The Application of Microsatellite Markers as Molecular Tools for Studying Genomic Variability in Vertebrate Populations
Abstract
1. Introduction
2. Microsatellite Markers: Their Features and PCR-Based Approach Used for Their Identification
2.1. What, Molecularly, Are Microsatellites and Why Are They Useful?
- Their incredible level of polymorphism, which allows typing both different populations and single individuals with a high degree of probability [59].
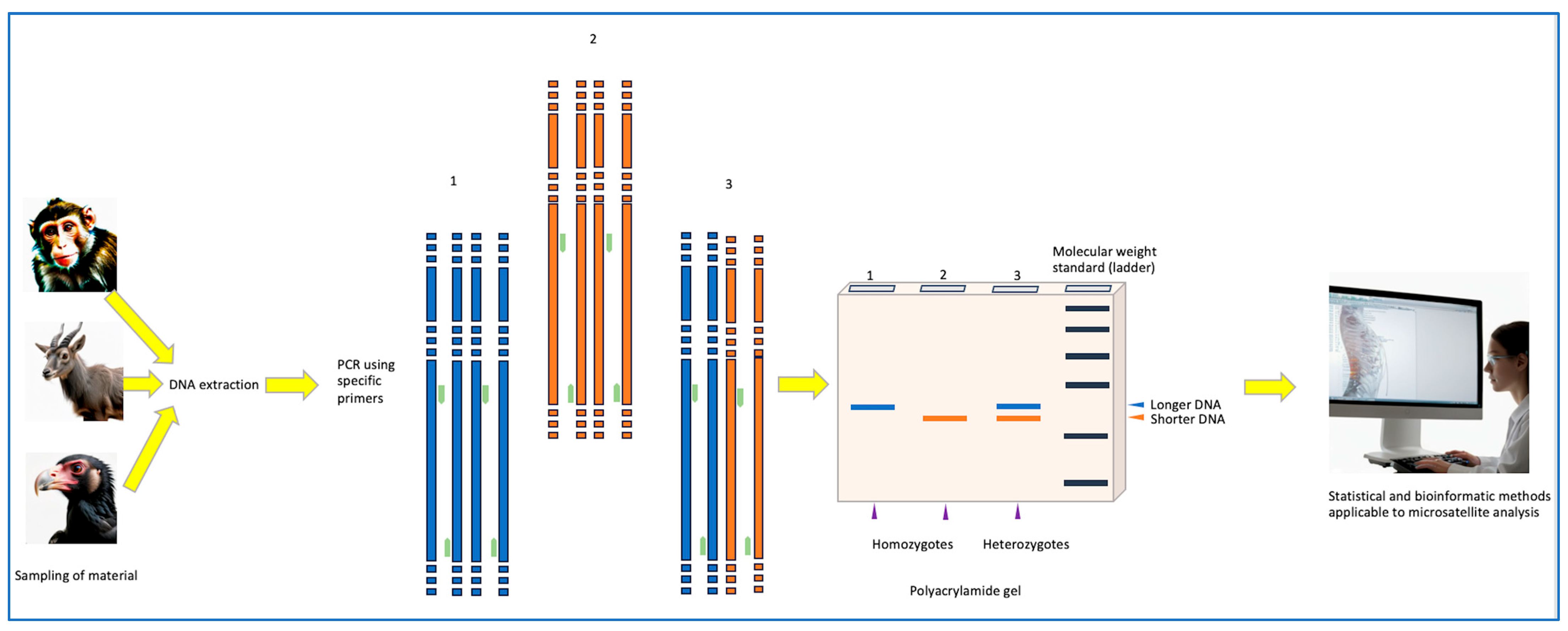
2.2. PCR Detection of Microsatellites
2.3. Genetic Diversity, Mutation Rates, and Heterozygosity: Microsatellites vs. SNPs
3. Statistical and Bioinformatic Methods Applicable to Microsatellite Analysis
3.1. Classical Statistics
3.2. Bioinformatic Software
4. Human and Livestock Microsatellite Studies as a “Road Map” for the Genetics, Breeding, and Conservation of Wildlife and Rare Breeds
4.1. Humans and Other Primates
4.2. Cattle and Other Artiodactyla
4.3. Perissodactyla
4.4. Chickens
4.5. Other Birds
4.6. Dogs
4.7. Cats
4.8. Elephantidae
4.9. Reptiles
4.10. Amphibians
4.11. Fish
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACO | ant colony optimization |
| dNTP | deoxynucleoside triphosphate |
| EST | expressed sequence-tag |
| FAO | Food and Agriculture Organization of the United Nations |
| FIS | inbreeding coefficient of individuals in a subpopulation |
| FISH | fluorescence in situ hybridization |
| FIT | inbreeding coefficient of individuals in the population as a whole |
| FST | inbreeding coefficient of the subpopulation relative to the entire population |
| GC-content | guanine-cytosine content |
| He | expected heterozygosity |
| Ho | observed heterozygosity |
| HRM | high-resolution melting |
| ISAG | International Society of Animal Genetics |
| ISSR | inter simple sequence repeat |
| LINEs | long interspersed nuclear elements |
| MAS | marker-assisted selection |
| mtDNA | mitochondrial DNA |
| PCR | polymerase chain reaction |
| PIC | polymorphic information content |
| POT1 | protection of telomeres 1 |
| QTLs | quantitative trait loci |
| RAPD | random amplified polymorphic DNA |
| RFLP | restriction fragment length polymorphism |
| SNP | single nucleotide polymorphism |
| SSR | simple sequence repeat |
| STRs | short tandem repeats |
| TRF1 | telomeric repeat binding factor 1 |
| TRF2 | telomeric repeat binding factor 2 |
References
- Dodgson, J.B.; Cheng, H.H.; Okimoto, R. DNA marker technology: A revolution in animal genetics. Poult. Sci. 1997, 76, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- Mäki-Tanila, A. Animal molecular genetics from major genes to genomics. In Sustainable Food Production; Christou, P., Savin, R., Costa-Pierce, B.A., Misztal, I., Whitelaw, C.B.A., Eds.; Springer: New York, NY, USA, 2013; pp. 127–151. [Google Scholar] [CrossRef]
- Andersson, L.; Purugganan, M. Molecular genetic variation of animals and plants under domestication. Proc. Natl. Acad. Sci. USA 2022, 119, e2122150119. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.L.; May, P.E. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am. J. Hum. Genet. 1989, 44, 388–396. [Google Scholar] [PubMed] [PubMed Central]
- Liu, Z.J.; Cordes, J.F. DNA marker technologies and their applications in aquaculture genetics. Aquaculture 2004, 238, 1–37. [Google Scholar] [CrossRef]
- Mohamed, L. DNA markers and their application in animal genetics: An overview. Sudan J. Vet. Res. 2006, 21, 1–13. Available online: https://sudanjvr.net/journal/29.pdf (accessed on 30 December 2024).
- Teneva, A. Molecular markers in animal genome analysis. Biotechnol. Anim. Husb. 2009, 25, 1267–1284. Available online: https://www.researchgate.net/publication/267403087 (accessed on 30 December 2024).
- Weigend, S.; Romanov, M.N.; Ben-Ari, G.; Hillel, J. Overview on the Use of Molecular Markers to Characterize Genetic Diversity in Chickens. In Proceedings of the XXII World’s Poultry Congress & Exhibition, Participant List & Full Text CD + Book of Abstracts, Istanbul, Turkey, 8–13 June 2004; WPSA—Turkish Branch: Istanbul, Turkey, 2004; p. 192. Available online: https://www.researchgate.net/publication/372751440 (accessed on 30 December 2024).
- Moiseeva, I.G.; Volokhovich, V.A.; Raetskii, A.V.; Mamontova, I.V.; Kostriukov, G.N. Genetic structure of Moskovskaia chickens of M5 line by biochemical markers. In Teoriia i praktika razvedeniia sel’skokhoziaistvennykh zhivotnykh [Theory and Practice of Breeding Farm Animals]; Puponin, A.I., Ed.; TSKhA: Moscow, Russia, 1981; pp. 84–88. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=tFr9MGIAAAAJ:ns9cj8rnVeAC (accessed on 30 December 2024). (In Russian)
- Moiseeva, I.G.; Volokhovich, V.A.; Tolokonnikova, E.V.; Altukhov, Y.P. Differentiation of fowl breeds for biochemical marker genes. Genetika 1984, 20, 672–681. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19840180407 (accessed on 30 December 2024).
- Romanov, M.N.; Chernikov, V.F. Polymorphic Ovoproteins As Markers of Intra-population Variability in Black-headed Gull. In Proceedings of the 1st International Conference “Molecular-Genetic Markers of Animals”, Kyiv, Ukraine, 27–29 January 1994; Zubets, M.V., Ed.; UAAS. Agrarna nauka: Kyiv, Ukraine, 1994; pp. 35–36. Available online: https://kar.kent.ac.uk/46307/ (accessed on 30 December 2024). (In Russian).
- Kutnyuk, P.I.; Gadyuchko, O.T.; Bondarenko, Y.V. Locus Om As Body Weight Marker in Process of Frequent-dependent Selection of Turkeys. In Proceedings of the 10th European Poultry Conference “The Poultry Industry Towards the 21st Century”, Jerusalem, Israel, 21–26 June 1998; World’s Poultry Science Association: Jerusalem, Israel, 1998; p. 67. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=c1PnDYEAAAAJ:mlAyqtXpCwEC (accessed on 30 December 2024).
- Semyenova, S.K.; Moiseev, I.G.; Vasil’ev, V.A.; Filenko, A.L.; Nikiforov, A.A.; Sevast’yanova, A.A.; Ryskov, A.P. Genetic Polymorphisms by DNA and Biochemical Markers in Old Russian Chicken Breeds. In Proceedings of the Biodiversity and Dynamics of Ecosystems in North Eurasia: First International Conference, Novosibirsk, Russia, 21–26 August 2000; Russian Academy of Sciences, Siberian Branch: Novosibirsk, Russia, 2000; Volume 1, Pt. 3. pp. 101–102. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=tFr9MGIAAAAJ:URolC5Kub84C (accessed on 30 December 2024). (In Russian).
- Semenova, S.K.; Moiseeva, I.G.; Vasil’ev, V.A.; Filenko, A.L.; Nikiforov, A.A.; Sevast’ianova, A.A.; Ryskov, A.P. Geneticheskiĭ polimorfizm russkikh, evropeĭskikh i aziatskikh porod kur, vyiavliaemyĭ s pomoshch’iu DNK i belkovykh markerov [Genetic polymorphism of Russian, European, and Asian chicken breeds as revealed with DNA and protein markers]. Genetika 2002, 38, 1304–1308, (In Russian with English Summary). [Google Scholar] [PubMed]
- Moiseyeva, I.G.; Nikiforov, A.A.; Sevast’yanova, A.A.; Semyenova, S.K. [Estimation of the level of genetic variability of chicken populations using different genetic markers]. BIO 2006, 1, 14. Available online: https://web.archive.org/web/20121221052719/http://lab-cga.ru/articles/Jornal02/Statia3.htm (accessed on 21 December 2012). (In Russian).
- Weigend, S.; Romanov, M.N. Genetische Diversitätsanalysen bei Hühnern mit Hilfe molekularer Marker—Assessment of genetic diversity in chickens using molecular markers. In Jahresbericht 2001; Bundesforschungsanstalt für Landwirtschaft (FAL): Braunschweig, Germany, 2002; p. 67. Available online: https://www.researchgate.net/publication/371722377 (accessed on 30 December 2024).
- Dementeva, N.V.; Romanov, M.N.; Kudinov, A.A.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Terletsky, V.P.; Fedorova, E.S.; Nikitkina, E.V.; Plemyashov, K.V. Studying the structure of a gene pool population of the Russian White chicken breed by genome-wide SNP scan. Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 2017, 52, 1166–1174. [Google Scholar] [CrossRef]
- Deniskova, T.E.; Dotsev, A.V.; Abdelmanova, A.S.; Petrov, S.N.; Frolov, A.N.; Platonov, S.A.; Gladyr, E.A.; Gusev, I.V.; Selionova, M.I.; Rodionov, A.N.; et al. Genetic diversity in the Orenburg goat breed revealed by single-nucleotide polymorphism (SNP) analysis: Initial steps in saving a threatened population. Genes 2024, 15, 1375. [Google Scholar] [CrossRef] [PubMed]
- Volkova, N.A.; Romanov, M.N.; Abdelmanova, A.S.; Larionova, P.V.; German, N.Y.; Vetokh, A.N.; Shakhin, A.V.; Volkova, L.A.; Sermyagin, A.A.; Anshakov, D.V.; et al. Genome-wide association study revealed putative SNPs and candidate genes associated with growth and meat traits in Japanese quail. Genes 2024, 15, 294. [Google Scholar] [CrossRef] [PubMed]
- Volkova, N.A.; Kotova, T.O.; Vetokh, A.N.; Larionova, P.V.; Volkova, L.A.; Romanov, M.N.; Zinovieva, N.A. Genome-wide association study of testes development indicators in roosters (Gallus gallus L.). Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 2024, 59, 649–657, (In Russian with English Summary). [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed] [PubMed Central]
- Smith, J.S.C.; Chin, E.C.L.; Shu, H.; Smith, O.S.; Wall, S.J.; Senior, M.L.; Mitchell, S.E.; Kresovich, S.; Ziegle, J. An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): Comparisons with data from RFLPS and pedigree. Theor. Appl. Genet. 1997, 95, 163–173. [Google Scholar] [CrossRef]
- Dementieva, N.V.; Shcherbakov, Y.S.; Tyshchenko, V.I.; Terletsky, V.P.; Vakhrameev, A.B.; Nikolaeva, O.A.; Ryabova, A.E.; Azovtseva, A.I.; Mitrofanova, O.V.; Peglivanyan, G.K.; et al. Comparative analysis of molecular RFLP and SNP markers in assessing and understanding the genetic diversity of various chicken breeds. Genes 2022, 13, 1876. [Google Scholar] [CrossRef]
- Romanov, M.N.; Weigend, S. Using RAPD markers for assessment of genetic diversity in chickens. Arch. Geflugelkd. 2001, 65, 145–148. Available online: https://kar.kent.ac.uk/46415/ (accessed on 30 December 2024). [CrossRef]
- Semyenova, S.K.; Moiseev, I.G.; Vasil’ev, V.A.; Filenko, A.L.; Nikiforov, A.A.; Sevast’yanova, A.A.; Ryskov, A.P. Genetic polymorphism of Russian, European, and Asian chicken breeds as revealed with DNA and protein markers. Russ. J. Genet. 2002, 38, 1109–1112. [Google Scholar] [CrossRef]
- Dehghanzadeh, H.; Mirhosseini, S.Z.; Romanov, M.N.; Ghorbani, A. Evaluation of genetic variability and distances among five Iranian native chicken populations using RAPD markers. Pak. J. Biol. Sci. 2009, 12, 866–871. [Google Scholar] [CrossRef]
- Buschiazzo, E.; Gemmell, N.J. Conservation of human microsatellites across 450 million years of evolution. Genome Biol. Evol. 2010, 2, 153–165. [Google Scholar] [CrossRef]
- Adams, R.H.; Blackmon, H.; Reyes-Velasco, J.; Schield, D.R.; Card, D.C.; Andrew, A.L.; Waynewood, N.; Castoe, T.A. Microsatellite landscape evolutionary dynamics across 450 million years of vertebrate genome evolution. Genome 2016, 59, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Fulton, J.E. Molecular genetics in a modern poultry breeding organization. Worlds Poult. Sci. J. 2008, 64, 171–176. [Google Scholar] [CrossRef]
- Ariyaraphong, N.; Ho My Nguyen, D.; Singchat, W.; Suksavate, W.; Panthum, T.; Langkaphin, W.; Chansitthiwet, S.; Angkawanish, T.; Promking, A.; Kaewtip, K.; et al. Standard identification certificate for legal legislation of a unique gene pool of Thai domestic elephants originating from a male elephant contribution to breeding. Sustainability 2022, 14, 15355. [Google Scholar] [CrossRef]
- Ariyaraphong, N.; Wongloet, W.; Wattanadilokchatkun, P.; Panthum, T.; Singchat, W.; Thong, T.; Lisachov, A.; Ahmad, S.F.; Muangmai, N.; Han, K.; et al. Should the identification guidelines for Siamese crocodiles be revised? Differing post-occipital scute scale numbers show phenotypic variation does not result from hybridization with saltwater crocodiles. Biology 2023, 12, 535. [Google Scholar] [CrossRef]
- Beuzen, N.D.; Stear, M.J.; Chang, K.C. Molecular markers and their use in animal breeding. Vet. J. 2000, 160, 42–52. [Google Scholar] [CrossRef]
- Podstreshnyi, O.P.; Tereshchenko, O.V.; Tkachyk, T.E.; Podstreshna, I.O.; Ishchenko, Y.B. Genetic Identification and Passportization of Poultry Breeds and Lines: Methodical Recommendations; Poultry Research Institute of the Ukrainian Academy of Agrarian Sciences: Birky, Ukraine, 2009; Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=deB5xCwAAAAJ:j8SEvjWlNXcC (accessed on 30 December 2024). (In Ukrainian)
- Duran, C.; Appleby, N.; Edwards, D.; Batley, J. Molecular genetic markers: Discovery, applications, data storage and visualization. Curr. Bioinform. 2009, 4, 16–27. [Google Scholar] [CrossRef]
- Wakchaure, R.; Ganguly, S.; Para, P.A.; Praveen, P.K.; Qadri, K. Molecular markers and their applications in farm animals: A review. Int. J. Recent Biotechnol. 2015, 3, 23–29. Available online: https://www.researchgate.net/publication/289602090 (accessed on 30 December 2024).
- Burkat, V.P.; Yefimenko, M.Y.; Podoba, B.Y.; Dzitsyuk, V.V. Scientific and applied aspects of genetic monitoring in livestock breeding. Vìsnik agrarnoï nauki [Bull. Agric. Sci.] 2003, 5, 32–39. Available online: https://lib.dsau.dp.ua/book/3885 (accessed on 30 December 2024).
- Naqvi, A.N. Application of molecular genetic technologies in livestock production: Potentials for developing countries. Adv. Biores. 2007, 1, 72–84. Available online: https://www.idosi.org/abr/1(3-4)/1.pdf (accessed on 30 December 2024).
- Kulibaba, R.O. Theoretical Justification and Practical Implementation of Marker-Associated Selection of Ukrainian Local Breeds of Chickens; NUBiP of Ukraine: Kyiv, Ukraine, 2021; ISBN 978-617-7878-71-0. Available online: https://dglib.nubip.edu.ua/server/api/core/bitstreams/b46b64c9-0f91-4078-b35b-569dd4229ecb/content (accessed on 30 December 2024).
- Kutnyuk, P.I.; Kotik, A.I.; Trufanova, V.A.; Bondarenko, Y.V. A Search for Genetic-biological Markers of Resistance to Mycotoxin T-2 in Fowls. In Nauch.-Proizv. Konf. “Nov. Metody Selektsii i Biotekhnol. v Zhivotnovod.”, Ch. 2. Reprod., Populyats. Genet. i Biotekhnol. [Scientific and Production Conference “New Methods of Selection and Biotechnology in Animal Husbandry”, Ch. 2. Reproduction, Population Genetics and Biotechnology]; Kiev; Resp. Proizv.-Nauch. Assots. po Vnedreniyu Nauch.-Tekhn. Progressa v Zhivotnovod.: Kiev, Ukraine, 1991; pp. 108–109. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19920198531 (accessed on 30 December 2024). (In Russian)
- Cregan, P.B.; Bhagwat, A.A.; Akkaya, M.S.; Rongwen, J. Microsatellite fingerprinting and mapping of soybean. Methods Mol. Cell Biol. 1994, 5, 49–61. Available online: https://scholar.google.com/scholar?q=”Microsatellite+fingerprinting+and+mapping+of+soybean” (accessed on 30 December 2024).
- Bumstead, N. Genomic mapping of resistance to Marek’s disease. Avian Pathol. 1998, 27 (Suppl. 1), S78–S81. [Google Scholar] [CrossRef]
- Menotti-Raymond, M.; David, V.A.; Lyons, L.A.; Schäffer, A.A.; Tomlin, J.F.; Hutton, M.K.; O’Brien, S.J. A genetic linkage map of microsatellites in the domestic cat (Felis catus). Genomics 1999, 57, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Wardecka, B.; Olszewski, R.; Jaszczak, K.; Zieba, G.; Pierzchała, M.; Wicińska, K. Relationship between microsatellite marker alleles on chromosomes 1-5 originating from the Rhode Island Red and Green-legged Partrigenous breeds and egg production and quality traits in F2 mapping population. J. Appl. Genet. 2002, 43, 319–329. Available online: https://www.researchgate.net/publication/11208879 (accessed on 30 December 2024). [PubMed]
- Romanov, M.N.; Da, Y.; Chemnick, L.G.; Thomas, S.M.; Dandekar, S.S.; Papp, J.C.; Ryder, O.A. Towards a genetic linkage map of the California condor, an endangered New World vulture species. Animals 2022, 12, 3266. [Google Scholar] [CrossRef]
- Eggert, L.S.; Rasner, C.A.; Woodruff, D.S. The evolution and phylogeography of the African elephant inferred from mitochondrial DNA sequence and nuclear microsatellite markers. Proc. R. Soc. Lond. B Biol. Sci. 2002, 269, 1993–2006. [Google Scholar] [CrossRef]
- Ali, F.; Suhail, S.M.; Khan, F.A.; Ahmad, I. Microsatellite markers & mitochondrial D-loop based phylogenetic and diversity analysis in Gabrali cattle. Trop. Anim. Health Prod. 2024, 56, 380. [Google Scholar] [CrossRef]
- Senan, S.; Kizhakayil, D.; Sasikumar, B.; Sheeja, T.E. Methods for development of microsatellite markers: An overview. Not. Sci. Biol. 2014, 6, 1–13. [Google Scholar] [CrossRef]
- Schlötterer, C. The evolution of molecular markers—Just a matter of fashion? Nat. Rev. Genet. 2004, 5, 63–69. [Google Scholar] [CrossRef]
- Arif, I.A.; Khan, H.A.; Shobrak, M.; Al Homaidan, A.A.; Al Sadoon, M.; Al Farhan, A.H.; Bahkali, A.H. Interpretation of electrophoretograms of seven microsatellite loci to determine the genetic diversity of the Arabian Oryx. Genet. Mol. Res. 2010, 9, 259–265. [Google Scholar] [CrossRef]
- Zhivotovsky, L.A. Microsatellite variation in human populations and the methods of their analysis. Vestnik VOGiS [Bull. Vavilov Soc. Genet. Breed.] 2006, 10, 74–96. Available online: https://www.elibrary.ru/item.asp?id=9573138 (accessed on 30 December 2024). (In Russian with English Summary).
- Hartley, G.; O’Neill, R.J. Centromere repeats: Hidden gems of the genome. Genes 2019, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Beklemisheva, V.R.; Lemskaya, N.A.; Prokopov, D.Y.; Perelman, P.L.; Romanenko, S.A.; Proskuryakova, A.A.; Serdyukova, N.A.; Utkin, Y.A.; Nie, W.; Ferguson-Smith, M.A.; et al. Maps of constitutive-heterochromatin distribution for four Martes species (Mustelidae, Carnivora, Mammalia) show the formative role of macrosatellite repeats in interspecific variation of chromosome structure. Genes 2023, 14, 489. [Google Scholar] [CrossRef] [PubMed]
- Miret, J.J.; Pessoa-Brandão, L.; Lahue, R.S. Instability of CAG and CTG trinucleotide repeats in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997, 17, 3382–3387. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Y.C.; Korol, A.B.; Fahima, T.; Nevo, E. Microsatellites within genes: Structure, function, and evolution. Mol. Biol. Evol. 2004, 21, 991–1007. [Google Scholar] [CrossRef]
- Rajendrakumar, P.; Biswal, A.K.; Balachandran, S.M.; Srinivasarao, K.; Sundaram, R.M. Simple sequence repeats in organellar genomes of rice: Frequency and distribution in genic and intergenic regions. Bioinformatics 2007, 23, 1–4. [Google Scholar] [CrossRef]
- FAO. Measurement of Domestic Animal Diversity (MoDAD): New Recommended Microsatellite Markers. New Microsatellite Marker Sets—Recommendations of Joint ISAG/FAO Standing Committee; Food and Agriculture Organization of the United Nations: Rome, Italy, 2004; Available online: http://www.fao.org/3/a-aq569e.pdf (accessed on 30 December 2024).
- FAO. Molecular Genetic Characterization of Animal Genetic Resources; FAO Animal Production and Health Guidelines, No. 9. Commission on Genetic Resources for Food and Agriculture; Food and Agriculture Organization of the United Nations: Rome, Italy, 2011; Available online: https://web.archive.org/web/20240214081209/https://www.fao.org/3/i2413e/i2413e00.pdf (accessed on 14 February 2024).
- Romanov, M.N.; Betuel, A.M.; Chemnick, L.G.; Ryder, O.A.; Kulibaba, R.O.; Tereshchenko, O.V.; Payne, W.S.; Delekta, P.C.; Dodgson, J.B.; Tuttle, E.M.; et al. Widely applicable PCR markers for sex identification in birds. Russ. J. Genet. 2019, 55, 220–231. [Google Scholar] [CrossRef]
- Nyström, V.; Humphrey, J.; Skoglund, P.; McKeown, N.J.; Vartanyan, S.; Shaw, P.W.; Lidén, K.; Jakobsson, M.; Barnes, I.; Angerbjörn, A.; et al. Microsatellite genotyping reveals end-Pleistocene decline in mammoth autosomal genetic variation. Mol. Ecol. 2012, 21, 3391–3402. [Google Scholar] [CrossRef]
- Hata, A.; Nunome, M.; Suwanasopee, T.; Duengkae, P.; Chaiwatana, S.; Chamchumroon, W.; Suzuki, T.; Koonawootrittriron, S.; Matsuda, Y.; Srikulnath, K. Origin and evolutionary history of domestic chickens inferred from a large population study of Thai red junglefowl and indigenous chickens. Sci. Rep. 2021, 11, 2035. [Google Scholar] [CrossRef]
- Papežík, P.; Sciberras, A.; Benovics, M.; Sciberras, J.; Deidun, A.; Mikulíček, P. Far from home: Tracing the origin of non-native water frogs (genus Pelophylax) in Malta by molecular markers. Biol. Invasions 2024, 26, 1045–1059. [Google Scholar] [CrossRef]
- Budi, T.; Singchat, W.; Tanglertpaibul, N.; Wongloet, W.; Chaiyes, A.; Ariyaraphong, N.; Thienpreecha, W.; Wannakan, W.; Mungmee, A.; Thong, T.; et al. Thai local chicken breeds, Chee Fah and Fah Luang, originated from Chinese black-boned chicken with introgression of red junglefowl and domestic chicken breeds. Sustainability 2023, 15, 6878. [Google Scholar] [CrossRef]
- Jaito, W.; Singchat, W.; Patta, C.; Thatukan, C.; Kumnan, N.; Chalermwong, P.; Budi, T.; Panthum, T.; Wongloet, W.; Wattanadilokchatkun, P.; et al. Shared alleles and genetic structures in different Thai domestic cat breeds: The possible influence of common racial origins. Genom. Inform. 2024, 22, 12. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Choi, S.K.; Sommer, J.; Louis Jr, E.; Brenneman, R.; Zemanová, B.; Hájková, P.; Park, G.; Min, M.S.; Kim, K.S.; et al. A core set of microsatellite markers for conservation genetics studies of Korean goral (Naemorhedus caudatus) and its cross-species amplification in Caprinae species. J. Vet. Sci. 2010, 11, 351. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; O’Meally, D.; Azad, B.; Georges, A.; Sarre, S.D.; Graves, J.A.M.; Matsuda, Y.; Ezaz, T. Amplification of microsatellite repeat motifs is associated with the evolutionary differentiation and heterochromatinization of sex chromosomes in Sauropsida. Chromosoma 2016, 125, 111–123. [Google Scholar] [CrossRef]
- Kayser, M.; Caglià, A.; Corach, D.; Fretwell, N.; Gehrig, C.; Graziosi, G.; Heidorn, F.; Herrmann, S.; Herzog, B.; Hidding, M.; et al. Evaluation of Y-chromosomal STRs: A multicenter study. Int. J. Legal Med. 1997, 110, 125–133. [Google Scholar] [CrossRef]
- Nyakaana, S.; Arctander, P. Isolation and characterization of microsatellite loci in the African elephant, Loxodonta africana. Mol. Ecol. 1998, 7, 1436–1437. Available online: https://onlinelibrary.wiley.com/doi/10.1111/j.1365-294X.1998.00445.x (accessed on 30 December 2024). [PubMed]
- Velasco, V.V.; Tsudzuki, M.; Hashimoto, N.; Goto, N.; Ishikawa, A. Genetic diversity, runs of homozygosity, and selection signatures in native Japanese chickens: Insights from single-nucleotide polymorphisms. Animals 2024, 14, 3341. [Google Scholar] [CrossRef]
- Zimmerman, S.J.; Aldridge, C.L.; Oyler-McCance, S.J. An empirical comparison of population genetic analyses using microsatellite and SNP data for a species conservation concern. BMC Genom. 2020, 21, 382. [Google Scholar] [CrossRef]
- Kutnyuk, P.I.; Moiseeva, I.G. Information and Statistical Aspects of Modeling Population Processes Using Molecular Genetic Markers. In Proceedings of the 1st International Conference “Molecular-Genetic Markers of Animals”, Kyiv, Ukraine, 27–29 January 1994; Zubets, M.V., Ed.; UAAS. Agrarna nauka: Kyiv, Ukraine, 1994; pp. 132–133. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=tFr9MGIAAAAJ:7T2F9Uy0os0C (accessed on 30 December 2024). (In Russian).
- Hamilton, M.B. Population Genetics; Wiley-Blackwell: Chichester, UK, 2009; Available online: https://books.google.com/books?id=jc0cEAAAQBAJ (accessed on 30 December 2024).
- Wright, S. Evolution and the Genetics of Populations. Vol. 4. Variability Within and Among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978; Available online: https://books.google.com/books?id=q0RDJf3K_aUC (accessed on 30 December 2024).
- Nei, M.; Chesser, R.K. Estimation of fixation indices and gene diversities. Ann. Hum. Genet. 1983, 47, 253–259. [Google Scholar] [CrossRef]
- Chesnokov, Y.V.; Artemyeva, A.M. Evaluation of the measure of polymorphism information of genetic diversity. Sel’skokhozyaistvennaya Biol. [Agric. Biol.] 2015, 50, 571–578. [Google Scholar] [CrossRef]
- Serrote, C.M.L.; Reiniger, L.R.S.; Silva, K.B.; dos Santos Rabaiolli, S.M.; Stefanel, C.M. Determining the Polymorphism Information Content of a molecular marker. Gene 2020, 726, 144175. [Google Scholar] [CrossRef]
- Roy, P.; Sogir, S.B.; Basak, T. On the Polymorphism Information Content (PIC)—A practical application for the DNA sequencing data. Eur. J. Med. Health Res. 2023, 1, 21–29. [Google Scholar] [CrossRef]
- Hildebrand, C.E.; Torney, D.C.; Wagner, R.P. Informativeness of polymorphic DNA markers. Los Alamos Sci. 1992, 20, 100–102. Available online: https://sgp.fas.org/othergov/doe/lanl/pubs/00326695.pdf (accessed on 30 December 2024).
- Elston, R.C. Polymorphism information content. In Encyclopedia of Biostatistics, 2nd ed.; Armitage, P., Colton, T., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2005; Volume 2. [Google Scholar] [CrossRef]
- Rasoarahona, R.; Wattanadilokchatkun, P.; Panthum, T.; Thong, T.; Singchat, W.; Ahmad, S.F.; Chaiyes, A.; Han, K.; Kraichak, E.; Muangmai, N.; et al. Optimizing microsatellite marker panels for genetic diversity and population genetic studies: An ant colony algorithm approach with polymorphic information content. Biology 2023, 12, 1280. [Google Scholar] [CrossRef] [PubMed]
- Waits, L.P.; Luikart, G.; Taberlet, P. Estimating the probability of identity among genotypes in natural populations: Cautions and guidelines. Mol. Ecol. 2001, 10, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Zhivotovsky, L.A.; Feldman, M.W. Microsatellite variability and genetic distances. Proc. Natl. Acad. Sci. USA 1995, 92, 11549–11552. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT Computer Package for PCs; Institute of Ecology, UNIL: Lausanne, Switzerland, 2002; Available online: https://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 30 December 2024).
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Rousset, F. GENEPOP’007: A complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. GENETIX 4.05, Logiciel sous WindowsTM pour la Génétique des Populations; Laboratoire Génome, Populations, Interactions, CNRS UMR 5000; Université de Montpellier II: Montpellier, France, 2004; Available online: https://kimura.univ-montp2.fr/genetix/ (accessed on 30 December 2024). (In French)
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- MEGA: Molecular Evolutionary Genetics Analysis, version 12; Temple University College of Science and Technology: Philadelphia, PA, USA. Available online: https://www.megasoftware.net/ (accessed on 30 December 2024).
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods); Version 4; Sinauer Associates: Sunderland, MA, USA, 2003; Available online: https://paup.phylosolutions.com/ (accessed on 30 December 2024).
- Felsenstein, J. Notices: PHYLIP–Phylogeny Inference Package (Version 3.2). Cladistics 1989, 5, 164–166. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Park, S.D.E. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection. Ph.D. Thesis, University of Dublin, Dublin, Ireland, 2001. Available online: https://web.archive.org/web/20110410053714/http://www.animalgenomics.ucd.ie/sdepark/ms-toolkit/ (accessed on 10 April 2011).
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Beaumont, M.A. Detecting population expansion and decline using microsatellites. Genetics 1999, 153, 2013–2029. [Google Scholar] [CrossRef]
- Colorni, A.; Dorigo, M.; Maniezzo, V. Distributed optimization by ant colonies. In Proceedings of the First European Conference on Artificial Life, Paris, France, 11–13 December 1991; Elsevier: Amsterdam, The Netherlands, 1991; Volume 142, pp. 134–142. Available online: https://faculty.washington.edu/paymana/swarm/colorni92-ecal.pdf (accessed on 30 December 2024).
- Al Salami, N.M.A. Ant colony optimization algorithm. UbiCC J. 2009, 4, 823–826. Available online: https://web.archive.org/web/20140713122934/http://www.ubicc.org/files/pdf/10_336.pdf (accessed on 13 July 2014).
- Yu, H.; Gu, G.; Liu, H.; Shen, J.; Zhao, J. A modified ant colony optimization algorithm for tumor marker gene selection. Genom. Proteom. Bioinform. 2009, 7, 200–208. [Google Scholar] [CrossRef]
- Rasoarahona, R.; Wattanadilokchatkun, P.; Panthum, T.; Jaisamut, K.; Lisachov, A.; Thong, T.; Singchat, W.; Ahmad, S.F.; Han, K.; Kraichak, E.; et al. MicrosatNavigator: Exploring nonrandom distribution and lineage-specificity of microsatellite repeat motifs on vertebrate sex chromosomes across 186 whole genomes. Chromosome Res. 2023, 31, 29. [Google Scholar] [CrossRef]
- Al-Samarai, F.R.; Al-Kazaz, A.A. Molecular markers and its applications in animal breeding: A review. Am. J. Appl. Sci. Res. 2015, 1, 1–5. Available online: https://www.researchgate.net/publication/338840588 (accessed on 30 December 2024). [CrossRef]
- Paetkau, D.; Calvert, W.; Stirling, I.; Strobeck, C. Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol. 1995, 4, 347–354. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Hellemans, B.; Volckaert, F.A. Microsatellites and their genomic distribution, evolution, function and applications: A review with special reference to fish genetics. Aquaculture 2006, 255, 1–29. [Google Scholar] [CrossRef]
- Kalia, R.K.; Rai, M.K.; Kalia, S.; Singh, R.; Dhawan, A.K. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- Rubtsova, G.A.; Ponomareva, E.V.; Afanasiev, K.I.; Shaikhaev, E.G.; Kholodova, M.V.; Pavlov, S.D.; Zhivotovsky, L.A. A detection of allelic variants at microsatellite markers by using capillary and traditional electrophoresis. Russ. J. Genet. 2016, 52, 423–427. [Google Scholar] [CrossRef]
- Chokan, T.; Radko, A.; Tarasjuk, S.; Szumiec, A.; Rubiś, D. Genetic structure of Ukrainian Mountain Carpathian sheep by use of microsatellite loci. Rozvedennâ ì genetika tvarin [Anim. Breed. Genet.] 2016, 51, 225–230. Available online: https://digest.iabg.org.ua/genetics/item/724-51-030 (accessed on 30 December 2024). (In Ukrainian with English Summary).
- Gholizadeh, M.; Mianji, G.R. Use of microsatellite markers in poultry research. Int. J. Poult. Sci. 2007, 6, 145–153. [Google Scholar] [CrossRef][Green Version]
- Muchadeyi, F.C.; Eding, H.; Wollny, C.B.A.; Groeneveld, E.; Makuza, S.M.; Shamseldin, R.; Simianer, H.; Weigend, S. Absence of population substructuring in Zimbabwe chicken ecotypes inferred using microsatellite analysis. Anim. Genet. 2007, 38, 332–339. [Google Scholar] [CrossRef]
- Sazanov, A.A.; Romanov, M.N.; Wardecka, B.; Sazanova, A.L.; Korczak, M.; Stekol’nikova, V.A.; Kozyreva, A.A.; Smirnov, A.F.; Dodgson, J.B.; Jaszczak, K. Chromosomal localization of GGA4 BACs containing QTL-linked microsatellites. Cytogenet. Genome Res. 2004, 106, 19. [Google Scholar] [CrossRef]
- Loywyck, V.; Bed’hom, B.; Pinard-van der Laan, M.H.; Pitel, F.; Verrier, É.; Bijma, P. Evolution of the polymorphism at molecular markers in QTL and non-QTL regions in selected chicken lines. Genet. Sel. Evol. 2008, 40, 639–661. [Google Scholar] [CrossRef]
- McElroy, J.P.; Dekkers, J.C.; Fulton, J.E.; O’Sullivan, N.P.; Soller, M.; Lipkin, E.; Zhang, W.; Koehler, K.J.; Lamont, S.J.; Cheng, H.H. Microsatellite markers associated with resistance to Marek’s disease in commercial layer chickens. Poult. Sci. 2005, 84, 1678–1688. [Google Scholar] [CrossRef]
- Sazanov, A.A.; Romanov, M.N.; Wardecka, B.; Sazanova, A.L.; Korczak, M.; Stekol’nikova, V.A.; Kozyreva, A.A.; Smirnov, A.F.; Jaszczak, K.; Dodgson, J.B. Chromosomal localization of 15 large insert BAC clones containing three microsatellites on chicken chromosome 4 (GGA4) which refine its centromere position. Anim. Genet. 2005, 36, 161–163. [Google Scholar] [CrossRef]
- Elzarei, M.F.; Alhasyani, M.S.; Al-Sharari, S.A.; Alodhiby, S.I.; Mousa, E.F. Associations between microsatellites markers and growth traits in goat. J. Agric. Sci. 2024, 16, 53–59. [Google Scholar] [CrossRef]
- Romanov, M.N. [Population and Genetic Characterization of Poultry for Autosexing and Other Marker Traits]. Author’s Abstract of the Candidate of Biological Sciences Thesis; Kharkiv State University: Kharkiv, Ukraine, 1996. Available online: https://kar.kent.ac.uk/46333/ (accessed on 30 December 2024). (In Russian with English Summary).
- Romanov, M.N.; Weigend, S. Genetic Diversity in Chicken Populations Based on Microsatellite Markers. In Proceedings of the Conference “From Jay Lush to Genomics: Visions for Animal Breeding and Genetics”, Ames, IA, USA, 16–18 May 1999; Dekkers, J.C.M., Lamont, S.J., Rothschild, M.F., Eds.; Department of Animal Science, Iowa State University: Ames, IA, USA, 1999; p. 174. Available online: https://web.archive.org/web/20050314091227/http://www.agbiotechnet.com/proceedings/jaylush.asp#34 (accessed on 14 March 2005).
- Garza, J.C.; Williamson, E.G. Detection of reduction in population size using data from microsatellite loci. Mol. Ecol. 2001, 10, 305–318. [Google Scholar] [CrossRef]
- Wang, R.; Zheng, L.; Touré, Y.T.; Dandekar, T.; Kafatos, F.C. When genetic distance matters: Measuring genetic differentiation at microsatellite loci in whole-genome scans of recent and incipient mosquito species. Proc. Natl. Acad. Sci. USA 2001, 98, 10769–10774. [Google Scholar] [CrossRef]
- Chapuis, M.P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef]
- Brown, W.R.A.; MacKinnon, P.J.; Villasanté, A.; Spurr, N.; Buckle, V.J.; Dobson, M.J. Structure and polymorphism of human telomere-associated DNA. Cell 1990, 63, 119–132. [Google Scholar] [CrossRef]
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef]
- Garza, J.C.; Slatkin, M.; Freimer, N.B. Microsatellite allele frequencies in humans and chimpanzees, with implications for constraints on allele size. Mol. Biol. Evol. 1995, 12, 594–603. [Google Scholar] [CrossRef]
- Coote, T.; Bruford, M.W. Human microsatellites applicable for analysis of genetic variation in apes and Old World monkeys. J. Hered. 1996, 87, 406–410. [Google Scholar] [CrossRef][Green Version]
- Subramanian, S.; Mishra, R.K.; Singh, L. Genome-wide analysis of microsatellite repeats in humans: Their abundance and density in specific genomic regions. Genome Biol. 2003, 4, R13. [Google Scholar] [CrossRef]
- Blanquer-Maumont, A.; Crouau-Roy, B. Polymorphism, monomorphism, and sequences in conserved microsatellites in primate species. J. Mol. Evol. 1995, 41, 492–497. [Google Scholar] [CrossRef]
- Goossens, B.; Chikhi, L.; Utami, S.S.; De Ruiter, J.; Bruford, M.W. A multi-samples, multi-extracts approach for microsatellite analysis of faecal samples in an arboreal ape. Conserv. Genet. 2000, 1, 157–162. [Google Scholar] [CrossRef]
- Nair, S.; Ha, J.; Rogers, J. Nineteen new microsatellite DNA polymorphisms in pigtailed macaques (Macaca nemestrina). Primates 2000, 41, 343–350. [Google Scholar] [CrossRef]
- Winkler, L.A.; Zhang, X.; Ferrell, R.; Wagner, R.; Dahl, J.; Peter, G.; Sohn, R. Geographic microsatellite variability in Central American howling monkeys. Int. J. Primatol. 2004, 25, 197–210. [Google Scholar] [CrossRef]
- Clisson, I.; Lathuilliere, M.; Crouau-Roy, B. Conservation and evolution of microsatellite loci in primate taxa. Am. J. Primatol. 2000, 50, 205–214. [Google Scholar] [CrossRef]
- Oklander, L.I.; Steinberg, E.R.; Mudry, M.D. A new world monkey microsatellite (AP74) highly conserved in primates. Acta Biol. Colomb. 2012, 17, 93–101. Available online: https://web.archive.org/web/20170811191344/http://www.scielo.org.co/pdf/abc/v17n1/v17n1a7.pdf (accessed on 11 August 2017).
- Nergadze, S.G.; Rocchi, M.; Azzalin, C.M.; Mondello, C.; Giulotto, E. Insertion of telomeric repeats at intrachromosomal break sites during primate evolution. Genome Res. 2004, 14, 1704–1710. [Google Scholar] [CrossRef]
- O’Sullivan, R.J.; Karlseder, J. Telomeres: Protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010, 11, 171–181. [Google Scholar] [CrossRef]
- Bandaria, J.N.; Qin, P.; Berk, V.; Chu, S.; Yildiz, A. Shelterin protects chromosome ends by compacting telomeric chromatin. Cell 2016, 164, 735–746. [Google Scholar] [CrossRef]
- Wyatt, H.D.M.; West, S.C.; Beattie, T.L. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010, 38, 5609–5622. [Google Scholar] [CrossRef]
- Maddar, H.; Ratzkovsky, N.; Krauskopf, A. Role for telomere cap structure in meiosis. Mol. Biol. Cell 2001, 12, 3191–3203. [Google Scholar] [CrossRef]
- Jurka, J.; Pethiyagoda, C. Simple repetitive DNA sequences from primates: Compilation and analysis. J. Mol. Evol. 1995, 40, 120–126. [Google Scholar] [CrossRef]
- Srikulnath, K.; Ahmad, S.F.; Panthum, T.; Malaivijitnond, S. Importance of Thai macaque bioresources for biological research and human health. J. Med. Primatol. 2022, 51, 62–72. [Google Scholar] [CrossRef]
- Satkoski, J.; George, D.; Smith, D.G.; Kanthaswamy, S. Genetic characterization of wild and captive rhesus macaques in China. J. Med. Primatol. 2008, 37, 67–80. [Google Scholar] [CrossRef]
- Koul, Y.; Karthickeyan, S.M.K.; Hepsibha, P.; Jeevan, C.; Jawahar, K.T.P.; Gopinathan, A. Microsatellite based molecular characterization of Nattukuttai–a unique short statured Bos indicus cattle population of southern India. Genetica 2025, 153, 1. [Google Scholar] [CrossRef]
- Ladyka, V.I.; Khmelnychyi, L.M.; Lyashenko, Y.V.; Kulibaba, R.O. Analysis of the genetic structure of a population of Lebedyn cattle by microsatellite markers. Regul. Mech. Biosyst. 2019, 10, 45–49. [Google Scholar] [CrossRef]
- Yang, J.; Zhu, G.F.; Jiang, J.; Xiang, C.L.; Gao, F.L.; Bao, W.D. Non-invasive genetic analysis indicates low population connectivity in vulnerable Chinese gorals: Concerns for segregated population management. Zool. Res. 2019, 40, 439. [Google Scholar] [CrossRef]
- Jangtarwan, K.; Kamsongkram, P.; Subpayakom, N.; Sillapaprayoon, S.; Muangmai, N.; Kongphoemph, A.; Wongsodchuen, A.; Intapan, S.; Chamchumroon, W.; Safoowong, M.; et al. Predictive genetic plan for a captive population of the Chinese goral (Naemorhedus griseus) and prescriptive action for ex situ and in situ conservation management in Thailand. PLoS ONE 2020, 15, e0234064. [Google Scholar] [CrossRef]
- Wongloet, W.; Kongthong, P.; Chaiyes, A.; Singchat, W.; Suksavate, W.; Ariyaraphong, N.; Panthum, T.; Lisachov, A.; Jaisamut, K.; Sonongbua, J.; et al. Genetic monitoring of the last captive population of greater mouse-deer on the Thai mainland and prediction of habitat suitability before reintroduction. Sustainability 2023, 15, 3112. [Google Scholar] [CrossRef]
- Shelyov, A.V.; Kopylov, K.V.; Kramarenko, S.S.; Kramarenko, A.S. Genetic structure of different equine breeds by microsatellite DNA loci. Agric. Sci. Pract. 2020, 7, 3–13. [Google Scholar] [CrossRef]
- Cho, G.J.; Cho, B.W. Microsatellite DNA typing using 16 markers for parentage verification of the Korean native horse. Anim. Biosci. 2004, 17, 750–754. [Google Scholar] [CrossRef]
- Orazymbetova, Z.; Ualiyeva, D.; Dossybayev, K.; Torekhanov, A.; Sydykov, D.; Mussayeva, A.; Baktybayev, G. Genetic diversity of Kazakhstani Equus caballus (Linnaeus, 1758) horse breeds inferred from microsatellite markers. Vet. Sci. 2023, 10, 598. [Google Scholar] [CrossRef]
- Sukri, A.; Dewi, I.N.; Primawati, S.N.; Wangiyana, I.G.A.S.; Muttaqin, Z.; Winaya, A. Revealing the genetic diversity of Sumbawa endemic horse using microsatellite-based DNA fingerprint. Biodiversitas 2022, 23, 837. [Google Scholar] [CrossRef]
- Kim, S.M.; Yun, S.W.; Cho, G.J. Assessment of genetic diversity using microsatellite markers to compare donkeys (Equus asinus) with horses (Equus caballus). Anim. Biosci. 2021, 34, 1460–1465. [Google Scholar] [CrossRef]
- Park, C.S.; Lee, S.Y.; Cho, G.J. Evaluation of recent changes in genetic variability in Thoroughbred horses based on microsatellite markers parentage panel in Korea. Anim. Biosci. 2022, 35, 527–532. [Google Scholar] [CrossRef]
- Wang, Y.; Hua, X.; Shi, X.; Wang, C. Origin, evolution, and research development of donkeys. Genes 2022, 13, 1945. [Google Scholar] [CrossRef]
- Narushin, V.G.; Romanov, M.N.; Sakhatsky, N.I. Modelling growth of chick embryo: Correction for egg weight [Modelowanie wzrostu zarodka kurzego z poprawką na masę jaja]. Zeszyty Naukowe. Przegląd Hodowlany [Anim. Prod. Rev., Appl. Sci. Rep.] 1997, 31, 55–57. Available online: https://www.researchgate.net/publication/355808171 (accessed on 30 December 2024).
- Narushin, V.G.; Laptev, G.Y.; Yildirim, E.A.; Ilina, L.A.; Filippova, V.A.; Kochish, I.I.; Gorfunkel, E.P.; Dubrovin, A.V.; Novikova, N.I.; Novikova, O.B.; et al. Modelling effects of phytobiotic administration on coherent responses to Salmonella infection in laying hens. Ital. J. Anim. Sci. 2020, 19, 282–287. [Google Scholar] [CrossRef]
- Artemenko, O.B.; Tagirov, M.T.; Baydevlyatova, O.M.; Shomina, N.V.; Tereshchenko, A.V. Study of the dynamics of changes in fatty acid composition of egg yolks during embryonic development of hens of different productivity types. Ptakhivnytstvo [Poult. Farming] 2014, 71, 7–17. Available online: https://www.researchgate.net/publication/342833358 (accessed on 30 December 2024). (In Ukrainian with English Summary).
- Singchat, W.; Chaiyes, A.; Wongloet, W.; Ariyaraphong, N.; Jaisamut, K.; Panthum, T.; Ahmad, S.F.; Chaleekarn, W.; Suksavate, W.; Inpota, M.; et al. Red junglefowl resource management guide: Bioresource reintroduction for sustainable food security in Thailand. Sustainability 2022, 14, 7895. [Google Scholar] [CrossRef]
- Canales Vergara, A.M.; Krupij, A.T.; Fresno Baquero, M.D.R.; Macrì, M.; Delgado Bermejo, J.V.; Martínez, A.M. Genetic diversity and population structure of Canarian chicken using microsatellite DNA markers. Ital. J. Anim. Sci. 2024, 23, 678–692. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Wen, X.; Falush, D. Documentation for Structure Software: Version 2.3; Division of the Biological Sciences, The University of Chicago: Chicago, IL, USA, 2010; Available online: https://web.archive.org/web/20130108025033/http://pritch.bsd.uchicago.edu/structure_software/release_versions/v2.3.3/structure_doc.pdf (accessed on 8 January 2013).
- Aoki, A.; Mori, Y.; Okamoto, Y.; Jinno, H. A high-resolution melting-based assay for discriminating a native Japanese chicken, the Nagoya breed, using the ABR0417 microsatellite marker. Eur. Food Res. Technol. 2024, 250, 745–750. [Google Scholar] [CrossRef]
- Podstreshny, A.P.; Bondarenko, Y.V.; Rozhkovskiy, A.V.; Gintovt, V.E. Ispolzovanie markernyih priznakov pri sozdanii perspektivnyih kombinatsiy yaichnyih kur [The use of marker traits in the creation of promising combinations of egg-laying hens]. Ptitsevodstvo [Poult. Farming] 1984, 37, 9–14. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=c1PnDYEAAAAJ:geHnlv5EZngC (accessed on 30 December 2024). (In Russian).
- Bondarenko, Y.V.; Podstreshny, O.P.; Kutnyuk, P.I.; Bogatyr, V.V. Assessment of the Hereditary Diversity of the Chicken Genetic Collection by Marker Traits. In Proceedings of the 1st Ukrainian Poultry Conference, Borky, Simferopol, Ukraine, 6–8 December 1993; World’s Poultry Science Association, Ukrainian Branch, Poultry Research Institute, UAAS: Kharkiv, Ukraine, 1993; pp. 28–29. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=c1PnDYEAAAAJ:XiVPGOgt02cC (accessed on 30 December 2024). (In Ukrainian with English Summary).
- Ostryakova, O.E.; Podstreshny, O.P.; Gadyuchko, O.T.; Bondarenko, Y.V. The use of genetic markers in the selection of ducks based on oomorphological indicators. Suchasne Ptakhivnytstvo [Mod. Poult. Farming] 2011, 5–6, 33–39. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=c1PnDYEAAAAJ:L7CI7m0gUJcC (accessed on 30 December 2024). (In Ukrainian).
- Kulibaba, R.O.; Sakhatskyi, M.I.; Griffin, D.K.; Romanov, M.N. Molecular diversity of Ukrainian native chicken breeds: A review. Worlds Poult. Sci. J. 2024, 80, 1265–1292. [Google Scholar] [CrossRef]
- Wongloet, W.; Singchat, W.; Chaiyes, A.; Ali, H.; Piangporntip, S.; Ariyaraphong, N.; Budi, T.; Thienpreecha, W.; Wannakan, W.; Mungmee, A.; et al. Environmental and socio–cultural factors impacting the unique gene pool pattern of Mae Hong-Son chicken. Animals 2023, 13, 1949. [Google Scholar] [CrossRef]
- Tanglertpaibul, N.; Budi, T.; Nguyen, C.P.T.; Singchat, W.; Wongloet, W.; Kumnan, N.; Chalermwong, P.; Luu, A.H.; Noito, K.; Panthum, T.; et al. Samae Dam chicken: A variety of the Pradu Hang Dam breed revealed from microsatellite genotyping data. Anim. Biosci. 2024, 37, 2033. [Google Scholar] [CrossRef]
- Wattanadilokcahtkun, P.; Chalermwong, P.; Singchat, W.; Wongloet, W.; Chaiyes, A.; Tanglertpaibul, N.; Budi, T.; Panthum, T.; Ariyaraphong, N.; Ahmad, S.F.; et al. Genetic admixture and diversity in Thai domestic chickens revealed through analysis of Lao Pa Koi fighting cocks. PLoS ONE 2023, 18, e0289983. [Google Scholar] [CrossRef]
- Budi, T.; Luu, A.H.; Singchat, W.; Wongloet, W.; Rey, J.; Kumnan, N.; Chalermwong, P.; Nguyen, C.P.T.; Panthum, T.; Tanglertpaibul, N.; et al. Purposive breeding strategies drive genetic differentiation in Thai fighting cock breeds. Genes Genom. 2024, 46, 1225–1237. [Google Scholar] [CrossRef]
- Luu, A.H.; Budi, T.; Singchat, W.; Nguyen, C.P.T.; Panthum, T.; Tanglertpaibul, N.; Thong, T.; Vangnai, K.; Chaiyes, A.; Yokthongwattana, C.; et al. Comparison of unique Dong Tao chickens from Vietnam and Thailand: Genetic background and differences for resource management. Genes Genom. 2025, 1–13. [Google Scholar] [CrossRef]
- Bei, Y.; Li, J.; Meng, S.; Li, G.; Sun, B. Isolation and characterization of 12 novel microsatellite loci in Hume’s Pheasant, Syrmaticus humiae. Conserv. Genet. Resour. 2013, 5, 19–21. [Google Scholar] [CrossRef]
- Bei, Y.; Lai, J.; Martin, K.; Chen, W. Landscape genetics of Hume’s Pheasant Syrmaticus humiae: Rivers act as potential genetic barriers. Ornithol. Sci. 2021, 20, 149–160. [Google Scholar] [CrossRef]
- Thintip, J.; Singchat, W.; Ahmad, S.F.; Ariyaraphong, N.; Muangmai, N.; Chamchumroon, W.; Pitiwong, K.; Suksavate, W.; Duangjai, S.; Duengkae, P.; et al. Reduced genetic variability in a captive-bred population of the endangered Hume’s pheasant (Syrmaticus humiae, Hume 1881) revealed by microsatellite genotyping and D-loop sequencing. PLoS ONE 2021, 16, e0256573. [Google Scholar] [CrossRef] [PubMed]
- Jangtarwan, K.; Koomgun, T.; Prasongmaneerut, T.; Thongchum, R.; Singchat, W.; Tawichasri, P.; Fukayama, T.; Sillapaprayoon, S.; Kraichak, E.; Muangmai, N.; et al. Take one step backward to move forward: Assessment of genetic diversity and population structure of captive Asian woolly-necked storks (Ciconia episcopus). PLoS ONE 2019, 14, e0223726. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.J.; Lee, M.Y.; Jeon, H.S.; An, J.; Yoon, J. Temporal changes in demography and genetic diversity of oriental storks at the stage of long-term captive propagation and reintroduction initiation. Zool. Sci. 2023, 40, 284–291. [Google Scholar] [CrossRef]
- Romanov, M.N.; Jones, K.C.; Chemnick, L.G.; Stremel-Mork, E.; Otten, C.; Da, Y.; Akhunov, E.D.; Ryder, O.A. California Condor Microsatellite-enriched Library As a Tool for Genetic and Genomic Studies in an Endangered Species. In Proceedings of the International Plant and Animal Genome XVII Conference, San Diego, CA, USA, 10–14 January 2009; Scherago International: San Diego, CA, USA, 2009; p. 107. Available online: https://kar.kent.ac.uk/46627/ (accessed on 30 December 2024).
- Moran, B.M.; Thomas, S.M.; Judson, J.M.; Navarro, A.; Davis, H.; Sidak-Loftis, L.; Korody, M.; Mace, M.; Ralls, K.; Callicrate, T.; et al. Correcting parentage relationships in the endangered California Condor: Improving mean kinship estimates for conservation management. Ornithol. Appl. 2021, 123, duab017. [Google Scholar] [CrossRef]
- Ryder, O.A.; Thomas, S.; Judson, J.M.; Romanov, M.N.; Dandekar, S.; Papp, J.C.; Sidak-Loftis, L.C.; Walker, K.; Stalis, I.H.; Mace, M.; et al. Facultative parthenogenesis in California condors. J. Hered. 2021, 112, 569–574. [Google Scholar] [CrossRef]
- Irion, D.N.; Schaffer, A.L.; Famula, T.R.; Eggleston, M.L.; Hughes, S.S.; Pedersen, N.C. Analysis of genetic variation in 28 dog breed populations with 100 microsatellite markers. J. Hered. 2003, 94, 81–87. [Google Scholar] [CrossRef]
- Singh, Y.; Kaur, B.; Kaur, M.; Yatish, H.M.; Mukhopadhyay, C. Microsatellite DNA analysis of genetic diversity and parentage testing in popular dog breeds in India. Preprints 2023, 2023111536. [Google Scholar] [CrossRef]
- .Patta, C.; Singchat, W.; Thatukan, C.; Jaito, W.; Kumnan, N.; Chalermwong, P.; Panthum, T.; Budi, T.; Wongloet, W.; Wattanadilokchatkun, P.; et al. Optimizing Bangkaew dog breed identification using DNA technology. Genes Genom. 2024, 46, 659–669. [Google Scholar] [CrossRef]
- Thatukan, C.; Patta, C.; Singchat, W.; Jaito, W.; Kumnan, N.; Chalermwong, P.; Panthum, T.; Wongloet, W.; Wattanadilokchatkun, P.; Thong, T.; et al. Small but mighty: Genetic diversity of the Thai Ridgeback dog population. Biochem. Genet. 2024. [Google Scholar] [CrossRef]
- Tahoor, A.; Khan, J.A.; Mahfooz, S. A comparative survey of microsatellites among wild and domestic cat provides valuable resources for marker development. Mol. Biol. Rep. 2019, 46, 3025–3033. [Google Scholar] [CrossRef]
- Rana, D.; Boyer, F.; Barba, M.D.; Taberlet, P.; Ramakrishnan, U. From single species to communities: Microsatellite amplicon sequencing to monitor felids using Feliplex. bioRxiv 2024. [Google Scholar] [CrossRef]
- Ubolrat, K.; Laopiem, S.; Nunklang, K.; Phavaphutanon, J. Genetic diversity and inbreeding situation of Korat and Siamese cats based on microsatellite markers. Vet. Integr. Sci. 2019, 17, 51–64. Available online: https://he02.tci-thaijo.org/index.php/vis/article/view/183616 (accessed on 30 December 2024).
- Lipinski, M.J.; Froenicke, L.; Baysac, K.C.; Billings, N.C.; Leutenegger, C.M.; Levy, A.M.; Longeri, M.; Niini, T.; Ozpinar, H.; Slater, M.R.; et al. The ascent of cat breeds: Genetic evaluations of breeds and worldwide random-bred populations. Genomics 2008, 91, 12–21. [Google Scholar] [CrossRef]
- Comstock, K.E.; Wasser, S.K.; Ostrander, E.A. Polymorphic microsatellite DNA loci identified in the African elephant (Loxodonta africana). Mol. Ecol. 2000, 9, 1004–1006. [Google Scholar] [CrossRef]
- Archie, E.A.; Moss, C.J.; Alberts, S.C. Characterization of tetranucleotide microsatellite loci in the African savannah elephant (Loxodonta africana africana). Mol. Ecol. Notes 2003, 3, 244–246. [Google Scholar] [CrossRef]
- Fickel, J.; Lieckfeldt, D.; Ratanakorn, P.; Pitra, C. Distribution of haplotypes and microsatellite alleles among Asian elephants (Elephas maximus) in Thailand. Eur. J. Wildl. Res. 2007, 53, 298–303. [Google Scholar] [CrossRef]
- Pasquesi, G.I.M.; Adams, R.H.; Card, D.C.; Schield, D.R.; Corbin, A.B.; Perry, B.W.; Reyes-Velasco, J.; Ruggiero, R.P.; Vandewege, M.W.; Shortt, J.A.; et al. Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals. Nat. Commun. 2018, 9, 2774. [Google Scholar] [CrossRef]
- Singh, L.; Purdom, I.F.; Jones, K.W. Sex chromosome associated satellite DNA: Evolution and conservation. Chromosoma 1980, 79, 137–157. [Google Scholar] [CrossRef]
- Matsubara, K.; Knopp, T.; Sarre, S.D.; Georges, A.; Ezaz, T. Karyotypic analysis and FISH mapping of microsatellite motifs reveal highly differentiated XX/XY sex chromosomes in the pink-tailed worm-lizard (Aprasia parapulchella, Pygopodidae, Squamata). Mol. Cytogenet. 2013, 6, 60. [Google Scholar] [CrossRef]
- Rovatsos, M.; Kratochvíl, L.; Altmanová, M.; Pokorná, M.J. Interstitial telomeric motifs in squamate reptiles: When the exceptions outnumber the rule. PLoS ONE 2015, 10, e0134985. [Google Scholar] [CrossRef] [PubMed]
- Rovatsos, M.; Altmanová, M.; Johnson Pokorná, M.; Augstenová, B.; Kratochvíl, L. Cytogenetics of the Javan file snake (Acrochordus javanicus) and the evolution of snake sex chromosomes. J. Zool. Syst. Evol. Res. 2018, 56, 117–125. [Google Scholar] [CrossRef]
- Augstenová, B.; Mazzoleni, S.; Kratochvíl, L.; Rovatsos, M. Evolutionary dynamics of the W chromosome in caenophidian snakes. Genes 2018, 9, 5. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Augstenová, B.; Clemente, L.; Auer, M.; Fritz, U.; Praschag, P.; Protiva, T.; Velenský, P.; Kratochvíl, L.; Rovatsos, M. Sex is determined by XX/XY sex chromosomes in Australasian side-necked turtles (Testudines: Chelidae). Sci. Rep. 2020, 10, 4276. [Google Scholar] [CrossRef]
- Neff, B.D.; Gross, M.R. Microsatellite evolution in vertebrates: Inference from AC dinucleotide repeats. Evolution 2001, 55, 1717–1733. [Google Scholar] [CrossRef]
- Alföldi, J.; Di Palma, F.; Grabherr, M.; Williams, C.; Kong, L.; Mauceli, E.; Russell, P.; Lowe, C.B.; Glor, R.E.; Jaffe, J.D.; et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 2011, 477, 587–591. [Google Scholar] [CrossRef]
- Castoe, T.A.; Hall, K.T.; Guibotsy Mboulas, M.L.; Gu, W.; Jason De Koning, A.P.; Fox, S.E.; Poole, A.W.; Vemulapalli, V.; Daza, J.M.; Mockler, T.; et al. Discovery of highly divergent repeat landscapes in snake genomes using high-throughput sequencing. Genome Biol. Evol. 2011, 3, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Castoe, T.A.; De Koning, A.P.J.; Hall, K.T.; Card, D.C.; Schield, D.R.; Fujita, M.K.; Ruggiero, R.P.; Degner, J.F.; Daza, J.M.; Gu, W.; et al. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc. Natl. Acad. Sci. USA 2013, 110, 20645–20650. [Google Scholar] [CrossRef]
- Giovannotti, M.; Nisi Cerioni, P.; Caputo, V.; Olmo, E. Characterisation of a GC-rich telomeric satellite DNA in Eumeces schneideri Daudin (Reptilia, Scincidae). Cytogenet. Genome Res. 2009, 125, 272–278. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Singchat, W.; Jehangir, M.; Panthum, T.; Srikulnath, K. Consequence of paradigm shift with repeat landscapes in reptiles: Powerful facilitators of chromosomal rearrangements for diversity and evolution. Genes 2020, 11, 827. [Google Scholar] [CrossRef]
- Ahmad, S.F.; Singchat, W.; Panthum, T.; Srikulnath, K. Impact of repetitive DNA elements on snake genome biology and evolution. Cells 2021, 10, 1707. [Google Scholar] [CrossRef] [PubMed]
- Le Chevalier, H.; Marí-Mena, N.; Carro, B.; Prunier, J.G.; Bossu, C.; Darnet, E.; Souchet, J.; Guillaume, O.; Calvez, O.; Bertrand, R. Isolation and characterization of fourteen polymorphic microsatellite markers in the viperine snake Natrix maura. Ecol. Evol. 2019, 9, 11227–11231. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Wang, Z.J.; Li, Q.Y.; Lian, J.M.; Zhou, Y.; Lu, B.Z.; Jin, L.J.; Qiu, P.X.; Zhang, P.; Zhu, W.B.; et al. Evolutionary trajectories of snake genes and genomes revealed by comparative analyses of five-pacer viper. Nat. Commun. 2016, 7, 13107. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H.; Chijiwa, T.; Oda-Ueda, N.; Nakamura, H.; Yamaguchi, K.; Hattori, S.; Matsubara, K.; Matsuda, Y.; Yamashita, A.; Isomoto, A.; et al. The habu genome reveals accelerated evolution of venom protein genes. Sci. Rep. 2018, 8, 11300. [Google Scholar] [CrossRef]
- Singchat, W.; Panthum, T.; Ahmad, S.F.; Baicharoen, S.; Muangmai, N.; Duengkae, P.; Griffin, D.K.; Srikulnath, K. Remnant of unrelated amniote sex chromosomal linkage sharing on the same chromosome in house gecko lizards, providing a better understanding of the ancestral super-sex chromosome. Cells 2021, 10, 2969. [Google Scholar] [CrossRef]
- Panthum, T.; Ariyaraphong, N.; Wongloet, W.; Wattanadilokchatkun, P.; Laopichienpong, N.; Rasoarahona, R.; Singchat, W.; Ahmad, S.F.; Kraichak, E.; Muangmai, N.; et al. Preserving pure Siamese crocodile populations: A comprehensive approach using multi-genetic tools. Biology 2023, 12, 1428. [Google Scholar] [CrossRef]
- Wongtienchai, P.; Lapbenjakul, S.; Jangtarwan, K.; Areesirisuk, P.; Mahaprom, R.; Subpayakom, N.; Singchat, W.; Sillapaprayoon, S.; Muangmai, N.; Songchan, R.; et al. Genetic management of a water monitor lizard (Varanus salvator macromaculatus) population at Bang Kachao Peninsula as a consequence of urbanization with Varanus Farm Kamphaeng Saen as the first captive research establishment. J. Zool. Syst. Evol. Res. 2021, 59, 484–497. [Google Scholar] [CrossRef]
- Panthum, T.; Singchat, W.; Laopichienpong, N.; Ahmad, S.F.; Kraichak, E.; Duengkae, P.; Muangmai, N.; Kitana, N.; Srikulnath, K. Genome-wide SNP analysis of male and female rice field frogs, Hoplobatrachus rugulosus, supports a non-genetic sex determination system. Diversity 2021, 13, 501. [Google Scholar] [CrossRef]
- Ergül Kalaycı, T.; Gül, Ç. Genetic variability and population genetic structure in the Caucasian Parsley Frog, Pelodytes caucasicus (Boulenger, 1896) based on microsatellite markers. Zool. Middle East 2024, 70, 226–235. [Google Scholar] [CrossRef]
- Winters, D.M.; Wilson, E.; Coster, S.S.; Rothenberger, M.B. Integrating population genetics with long-term environmental monitoring to evaluate and guide vernal pool creation for amphibian conservation. Ecol. Evol. 2024, 14, e70431. [Google Scholar] [CrossRef]
- Chen, J.A.; Yu, P.J.; Jheng, S.W.; Lin, Y.Z.; Sun, P.W.; Ko, W.Y.; Lin, C.F.; Ju, Y.T. Mining expressed sequence tag (EST) microsatellite markers to assess the genetic differentiation of five Hynobius species endemic to Taiwan. Sci. Rep. 2024, 14, 20898. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Ye, H.; Song, X.; Fan, J.; Li, J.; Shao, J.; Wang, Y.; Lin, D.; Yue, H.; Ruan, R.; et al. Genetic diversity and population structure of Chinese longsnout catfish (Leiocassis longirostris) using microsatellite DNA markers. Fishes 2024, 9, 35. [Google Scholar] [CrossRef]
- Peng, C.; Luo, C.; Xiang, G.; Huang, J.; Shao, L.; Huang, H.; Fan, S. Genome-wide microsatellites in Acanthopagrus latus: Development, distribution, characterization, and polymorphism. Animals 2024, 14, 3709. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-R.; Sung, M.-S.; Hwang, Y.; Jeong, J.H.; Yu, J.-N. Assessment of the genetic diversity and structure of the Korean endemic freshwater fish Microphysogobio longidorsalis (Gobioninae) using microsatellite markers: A first glance from population genetics. Genes 2024, 15, 69. [Google Scholar] [CrossRef]
- Adyrbekova, K.; Perea, S.; Doadrio, I. Development and characterization of fifteen polymorphic microsatellite loci for rare and endangered species within Luciobarbus Heckel, 1843 genus in the Aral basin and their conservation application. Fishes 2024, 9, 169. [Google Scholar] [CrossRef]
- Chailertrit, V.; Swatdipong, A.; Peyachoknagul, S.; Salaenoi, J.; Srikulnath, K. Isolation and characterization of novel microsatellite markers from Siamese fighting fish (Betta splendens, Osphronemidae, Anabantoidei) and their transferability to related species, B. smaragdina and B. imbellis. Genet. Mol. Res. 2014, 13, 7157–7162. [Google Scholar] [CrossRef]
- Wattanadilokchatkun, P.; Chaiyes, A.; Ariyaraphong, N.; Wongloet, W.; Suksavate, W.; Thatukan, C.; Kumnan, N.; Panthum, T.; Thong, T.; Singchat, W.; et al. Integrative approach for landscape demography analysis of Plakad-Pa Pak-Tawan-Ok (Betta siamorientalis): Deciphering genetic and environmental factors in Eastern Thailand’s conservation efforts. Glob. Ecol. Conserv. 2024, 51, e02870. [Google Scholar] [CrossRef]
- Wattanadilokchatkun, P.; Panthum, T.; Jaisamut, K.; Ahmad, S.F.; Dokkaew, S.; Muangmai, N.; Duengkae, P.; Singchat, W.; Srikulnath, K. Characterization of microsatellite distribution in Siamese fighting fish genome to promote conservation and genetic diversity. Fishes 2022, 7, 251. [Google Scholar] [CrossRef]
- Suntronpong, A.; Thapana, W.; Twilprawat, P.; Prakhongcheep, O.; Somyong, S.; Muangmai, N.; Peyachoknagul, S.; Srikulnath, K. Karyological characterization and identification of four repetitive element groups (the 18S–28S rRNA gene, telomeric sequences, microsatellite repeat motifs, Rex retroelements) of the Asian swamp eel (Monopterus albus). Comp. Cytogenet. 2017, 11, 435. [Google Scholar] [CrossRef]
- Panthum, T.; Wattanadilokchatkun, P.; Jaisamut, K.; Singchat, W.; Ahmad, S.F.; Muangmai, N.; Duengkae, P.; Antunes, A.; Srikulnath, K. In silico chromosome mapping of the male-specific/linked loci in the jade perch (Scortum barcoo) suggests chromosome 19 as the putative Y sex chromosome. Fishes 2023, 8, 482. [Google Scholar] [CrossRef]
- Deakin, J.E.; Ezaz, T. Understanding the evolution of reptile chromosomes through applications of combined cytogenetics and genomics approaches. Cytogenet. Genome Res. 2019, 157, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Maneechot, N.; Yano, C.F.; Bertollo, L.A.C.; Getlekha, N.; Molina, W.F.; Ditcharoen, S.; Tengjaroenkul, B.; Supiwong, W.; Tanomtong, A.; de Bello Cioffi, M. Genomic organization of repetitive DNAs highlights chromosomal evolution in the genus Clarias (Clariidae, Siluriformes). Mol. Cytogenet. 2016, 9, 4. [Google Scholar] [CrossRef]
- Lisachov, A.; Panthum, T.; Dedukh, D.; Singchat, W.; Ahmad, S.F.; Wattanadilokcahtkun, P.; Thong, T.; Srikampa, P.; Noito, K.; Rasoarahona, R.; et al. Genome-wide sequence divergence of satellite DNA could underlie meiotic failure in male hybrids of bighead catfish and North African catfish (Clarias, Clariidae). Genomics 2024, 116, 110868. [Google Scholar] [CrossRef] [PubMed]
- Patta, C.; Panthum, T.; Thatukan, C.; Wongloet, W.; Chalermwong, P.; Wattanadilokchatkun, P.; Thong, T.; Srikampa, P.; Singchat, W.; Ahmad, S.F.; et al. Questioning inbreeding: Could outbreeding affect productivity in the North African catfish in Thailand? PLoS ONE 2024, 19, e0302584. [Google Scholar] [CrossRef]
- Wachirachaikarn, A.; Na-Nakorn, U. Genetic diversity of the North African catfish, Clarias gariepinus (Burchell, 1822) hatchery stocks in Thailand. ScienceAsia 2019, 45, 301–308. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Na-Nakorn, U. Genetic impacts of translocations on biodiversity of aquatic species with particular reference to Asian countries. Aquac. Asia 2004, 9, 4–7. Available online: https://www.researchgate.net/publication/254965380 (accessed on 30 December 2024).
- Wachirachaikarn, A.; Rungsin, W.; Srisapoome, P.; Na-Nakorn, U. Crossing of African catfish, Clarias gariepinus (Burchell, 1822), strains based on strain selection using genetic diversity data. Aquaculture 2009, 290, 53–60. [Google Scholar] [CrossRef]
- Lawonyawut, K. Hybridization and Genetic Manipulation in Clarias Catfish. Ph.D. Thesis, University of Sterling, Sterling, UK, 1996. Available online: https://dspace.stir.ac.uk/handle/1893/21426 (accessed on 30 December 2024).
- Barasa, J.E.; Abila, R.; Grobler, J.P.; Agaba, M.; Chemoiwa, E.J.; Kaunda-Arara, B. High genetic diversity and population differentiation in Clarias gariepinus of Yala Swamp: Evidence from mitochondrial DNA sequences. J. Fish Biol. 2016, 89, 2557–2570. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.D.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef]
- Moniruzzaman, M.; Khatun, R.; Yaakob, Z.; Khan, M.S.; Mintoo, A.A. Development of microsatellites: A powerful genetic marker. Agriculturists 2015, 13, 152–172. [Google Scholar] [CrossRef]
- Sukumaran, S. Molecular tools and techniques in marine fish identification. In Training Manual on “Know Your Marine Biodiversity and Environment (MarBiE 1)”; Sreeram, M.P., George, G., Eds.; CMFRI Training Manual Series; ICAR-Central Marine Fisheries Research Institute: Kochi, India, 2024; Volume 42, pp. 185–189. Available online: https://eprints.cmfri.org.in/18504/ (accessed on 30 December 2024).
- Väli, Ü.; Einarsson, A.; Waits, L.; Ellegren, H. To what extent do microsatellite markers reflect genome-wide genetic diversity in natural populations? Mol. Ecol. 2008, 17, 3808–3817. [Google Scholar] [CrossRef] [PubMed]
- Payseur, B.A.; Jing, P.; Haasl, R.J. A genomic portrait of human microsatellite variation. Mol. Biol. Evol. 2011, 28, 303–312. [Google Scholar] [CrossRef]
- Weising, K.; Winter, P.; Hüttel, B.; Kahl, G. Microsatellite markers for molecular breeding. J. Crop Prod. 1997, 1, 113–143. [Google Scholar] [CrossRef]
- Legendre, M.; Pochet, N.; Pak, T.; Verstrepen, K.J. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 2007, 17, 1787–1796. [Google Scholar] [CrossRef]
- Pemberton, T.J.; Sandefur, C.I.; Jakobsson, M.; Rosenberg, N.A. Sequence determinants of human microsatellite variability. BMC Genom. 2009, 10, 612. [Google Scholar] [CrossRef] [PubMed]
- Al-Samarai, F.R.; Al-Kazaz, A.A. Molecular markers: An introduction and applications. Eur. J. Mol. Biotechnol. 2015, 9, 118–130. [Google Scholar] [CrossRef]
- Hoshino, A.A.; Bravo, J.P.; Nobile, P.M.; Morelli, K.A. Microsatellites as tools for genetic diversity analysis. In Genetic Diversity in Microorganisms; Çalışkan, M., Ed.; InTech Europe: Rijeka, Croatia, 2012; pp. 149–170. [Google Scholar] [CrossRef]
- Grover, A.; Sharma, P.C. Development and use of molecular markers: Past and present. Crit. Rev. Biotechnol. 2016, 36, 290–302. [Google Scholar] [CrossRef]
- Wright, J.M.; Bentzen, P. Microsatellites: Genetic markers for the future. In Molecular Genetics in Fisheries; Carvalho, G.R., Pitcher, T.J., Eds.; Springer: Dordrecht, The Netherlands, 1995; pp. 117–121. [Google Scholar] [CrossRef]
- Debrauwere, H.; Gendrel, C.G.; Lechat, S.; Dutreix, M. Differences and similarities various tandem repeat sequences: Minisatellites and microsatellites. Biochimie 1997, 79, 577–586. [Google Scholar] [CrossRef]
- Khlestkina, E.K. Molecular markers in genetic studies and breeding. Russ. J. Genet. Appl. Res. 2014, 4, 236–244. [Google Scholar] [CrossRef]
- Mishra, S.P.; Mishra, C.; Mishra, D.P.; Rosalin, B.P.; Bhuyan, C. Application of advanced molecular marker technique for improvement of animal: A critical review. J. Entomol. Zool. Stud. 2017, 5, 1283–1295. Available online: https://www.entomoljournal.com/archives/?year=2017&vol=5&issue=5&part=Q&ArticleId=2467 (accessed on 30 December 2024).
- Sheriff, O.; Alemayehu, K. Genetic diversity studies using microsatellite markers and their contribution in supporting sustainable sheep breeding programs: A review. Cogent Food Agric. 2018, 4, 1459062. [Google Scholar] [CrossRef]
- Serrano, M.; Calvo, J.H.; Martínez, M.; Marcos-Carcavilla, A.; Cuevas, J.; González, C.; Jurado, J.J.; de Tejada, P.D. Microsatellite based genetic diversity and population structure of the endangered Spanish Guadarrama goat breed. BMC Genet. 2009, 10, 61. [Google Scholar] [CrossRef]
- Peralta, W.; Nestares, A.; Gamarra, J.; Rojas, M.; Sullca, J.; Estrada, R. Genetic diversity and population structure of alpacas (Vicugna pacos) in Peru: A microsatellite analysis. Diversity 2025, 17, 353. [Google Scholar] [CrossRef]
- Li, M.H.; Strandén, I.; Tiirikka, T.; Sevón-Aimonen, M.L.; Kantanen, J. A comparison of approaches to estimate the inbreeding coefficient and pairwise relatedness using genomic and pedigree data in a sheep population. PLoS ONE 2011, 6, e26256. [Google Scholar] [CrossRef]
- Daw, E.W.; Heath, S.C.; Lu, Y. Single-nucleotide polymorphism versus microsatellite markers in a combined linkage and segregation analysis of a quantitative trait. BMC Genet. 2005, 6 (Suppl 1). [Google Scholar] [CrossRef] [PubMed][Green Version]
- Flanagan, S.P.; Jones, A.G. The future of parentage analysis: From microsatellites to SNPs and beyond. Mol. Ecol. 2019, 28, 544–567. [Google Scholar] [CrossRef]
- Ball, A.D.; Stapley, J.; Dawson, D.A.; Birkhead, T.R.; Burke, T.; Slate, J. A comparison of SNPs and microsatellites as linkage mapping markers: Lessons from the zebra finch (Taeniopygia guttata). BMC Genom. 2010, 11, 218. [Google Scholar] [CrossRef]
- Tokarska, M.; Marshall, T.; Kowalczyk, R.; Wójcik, J.M.; Pertoldi, C.; Kristensen, T.N.; Loeschcke, V.; Gregersen, V.R.; Bendixen, C. Effectiveness of microsatellite and SNP markers for parentage and identity analysis in species with low genetic diversity: The case of European bison. Heredity 2009, 103, 326–332. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Coates, B.S.; Sumerford, D.V.; Miller, N.J.; Kim, K.S.; Sappington, T.W.; Siegfried, B.D.; Lewis, L.C. Comparative performance of single nucleotide polymorphism and microsatellite markers for population genetic analysis. J. Hered. 2009, 100, 556–564. [Google Scholar] [CrossRef]
- Elbers, J.P.; Clostio, R.W.; Taylor, S.S. Population genetic inferences using immune gene SNPs mirror patterns inferred by microsatellites. Mol. Ecol. Res. 2017, 17, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Osborne, M.J.; Caeiro-Dias, G.; Turner, T.F. Transitioning from microsatellites to SNP-based microhaplotypes in genetic monitoring programmes: Lessons from paired data spanning 20 years. Mol. Ecol. 2023, 32, 316–334. [Google Scholar] [CrossRef] [PubMed]
- Puckett, E.E. Variability in total project and per sample genotyping costs under varying study designs including with microsatellites or SNPs to answer conservation genetic questions. Conserv. Genet. Resour. 2017, 9, 289–304. [Google Scholar] [CrossRef]
- Roques, S.; Chancerel, E.; Boury, C.; Pierre, M.; Acolas, M.L. From microsatellites to single nucleotide polymorphisms for the genetic monitoring of a critically endangered sturgeon. Ecol. Evol. 2019, 9, 7017–7029. [Google Scholar] [CrossRef]
- Campbell, M.R.; Vu, N.V.; LaGrange, A.P.; Hardy, R.S.; Ross, T.J.; Narum, S.R. Development and application of single-nucleotide polymorphism (SNP) genetic markers for conservation monitoring of Burbot populations. Trans. Am. Fish. Soc. 2019, 148, 661–670. [Google Scholar] [CrossRef]
- Fabbri, E.; Caniglia, R.; Mucci, N.; Thomsen, H.P.; Krag, K.; Pertoldi, C.; Loeschcke, V.; Randi, E. Comparison of single nucleotide polymorphisms and microsatellites in non-invasive genetic monitoring of a wolf population. Arch. Biol. Sci. 2012, 64, 321–335. [Google Scholar] [CrossRef]
- Carroll, E.L.; Bruford, M.W.; DeWoody, J.A.; Leroy, G.; Strand, A.; Waits, L.; Wang, J. Genetic and genomic monitoring with minimally invasive sampling methods. Evol. Appl. 2018, 11, 1094–1119. [Google Scholar] [CrossRef]
- Lapian, H.F.N. Molecular genetic approaches on cattle and chicken breeding: A review. Zootec 2023, 43, 254–263. Available online: https://ejournal.unsrat.ac.id/v3/index.php/zootek/article/view/48397/43755 (accessed on 30 December 2024).
| Name | Function | Reference |
|---|---|---|
| MICRO-CHECKER | Investigates microsatellite data, calculates simple summary statistics, and shows the potential for mistyped and null alleles | [93] |
| Microsatellite Toolkit | A practical Excel microsatellite data handling tool that offers summary statistics (the number of alleles observed and expected heterozygosity and allele frequencies) and verifies the dataset for errors | [94] |
| Power Marker | A feature-rich Windows application that offers a variety of summary statistics, genetic distances and bootstrapped phylogenetic trees for microsatellites, SNPs, and other biallelic data | [95] |
| Msvar | Uses microsatellite frequencies to identify a previous population expansion or decline | [96] |
| Type/Species | Subtype/Breed | Types of Study | Main Findings | References |
|---|---|---|---|---|
| Humans/other primates | Apes, baboons, macaques, and certain platyrrhine monkeys | Mostly telomere repeats | Weak conservation among monkey lineages; humans/monkeys have similar sequence lengths | [119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137] |
| Cattle | Short statured Nattukuttai | Bottleneck analysis | No population decline | [138] |
| Gabrali | Genetic diversity, 12 loci | Substantial genetic diversity and does not face threats of inbreeding/bottlenecks | [46] | |
| Lebedyn | 10 FAO-ISAG-recommended loci | Genetic equilibrium and propensity for inbreeding in some breeds | [139] | |
| Other Artiodactyla | Chinese goral | Population genetics; 11 loci | Low genetic variation due to inbreeding and a small effective population size in captivity; management as evolutionarily significant units recommended | [141] |
| Mouse-deer | Genotyping and demographic analysis | No historical bottleneck, a reduced effective population size, and inbreeding, raising extinction risk. Improved care boosted population growth | [142] | |
| Horses | Korean native horse | Parentage verification | Early application of 16 STRs for pedigree control; foundation for the national ID system | [144] |
| Thoroughbred, Jeju, Sumbawa, and Kazakh | Parentage testing, breed certification, diversity, and conservation | High heterozygosity in Thoroughbreds; low diversity in Sumbawa; admixture in Kazakh horses; ISAG STR panel validated | [145,146,148] | |
| Donkeys | Korean domestic donkeys | Genetic diversity, breed identification, and conservation | Lower heterozygosity than horses; clear species distinction; 9 STR loci validated | [147] |
| Mediterranean and Asian breeds | Genetic structure and conservation status | Moderate diversity; breed-specific structure (e.g., Pantesco); highlights importance of structured conservation programs | [149] | |
| Chickens | Canarian population | Genetic variation | High variation and did not cluster with other Spanish breeds | [154,155] |
| Nagoya breed | Breed discrimination | 4 Nagoya breeds identical to the ABR0417 reference sequence | [156] | |
| Ukrainian breeds | Genetic variation | Largest genetic differences found between Plymouth Rock White and Rhode Island Red and smallest between the Plymouth Rock White and Poltava Clay breeds | [38,157,158,159,160] | |
| Indigenous/Red Jungle fowl | Genetic diversity and population structure; 28 ISAG-FAO loci | High genetic variability; evidence of genetic introgression; selection pressures in fighting cocks; distinct clustering of Thai local breeds; importance of red junglefowl gene pool for reintroduction | [60,62,79,153,161,162,163,164] | |
| Lao Pa Koi | Genetic admixture and diversity; 28 loci | Shared partial gene pool with red junglefowl; high genetic diversity | [163] | |
| Lueng Hang Khao | Genetic admixture, diversity; 28 loci | Hitchhiking selection, indicating directional selection in fighting cocks | [164] | |
| Pradu Hang Dam | Genetic admixture, diversity; 28 loci | Partial gene pool overlap, suggesting that Samae Dam may be variety of Pradu Hang Dam | [162] | |
| Chinese black-boned chicken | Population structure | Originated from a native Chinese chicken with introgression from the red junglefowl and other domestic breeds | [62] | |
| Other birds | Hume’s pheasant | Genetic diversity and population structure | High genetic diversity in wild populations but low differentiation and inbreeding in Thai captive flocks; findings and conservation efforts | [166,167,168] |
| Asian woolly-necked storks | Genetic diversity, population structure, demographic history, and captive and reintroduced populations | Captive breeding caused inbreeding and a small effective population in one population, while another showed signs of a recent bottleneck; in oriental storks, prolonged captive propagation stabilized genetic diversity, highlighting the need for genetic assessments in reintroduction efforts | [169] | |
| California condor | Various factors/phenomena and variation | Established parentage, facultative parthenogenesis, and linkage map | [44,171,172,173] | |
| Dogs | Labrador, German Shepherd etc. | Parentage testing | No signs of inbreeding, sufficient for establishing dog parentage | [175] |
| Bangkaew and Thai Ridgeback | Genotyping and genetic diversity | Bangkaew dogs exhibit significant genetic variation with low inbreeding risk; Thai Ridgebacks maintain high genetic diversity with no bottlenecks | [176,177] | |
| American Kennel Club breeds | Differentiation within breeds | Breed-to-breed genetic relatedness less clear-cut; autosomal microsatellite set proved helpful in characterizing genetic variation within breeds | [174] | |
| Cats | Felis, Panthera and Prionailurus | Genotyping and comparative analysis | Domestic cats have a higher microsatellite frequency than wild cats, providing extensive genetic resources | [178,179] |
| Siamese and Korat | Genetic diversity and population structure | Moderate genetic diversity and high inbreeding; broader studies of Thai cat breeds reveal high genetic diversity and distinct gene pool patterns | [180] | |
| Elephants | African and Asian | Genotyping and population genetics | Cryptic speciation in Asian elephants; in captive Thai elephants, genetic diversity varied across populations and declined over 50 generations | [30,45,67,182,183,184] |
| Woolly mammoths | Demographic history until extinction | Reduction in genetic variation before extinction 4000 years ago | [59] | |
| Reptiles | General studies | Microsatellite identification | Remarkably abundant in squamate reptile genomes, majority of microsatellites distributed on sex chromosomes; lower abundance in geckos | [28,185] |
| Snakes | Abundance, distribution, and evolution | Particularly colubrids, highest density of microsatellites among vertebrates; most enriched on sex chromosomes; microsatellite expansion driven by transposable elements that are linked to venom gene duplication | [28,65,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201] | |
| Siamese/saltwater crocodiles | Endangered species genotyping, genetic variability, and population structure | Evidence of past genetic bottlenecks and hybridization. Identified hybrids for long-term conservation management to enhance conservation strategies | [31] | |
| Water monitors | Genetic diversity and population structure | Substantial genetic diversity despite habitat fragmentation due to urbanization; well-established captive population identified through genetic monitoring; future relocation efforts require allelic profile comparisons | [204] | |
| Amphibians | Rice field frogs | FISH mapping of repeat motifs and telomeric sequences | Identified 19 microsatellite repeat motifs and telomeric sequences; the absence of sex-specific signals suggests a non-genetic sex determination system | [205] |
| Caucasian parsley frog | Population studies | Population decline, rapid genetic drift, and moderate genetic differentiation | [206] | |
| Fish | Chinese longsnout catfish | Genetic variation and population structure | Substantial genetic variety; neither a systematic regional pattern of variation nor considerable genetic differentiation | [209] |
| Yellowfin seabream | Population studies; 19 loci | Low level of genetic divergence and diversity conservation measures needed | [210] | |
| Siamese fighting fish and related species | Development and characterization of markers | Ten new markers identified and characterized; they are used for hatchery breeding strategies, genetic diversity assessments, population monitoring, marker-assisted selection (MAS), and conservation efforts | [213,214,215] | |
| Asian swamp eel, Jade perch | Chromosomal mapping and distribution analysis | Eight microsatellite repeat motifs scattered across most chromosomes that are co-localized with retroelements, suggesting co-amplification during evolution; in Perch highly concentrated on chromosome 19, the putative Y chromosome | [216,217] | |
| Bighead catfish | Hybrid sterility and genetic diversity | (CA)n microsatellite-differential accumulation, possibly disrupting spermatogenesis. Low inbreeding but high genetic diversity, suggesting potential outbreeding depression | [219,220,221] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulibaba, R.O.; Srikulnath, K.; Singchat, W.; Liashenko, Y.V.; Griffin, D.K.; Romanov, M.N. The Application of Microsatellite Markers as Molecular Tools for Studying Genomic Variability in Vertebrate Populations. Curr. Issues Mol. Biol. 2025, 47, 447. https://doi.org/10.3390/cimb47060447
Kulibaba RO, Srikulnath K, Singchat W, Liashenko YV, Griffin DK, Romanov MN. The Application of Microsatellite Markers as Molecular Tools for Studying Genomic Variability in Vertebrate Populations. Current Issues in Molecular Biology. 2025; 47(6):447. https://doi.org/10.3390/cimb47060447
Chicago/Turabian StyleKulibaba, Roman O., Kornsorn Srikulnath, Worapong Singchat, Yuriy V. Liashenko, Darren K. Griffin, and Michael N. Romanov. 2025. "The Application of Microsatellite Markers as Molecular Tools for Studying Genomic Variability in Vertebrate Populations" Current Issues in Molecular Biology 47, no. 6: 447. https://doi.org/10.3390/cimb47060447
APA StyleKulibaba, R. O., Srikulnath, K., Singchat, W., Liashenko, Y. V., Griffin, D. K., & Romanov, M. N. (2025). The Application of Microsatellite Markers as Molecular Tools for Studying Genomic Variability in Vertebrate Populations. Current Issues in Molecular Biology, 47(6), 447. https://doi.org/10.3390/cimb47060447









