy Findings and Causality Relationship between Death and COVID-19 Vaccination: A Systematic Review. J. Clin. Med. 2021, 10, 5876. [Google Scholar] [CrossRef]
- Pomara, C.; Sessa, F.; Ciaccio, M.; Dieli, F.; Esposito, M.; Giammanco, G.M.; Garozzo, S.F.; Giarratano, A.; Prati, D.; Rappa, F.; et al. COVID-19 Vaccine and Death: Causality Algorithm According to the WHO Eligibility Diagnosis. Diagnostics 2021, 11, 955. [Google Scholar] [CrossRef]
- Schreckenberg, R.; Woitasky, N.; Itani, N.; Czech, L.; Ferdinandy, P.; Schulz, R. Cardiac side effects of RNA-based SARS-CoV-2 vaccines: Hidden cardiotoxic effects of mRNA-1273 and BNT162b2 on ventricular myocyte function and structure. Br. J. Pharmacol. 2024, 181, 345–361. [Google Scholar] [CrossRef] [PubMed]
- WHO. A Clinical Case Definition of Post COVID-19 Condition by a Delphi Consensus. Available online: https://www.who.int/publications/i/item/WHO-2019-nCoV-Post_COVID-19_condition-Clinical_case_definition-2021.1 (accessed on 21 April 2025).
- Schneider, A.; Hapfelmeier, A.; Greißel, A.; Limbach, M.; Schwarzl, G.; Ebert, F.; Huber, V.; Hayden, M.C. The implications of somatic symptom disorder on the impairment of daily life are greater in post-COVID syndrome than in asthma or COPD—Results of a cross-sectional study in a rehabilitation clinic. Sci. Rep. 2025, 15, 11719. [Google Scholar] [CrossRef]
- Gangi, S.; Bergantini, L.; Paggi, I.; Spalletti, M.; Cameli, P.; Bargagli, E.; d’Alessandro, M. Regulatory T Cell Phenotype Related to Cytokine Expression Patterns in Post-COVID-19 Pulmonary Fibrosis and Idiopathic Pulmonary Fibrosis. Immun. Inflamm. Dis. 2025, 13, e70123. [Google Scholar] [CrossRef]
- Tanabe, S.; Quader, S.; Ono, R.; Cabral, H.; Aoyagi, K.; Hirose, A.; Yokozaki, H.; Sasaki, H. Molecular network analysis of RNA viral infection pathway in diffuse- and intestinal-type gastric cancer. Fundam. Toxicol. Sci. 2022, 9, 37–46. [Google Scholar] [CrossRef]
- Tanabe, S.; Quader, S.; Ono, R.; Cabral, H.; Aoyagi, K.; Hirose, A.; Yokozaki, H.; Sasaki, H. Cell Cycle Regulation and DNA Damage Response Networks in Diffuse- and Intestinal-Type Gastric Cancer. Cancers 2021, 13, 5786. [Google Scholar] [CrossRef]
- Tanabe, S.; Quader, S.; Ono, R.; Cabral, H.; Aoyagi, K.; Hirose, A.; Perkins, E.J.; Yokozaki, H.; Sasaki, H. Regulation of Epithelial–Mesenchymal Transition Pathway and Artificial Intelligence-Based Modeling for Pathway Activity Prediction. Onco 2023, 3, 13–25. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.A.; Clarke, D.J.B.; Møller, R.; Han, Y.; Yang, L.; Wojciechowicz, M.L.; Lachmann, A.; Oguntuyo, K.Y.; Stevens, C.; Lee, B.; et al. Modulating the transcriptional landscape of SARS-CoV-2 as an effective method for developing antiviral compounds. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Hill, T.E.; Yoshikawa, N.; Popov, V.L.; Galindo, C.L.; Garner, H.R.; Peters, C.J.; Tseng, C.-T. Dynamic Innate Immune Responses of Human Bronchial Epithelial Cells to Severe Acute Respiratory Syndrome-Associated Coronavirus Infection. PLoS ONE 2010, 5, e8729. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.-C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Daamen, A.R.; Bachali, P.; Owen, K.A.; Kingsmore, K.M.; Hubbard, E.L.; Labonte, A.C.; Robl, R.; Shrotri, S.; Grammer, A.C.; Lipsky, P.E. Comprehensive transcriptomic analysis of COVID-19 blood, lung, and airway. Sci. Rep. 2021, 11, 7052. [Google Scholar] [CrossRef]
- Pérez-Bermejo, J.A.; Kang, S.; Rockwood, S.J.; Simoneau, C.R.; Joy, D.A.; Ramadoss, G.N.; Silva, A.C.; Flanigan, W.R.; Li, H.; Nakamura, K.; et al. SARS-CoV-2 infection of human iPSC-derived cardiac cells predicts novel cytopathic features in hearts of COVID-19 patients. bioRxiv 2020. [Google Scholar] [CrossRef]
- Tanabe, S.; Quader, S.; Cabral, H.; Perkins, E.J.; Yokozaki, H.; Sasaki, H. Master Regulators of Causal Networks in Intestinal- and Diffuse-Type Gastric Cancer and the Relation to the RNA Virus Infection Pathway. Int. J. Mol. Sci. 2024, 25, 8821. [Google Scholar] [CrossRef]
- Shimizu, H. Machine Learning in Python: Machine Learning of Life Science Data; Jikken Igaku Bessatsu Python De Jissen Seimeikagaku data no Kikaigakushu; Shimizu, H., Ed.; Yodosha: Tokyo, Japan, 2023. [Google Scholar]
- Hartman, E.; Scott, A.M.; Karlsson, C.; Mohanty, T.; Vaara, S.T.; Linder, A.; Malmström, L.; Malmström, J. Interpreting biologically informed neural networks for enhanced proteomic biomarker discovery and pathway analysis. Nat. Commun. 2023, 14, 5359. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Matsuura, Y.; Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 2025, 53, D672–D677. [Google Scholar] [CrossRef]
- Dhillon, S. Decitabine/Cedazuridine: First Approval. Drugs 2020, 80, 1373–1378. [Google Scholar] [CrossRef]
- Yabushita, T.; Chinen, T.; Nishiyama, A.; Asada, S.; Shimura, R.; Isobe, T.; Yamamoto, K.; Sato, N.; Enomoto, Y.; Tanaka, Y.; et al. Mitotic perturbation is a key mechanism of action of decitabine in myeloid tumor treatment. Cell Rep. 2023, 42, 113098. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Chen, Y.; Luo, D.; Zhuang, Z.; Jin, H.; Zhou, H.; Li, X.; Lin, H.; Zheng, X.; Zhang, J.; et al. Therapeutic potential of C1632 by inhibition of SARS-CoV-2 replication and viral-induced inflammation through upregulating let-7. Signal Transduct. Target. Ther. 2021, 6, 84. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Hirsch, H.A.; Struhl, K. An epigenetic switch involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009, 139, 693–706. [Google Scholar] [CrossRef]
- Coomes, E.A.; Haghbayan, H. Interleukin-6 in COVID-19: A systematic review and meta-analysis. Rev. Med. Virol. 2020, 30, 1–9. [Google Scholar] [CrossRef] [PubMed]

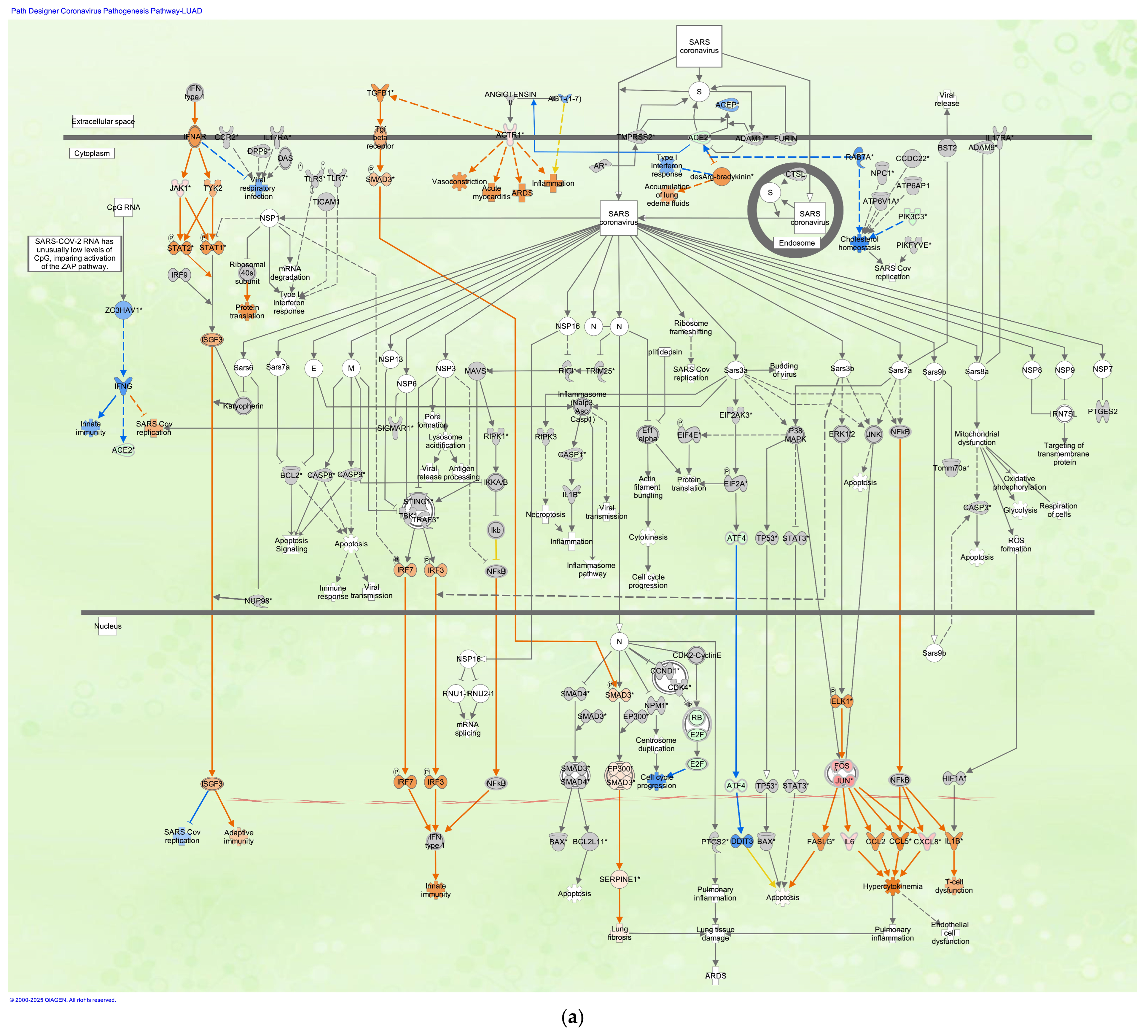
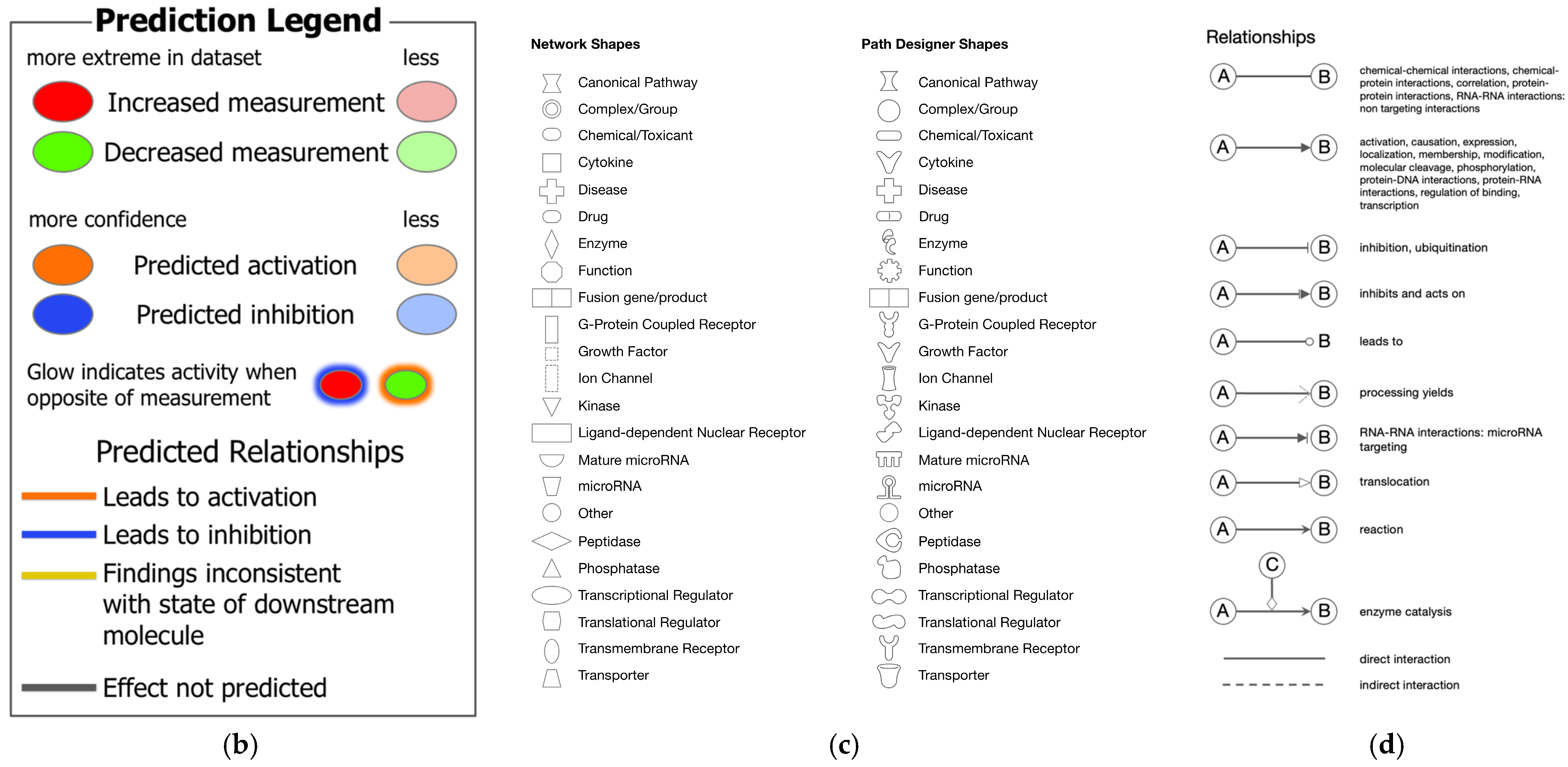
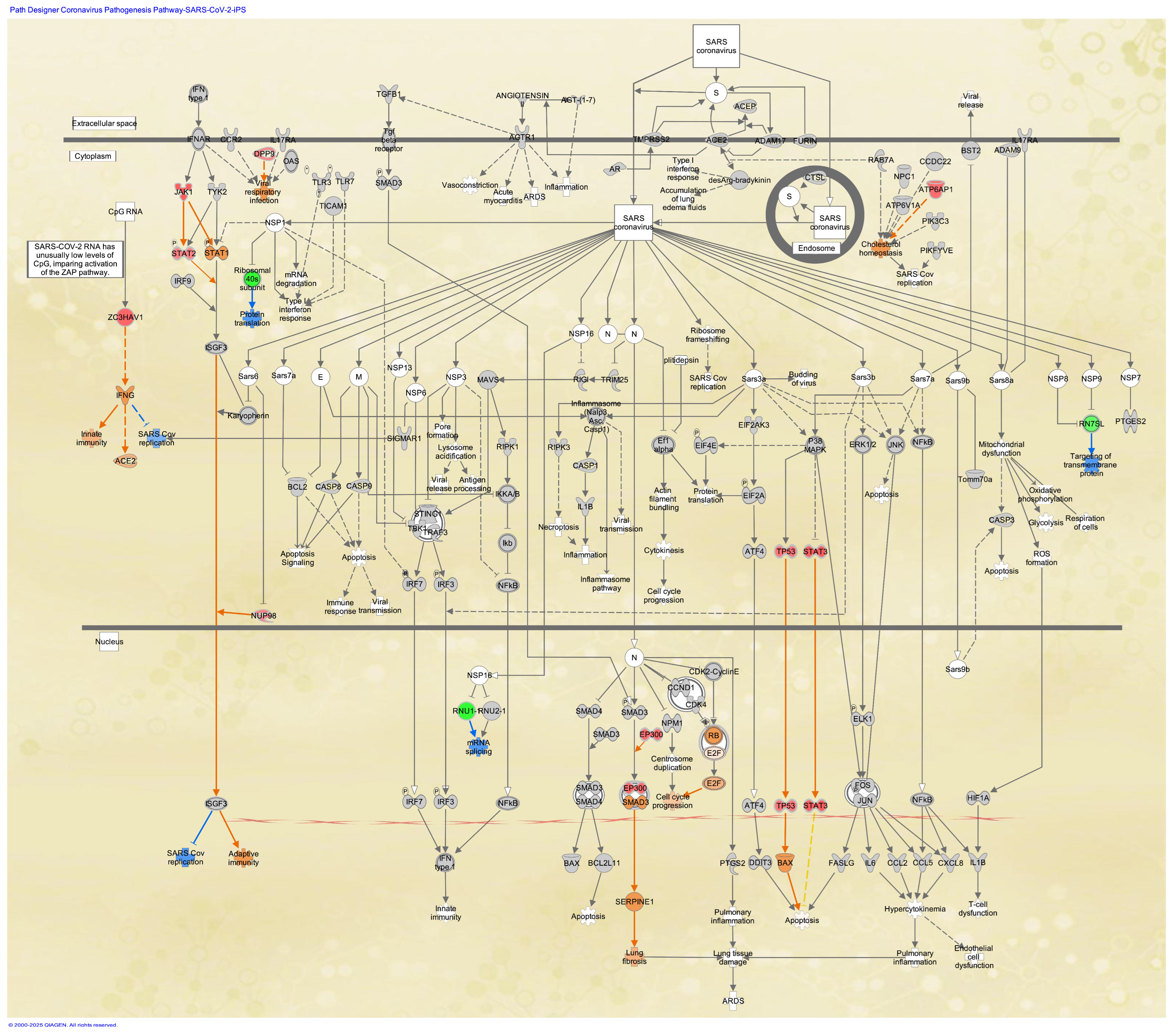
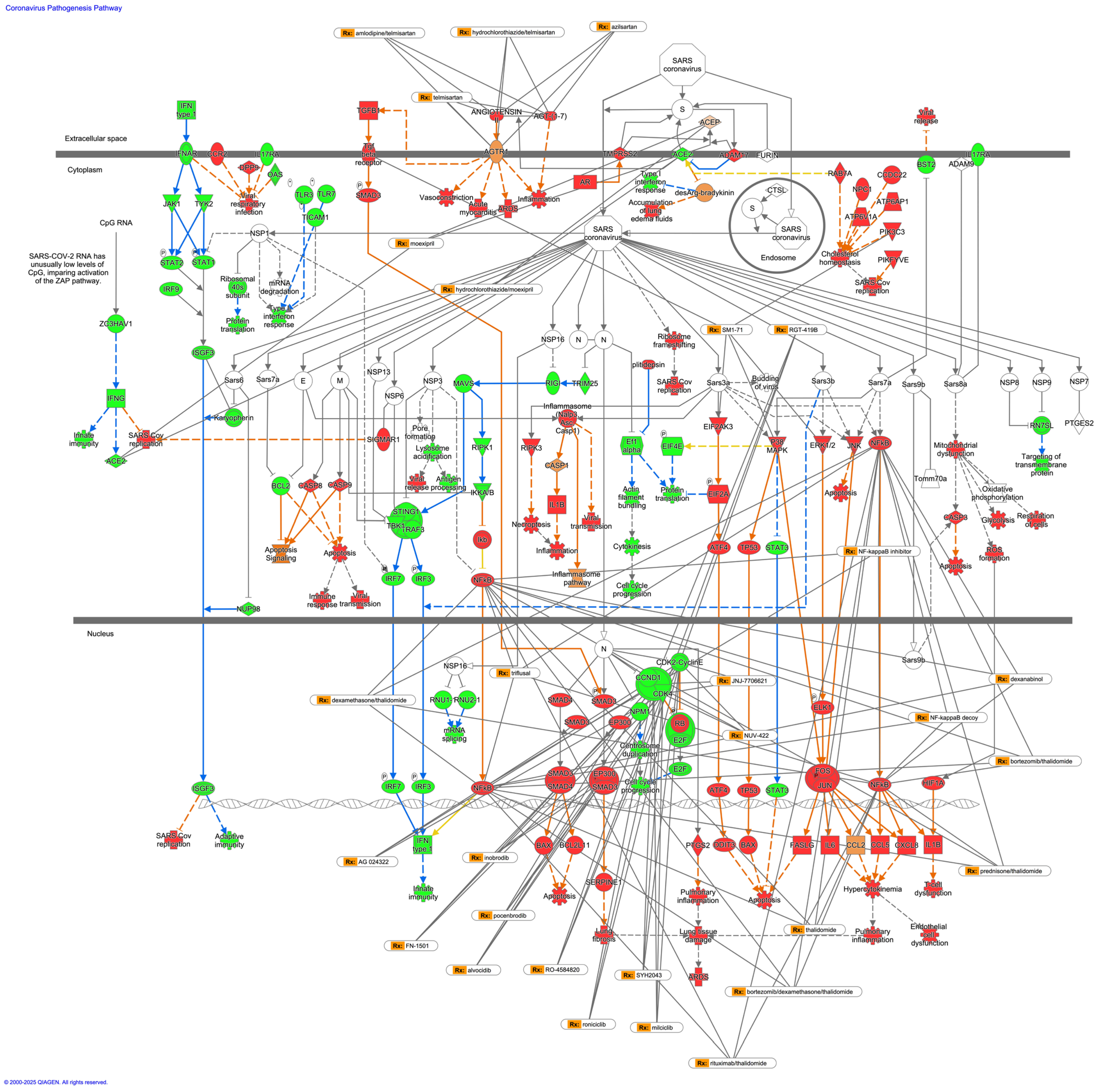
| Analysis Name | Activation z-Score of Coronavirus Pathogenesis Pathway * | Comparison Contrast |
|---|---|---|
| 2-lung adenocarcinoma (LUAD) alveoli 7103 | −1.706 | SARS-CoV-2-infected A549 cell line (MOI 0.2) vs. mock-infected A549 cell line |
| 3-lung adenocarcinoma (LUAD) alveoli 7109 | 3.464 | SARS-CoV-2-infected A549 cell line (MOI 2) vs. mock-infected A549 cell line |
| 7-lung adenocarcinoma (LUAD) alveoli 7113 | 1.147 | SARS-CoV-2-infected ACE2-transfected A549 cell line (MOI 0.2) vs. mock-infected ACE2-transfected A549 cell line |
| 22-lung adenocarcinoma (LUAD) alveoli DMSO 7106 | 0 | SARS-CoV-2-infected ACE2-transfected A549 cell line vs. mock-infected ACE2-transfected A549 cell line |
| 23-lung adenocarcinoma (LUAD) alveoli ruxolitinib 7107 | 1.941 | SARS-CoV-2-infected ACE2-transfected A549 cell line and ruxolitinib vs. mock-infected ACE2-transfected A549 cell line |
| 24-lung adenocarcinoma (LUAD) alveoli ruxolitinib 7108 | 1.732 | SARS-CoV-2-infected ACE2-transfected A549 cell line and ruxolitinib vs. SARS-CoV-2-infected ACE2-transfected A549 cell line |
| 4-lung adenocarcinoma (LUAD) alveoli 7110 | 3.742 | SARS-CoV-2-infected A549 cell line (MOI 2) vs. SARS-CoV-2 infected A549 cell line (MOI 0.2) |
| 8-lung adenocarcinoma (LUAD) bronchial epithelium 7114 | −0.2 | SARS-CoV-2-infected CALU3 cell line vs. mock-infected CALU-3 cell line |
| Entity Type | Entity Name | Diffuse-Type Gastric Cancer | iPSC-Derived Cardiomyocyte Infected with SARS-CoV-2 0.001 MOI vs. Mock | iPSC-Derived Cardiomyocyte Infected with SARS-CoV-2 0.01 MOI vs. Mock | iPSC-Derived Cardiomyocyte Infected with SARS-CoV-2 0.1 MOI vs. Mock | iPSC-Derived Cardiac Fibroblast Infected with SARS-CoV-2 0.006 MOI vs. Mock | iPSC Infected with SARS-CoV-2 0.006 MOI vs. Mock |
|---|---|---|---|---|---|---|---|
| DE | Organismal death | 6.09939477 | 0 | 0 | 0 | 0 | −10.332512 |
| DE | Morbidity or mortality | 6.0991701 | 0 | 0 | 0 | 0 | −10.254265 |
| UR | TP53 | 5.25209454 | −2.9576275 | −3.8544245 | 0 | −4.0046362 | 3.36923989 |
| UR | let-7a-5p (and other miRNAs w/seed GAGGUAG) | 2.95701052 | 2.98384345 | 3.24713706 | 2.75140771 | 0 | −3.6818652 |
| UR | let-7 | 5.88141247 | 3.36872653 | 3.07534027 | 2.76863583 | −0.7453134 | −2.628098 |
| UR | CDKN2A | 5.00037308 | 0.34050945 | 0.51898468 | 1.34164079 | −3.2237322 | 2.97677657 |
| UR | calcitriol | 5.35668014 | 0 | 0 | 1.23787842 | −1.587867 | 1.9593573 |
| CN | NUPR1 | 6.68503217 | 0 | 0 | 0 | −6.0621778 | 0.33752637 |
| CN | l-asparaginase | 7.00201178 | 0 | 0 | 0 | −4.2 | 0 |
| UR | l-asparaginase | 6.92462738 | 0 | 0 | 2.23606798 | −4.1949137 | 0 |
| UR | NUPR1 | 6.68503217 | 0 | 0 | 2.49615088 | −6.0621778 | −0.386494 |
| UR | SMARCB1 | 2.84931818 | −1.8898224 | −1.1338934 | −2 | −1.407767 | 2.57658201 |
| UR | MEF2D | 2.38560366 | −2.5729119 | −2.3785413 | −1.9249444 | −2.236068 | 0 |
| UR | Decitabine | 3.08835855 | −3.4575395 | −2.2066886 | −1.5180635 | 0 | −0.2058335 |
| UR | SPARC | 3.28571429 | −1.3516756 | −1.7509621 | −1.9686483 | 0 | 0 |
| DE | Growth failure or short stature | 3.70765671 | 0 | 0 | 0 | 0 | −5.311879 |
| UR | RB1 | 3.36893187 | 0 | 0 | −1.8347785 | −1.3252763 | −2.7095152 |
| CN | Osimertinib | 5.93335075 | 0 | 0 | 1 | 0 | −2.4596748 |
| Symbol | Entrez Gene Name | Drugs |
|---|---|---|
| ACE2 | angiotensin converting enzyme 2 | hydrochlorothiazide/moexipril, and moexipril |
| ACEP | angiotensin I converting enzyme | ceronapril, indolapril, pentopril, quinaprilat, perindoprilat, angiotensin I (1– 7), amlodipine/perindopril, benazeprilat, trandolaprilat, angiotensin-converting enzyme inhibitor, aspirin/lisinopril, amlodipine/benazepril, hydrochlorothiazide/lisinopril, benazepril, enalapril, perindopril, captopril, cilazapril, enalapril/felodipine, hydrochlorothiazide/moexipril, benazepril/hydrochlorothiazide, hydrochlorothiazide/quinapril, fosinopril/hydrochlorothiazide, captopril/hydrochlorothiazide, enalapril/hydrochlorothiazide, hydrochlorothiazide/trandolapril, ramipril, ramiprilat, moexipril, quinapril, amlodipine/indapamide/perindopril, lisinopril, enalaprilat, trandolapril, moexiprilat, idrapril, rentiapril, imidaprilat, gemopatrilat, zabiciprilat, libenzapril, fosinoprilat, zofenoprilat, trandolapril/verapamil, diltiazem/enalapril, fosinopril, and carvedilol/enalapril |
| ADAM17 | ADAM metallopeptidase domain 17 | aderbasib |
| ADAM9 | ADAM metallopeptidase domain 9 | IMGC936 |
| AGTR1 | angiotensin II receptor type 1 | caffeine/dextromethorphan/losartan/midazolam/omeprazole, amlodipine/olmesartan medoxomil, olmesartan, amlodipine/hydrochlorothiazide/valsartan, amlodipine/telmisartan, aliskiren/valsartan, azilsartan, azilsartan kamedoxomil, amlodipine/hydrochlorothiazide/olmesartan medoxomil, aspirin/dipyridamole/telmisartan, clopidogrel/telmisartan, azilsartan medoxomil/chlorthalidone, sacubitril/valsartan, amlodipine/valsartan, sparsentan, nebivolol/valsartan, hydrochlorothiazide/losartan, hydrochlorothiazide/valsartan, candesartan, candesartan cilexetil, olmesartan medoxomil, irbesartan, losartan potassium, telmisartan, eprosartan, candesartan cilexetil/hydrochlorothiazide, hydrochlorothiazide/irbesartan, eprosartan/hydrochlorothiazide, hydrochlorothiazide/telmisartan, hydrochlorothiazide/olmesartan medoxomil, amlodipine/ezetimibe/losartan/rosuvastatin, and valsartan |
| ANGIOTENSINII | angiotensinogen | amlodipine/telmisartan, azilsartan, telmisartan, and hydrochlorothiazide/telmisartan |
| AR | androgen receptor | TAS3681, ARV-110, ODM 204, bicalutamide/GnRH analog, CC-94676, CB0310, EPI-7386, AC176, estradiol valerate/testosterone enanthate, estradiol cypionate/testosterone cypionate, ARV-766, TQB3720, BMS-641988, cyproterone acetate/ethinyl estradiol, enzalutamide, galeterone, ostarine, 1-testosterone, clascoterone, flutamide/goserelin, nandrolone phenpropionate, androgen receptor antagonist, apalutamide, darolutamide, AZD3514, APC-100, EPI-506, bicalutamide/leuprolide, bicalutamide/goserelin, dexamethasone/enzalutamide, rezvilutamide, LY2452473, enzalutamide/exemestane, drospirenone/ethinyl estradiol, nilutamide, TRC253, bicalutamide, SXL01, proxalutamide, hydroxyflutamide, testolone, GSK2881078, flutamide, nandrolone decanoate, testosterone cypionate, deuterated enzalutamide, AZD5312, fluoxymesterone/tamoxifen, cyproterone acetate, nandrolone, drospirenone, medroxyprogesterone acetate, oxandrolone, danazol, dihydrotestosterone, fluoxymesterone, stanozolol, spironolactone, methyltestosterone, testosterone, oxymetholone, 7alpha-hydroxytestosterone, norgestimate, testosterone propionate, and testosterone enanthate |
| ATP6V1A | ATPase H+ transporting V1 subunit A | bafilomycin A1 and bafilomycin b1 |
| BCL2 | BCL2 apoptosis regulator | BP1002, S65487, rosomidnar, FCN-338, BGB-11417, LP-118, ZN-d5, oblimersen, ABBV-623, ABBV-453, TQB3909, rasagiline, (-)-gossypol, obatoclax, ABT-737, BCL-2 blocker, navitoclax, gemcitabine/paclitaxel, bortezomib/paclitaxel, venetoclax, paclitaxel/trastuzumab, paclitaxel/pertuzumab/trastuzumab, lapatinib/paclitaxel, doxorubicin/paclitaxel, epirubicin/paclitaxel, paclitaxel/ramucirumab, paclitaxel/topotecan, BCL201, pelcitoclax, paclitaxel/rituximab, afatinib/paclitaxel, doxorubicin/lapatinib/paclitaxel/trastuzumab, levodopa/rasagiline, everolimus/paclitaxel, lisaftoclax, SPC2996, paclitaxel/pembrolizumab/ramucirumab, paclitaxel, chelerythrine, AZD0466, and paclitaxel/pembrolizumab, LP-108 |
| CASP1 | caspase 1 | caspase 1 inhibitor |
| CASP3 | caspase 3 | caspase 3 inhibitor |
| CASP9 | caspase 9 | caspase-9 inhibitor |
| CCL2 | C-C motif chemokine ligand 2 | CNTO 888, mimosine |
| CCND1 | cyclin D1 | arsenic trioxide/tretinoin, arsenic trioxide/daunorubicin/tretinoin, arsenic trioxide/gemtuzumab ozogamicin/tretinoin, arsenic trioxide/idarubicin/tretinoin, arsenic trioxide/cytarabine/methotrexate, and arsenic trioxide |
| CCR2 | C-C motif chemokine receptor 2 | AZD2423, PF-4136309, MLN1202, BMS-813160, propagermanium, ilacirnon, and MK-0812 |
| CDK4 | cyclin dependent kinase 4 | XZP-3287, CINK4, PD 0183812, BPI-16350, PF-07220060, dalpiciclib, TQB3616, NUV-422, TQB3303, narazaciclib, CS3002, TY-302, RGT-419B, RO0506220, palbociclib, PRT3645, SYH2043, QLS12004, SPH4336, cyclin dependent kinase 4 inhibitor, riviciclib, AG 024322, milciclib, RO-4584820, ribociclib, voruciclib, abemaciclib, letrozole/palbociclib, roniciclib, FLX925, fulvestrant/palbociclib, trilaciclib, lerociclib, letrozole/ribociclib, abemaciclib/fulvestrant, anastrozole/palbociclib, anastrozole/ribociclib, exemestane/palbociclib, exemestane/ribociclib, abemaciclib/aromatase inhibitor, JNJ-7706621, fulvestrant/ribociclib, abemaciclib/exemestane, abemaciclib/anastrozole, abemaciclib/letrozole, ribociclib/tamoxifen, everolimus/ribociclib, fascaplysin, abemaciclib/fulvestrant/GnRH analog, abemaciclib/aromatase inhibitor/GnRH analog, HS-10342, FCN-437, FN-1501, alvocidib, GLR2007, BPI-1178 |
| CTSL | cathepsin L | pegulicianine, and cathepsin L inhibitor |
| CXCL8 | C-X-C motif chemokine ligand 8 | BMS-986253 |
| EIF2AK3 | eukaryotic translation initiation factor 2 alpha kinase 3 | HC-5404-FU, NMS-03597812, AMG44, GSK2656157, and SM1-71 |
| EIF4E | eukaryotic translation initiation factor 4E | ISIS 183750 |
| EP300 | E1A binding protein p300 | pocenbrodib and inobrodib |
| FASLG | Fas ligand | APG101 |
| FURIN | furin, paired basic amino acid cleaving enzyme | hexa-D-arginine, furin inhibitor, and nona-D-arginine amide |
| HIF1A | hypoxia inducible factor 1 subunit alpha | BAY 87-2243, PX 478, and EZN 2968 |
| IFNG | interferon gamma | emapalumab |
| IL17RA | interleukin 17 receptor A | brodalumab |
| IL1B | interleukin 1 beta | FL-101, anakinra, rilonacept, AK114, canakinumab, gevokizumab, canakinumab/INS, canakinumab/metformin, canakinumab/metformin/sulfonylurea, canakinumab/colchicine, canakinumab/methotrexate, anakinra/methotrexate, and gallium nitrate |
| IL6 | interleukin 6 | anti-IL-6 monoclonal antibody, tocilizumab, vamikibart, siltuximab, clazakizumab, interleukin-6 receptor inhibitor, and ziltivekimab |
| JAK1 | Janus kinase 1 | ivarmacitinib, solcitinib, deuruxolitinib, delgocitinib, momelotinib metabolite M21, tofacitinib, ruxolitinib, momelotinib, baricitinib, INCB-16562, filgotinib, oclacitinib, SAR-20347, itacitinib, INCB052793, methotrexate/tofacitinib, upadacitinib, brepocitinib, abrocitinib, baricitinib/methotrexate, pralsetinib, JAK inhibitor I, JAK1 inhibitor, AZD4205, povorcitinib, erlotinib/ruxolitinib, jaktinib, tinengotinib, and methotrexate/ruxolitinib/vincristine |
| JNK | JNK inhibitor, CC 401, SR-3562, and AS601245 | |
| NFkB | NF-kappaB inhibitor and dexanabinol | |
| P38MAPK | SB 220025, doramapimod, AZ10164773, PHA-666859, acumapimod, PD 169316, merck C, SC68376, SK & F 86002, SB 239063, SD-282, SB203580, RWJ 67657, and TAK715 | |
| PIK3C3 | phosphatidylinositol 3-kinase catalytic subunit type 3 | PIK-III, VPS34 inhibitor 1, SAR405, VPS34-IN1, and SM1-71 |
| PIKFYVE | phosphoinositide kinase, FYVE-type zinc finger containing | APY0201 and apilimod |
| PTGS2 | prostaglandin-endoperoxide synthase 2 | bupivacaine/meloxicam, acetaminophen/pentazocine, acetaminophen/clemastine/pseudoephedrine, aspirin/butalbital/caffeine, acetaminophen/caffeine/dihydrocodeine, aspirin/hydrocodone, aspirin/oxycodone, acetaminophen/aspirin/caffeine, aspirin/pravastatin, acetaminophen/dexbrompheniramine/pseudoephedrine, aspirin/meprobamate, aspirin/caffeine/propoxyphene, aspirin/butalbital/caffeine/codeine, aspirin/caffeine/dihydrocodeine, STP707, chlorpheniramine/ibuprofen/pseudoephedrine, licofelone, menatetrenone, polmacoxib, cotsiranib, enflicoxib, icosapent, ECP-1014, aspirin/caffeine/phenacetin, suprofen, lornoxicam, tiaprofenic acid, lumiracoxib, tenoxicam, naproxen/sumatriptan, apricoxib, parecoxib, ibuprofen/phenylephrine, acetaminophen/aspirin/codeine, esomeprazole/naproxen, aspirin/esomeprazole, aspirin/dipyridamole/telmisartan, famotidine/ibuprofen, aspirin/dabigatran etexilate, diclofenac/omeprazole, chlorpheniramine/ibuprofen/phenylephrine, dexamethasone/pomalidomide, sulindac/tamoxifen, sulindac/toremifene, raloxifene/sulindac, ketorolac/phenylephrine, aspirin/bivalirudin, diclofenac/hyaluronic acid, aspirin/clopidogrel, aspirin/omeprazole, aspirin/enoxaparin, aspirin/lisinopril, COX2 inhibitor, diclofenac/misoprostol, acetaminophen/butalbital/caffeine, hydrocodone/ibuprofen, acetaminophen/hydrocodone, acetaminophen/tramadol, acetaminophen/codeine, acetaminophen/oxycodone, acetaminophen/propoxyphene, niflumic acid, nitroaspirin, ketoprofen, diclofenac, etoricoxib, naproxen, meclofenamic acid, pomalidomide, meloxicam, celecoxib, ibuprofen/pseudoephedrine, diphenhydramine/ibuprofen, dipyrone, nimesulide, acetaminophen, mefenamic acid, bortezomib/dexamethasone/pomalidomide, diflunisal, ibuprofen, GW406381X, phenylbutazone, indomethacin, sulfasalazine, SB203580, piroxicam, valdecoxib, aspirin, carprofen, zomepirac, rofecoxib, sorafenib/sulindac/sunitinib, aspirin/caffeine/orphenadrine, acetaminophen/butalbital, balsalazide, aspirin/dipyridamole, acetaminophen/butalbital/caffeine/codeine, naproxen/pseudoephedrine, acetaminophen/diphenhydramine/prednisolone, acetaminophen/diphenhydramine/methylprednisolone, acetaminophen/diphenhydramine, acetaminophen/cetirizine/prednisolone, racemic flurbiprofen, phenacetin, sulindac, nabumetone, etodolac, tolmetin, amlodipine/celecoxib, aspirin/prasugrel, ketorolac, oxaprozin, mesalamine, salsalate, fenoprofen, salicylic acid, aspirin/rivaroxaban, aspirin/clopidogrel/rivaroxaban, aspirin/cangrelor, aspirin/rivaroxaban/ticlopidine, aspirin/ticagrelor, deracoxib, firocoxib, acetaminophen/ibuprofen, acetaminophen/caffeine/chlorpheniramine/hydrocodone/phenylephrine, and bromfenac |
| RIGI | RNA sensor RIG-I | MK-4621 |
| RIPK1 | receptor interacting serine/threonine kinase 1 | eclitasertib, GDC-8264, GSK2982772, GSK3145095, and GSK963 |
| RIPK3 | receptor interacting serine/threonine kinase 3 | N-[6-[3-[(3-bromophenyl)carbamoylamino]-4-fluorophenoxy]-1,3-benzothiazol-2-yl]cyclopropanecarboxamide, GSK843, GSK872, and GSK840 |
| SERPINE1 | serpin family E member 1 | TM5614, drotrecogin alfa, and ACT001 |
| SIGMAR1 | sigma non-opioid intracellular receptor 1 | caffeine/dextromethorphan/losartan/midazolam/omeprazole, acetaminophen/pentazocine, dihydrocodeine, dextromethorphan/morphine, dimemorfan, morphine/naltrexone, opipramol, dextromethorphan/quinidine, naloxone/pentazocine, bupropion/naltrexone, naltrexone/oxycodone, bupropion/dextromethorphan, etorphine, SA 4503, fenfluramine, hydromorphone, naltrexone, dextromethorphan, oxycodone, pentazocine, naloxone, SR 31747, brompheniramine/dextromethorphan/pseudoephedrine, chlorpheniramine/dextromethorphan/phenylephrine, carbinoxamine/dextromethorphan/pseudoephedrine, and dextromethorphan/promethazine |
| STAT3 | signal transducer and activator of transcription 3 | CAS3/SS3, KT-333, golotimod, OPB-31121, OPB-51602, danvatirsen, TTI-101, STAT3 inhibitor, and NT219 |
| STING1 | stimulator of interferon response cGAMP interactor 1 | CDK-002, E7766, dazostinag, SNX281, KL340399, ulevostinag, MIW815, GSK3745417, BMS-986301, MK-2118, SB 11285, IMSA101, and BI 1387446 |
| TBK1 | TANK binding kinase 1 | MRT-68601, 6-aminopyrazolopyrimidine derivative compound II, and BX-795 |
| TGFB1 | transforming growth factor beta 1 | SHR1701, HB-002T, STP707, cotsiranib, dalantercept, LY2109761, fresolimumab, LY3200882, MSB0011359C, NIS793, AVID200, YL-13027, SRK-181 |
| TLR3 | toll like receptor 3 | rintatolimod, and poly rI:rC-RNA |
| TLR7 | toll like receptor 7 | SHR2150, APR003, BDB018, enpatoran, vesatolimod, UC-1V150, PF-4878691, 5-fluorouracil/imiquimod, resiquimod, hydroxychloroquine, imiquimod, NKTR-262, LHC165, DSP-0509, BDC-1001, TQ-A3334, BNT411, RO7119929 |
| TP53 | tumor protein p53 | PC14586, eprenetapopt, cenersen, ALT-801, CGM097, kevetrin, azurin 50–77, COTI-2, and BI 907828 |
| TYK2 | tyrosine kinase 2 | deuruxolitinib, delgocitinib, ropsacitinib, zasocitinib, VTX958, momelotinib metabolite M21, tofacitinib, ruxolitinib, momelotinib, baricitinib, filgotinib, oclacitinib, SAR-20347, brepocitinib, deucravacitinib, baricitinib/methotrexate, and JAK inhibitor I |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanabe, S.; Quader, S.; Ono, R.; Tanaka, H.Y.; Yamamoto, A.; Kojima, M.; Perkins, E.J.; Cabral, H. Artificial Intelligence Approach in Machine Learning-Based Modeling and Networking of the Coronavirus Pathogenesis Pathway. Curr. Issues Mol. Biol. 2025, 47, 466. https://doi.org/10.3390/cimb47060466
Tanabe S, Quader S, Ono R, Tanaka HY, Yamamoto A, Kojima M, Perkins EJ, Cabral H. Artificial Intelligence Approach in Machine Learning-Based Modeling and Networking of the Coronavirus Pathogenesis Pathway. Current Issues in Molecular Biology. 2025; 47(6):466. https://doi.org/10.3390/cimb47060466
Chicago/Turabian StyleTanabe, Shihori, Sabina Quader, Ryuichi Ono, Hiroyoshi Y. Tanaka, Akihisa Yamamoto, Motohiro Kojima, Edward J. Perkins, and Horacio Cabral. 2025. "Artificial Intelligence Approach in Machine Learning-Based Modeling and Networking of the Coronavirus Pathogenesis Pathway" Current Issues in Molecular Biology 47, no. 6: 466. https://doi.org/10.3390/cimb47060466
APA StyleTanabe, S., Quader, S., Ono, R., Tanaka, H. Y., Yamamoto, A., Kojima, M., Perkins, E. J., & Cabral, H. (2025). Artificial Intelligence Approach in Machine Learning-Based Modeling and Networking of the Coronavirus Pathogenesis Pathway. Current Issues in Molecular Biology, 47(6), 466. https://doi.org/10.3390/cimb47060466








