Potential Resistance to Oxaliplatin-Based Regimens in Gastric Cancer Patients with ERBB2 R678Q Mutation: Evidence from a National Genomic Database
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort and Data Acquisition
2.2. Clinical Information Gathering and Evaluation of the Outcomes
2.3. Statistical Analysis
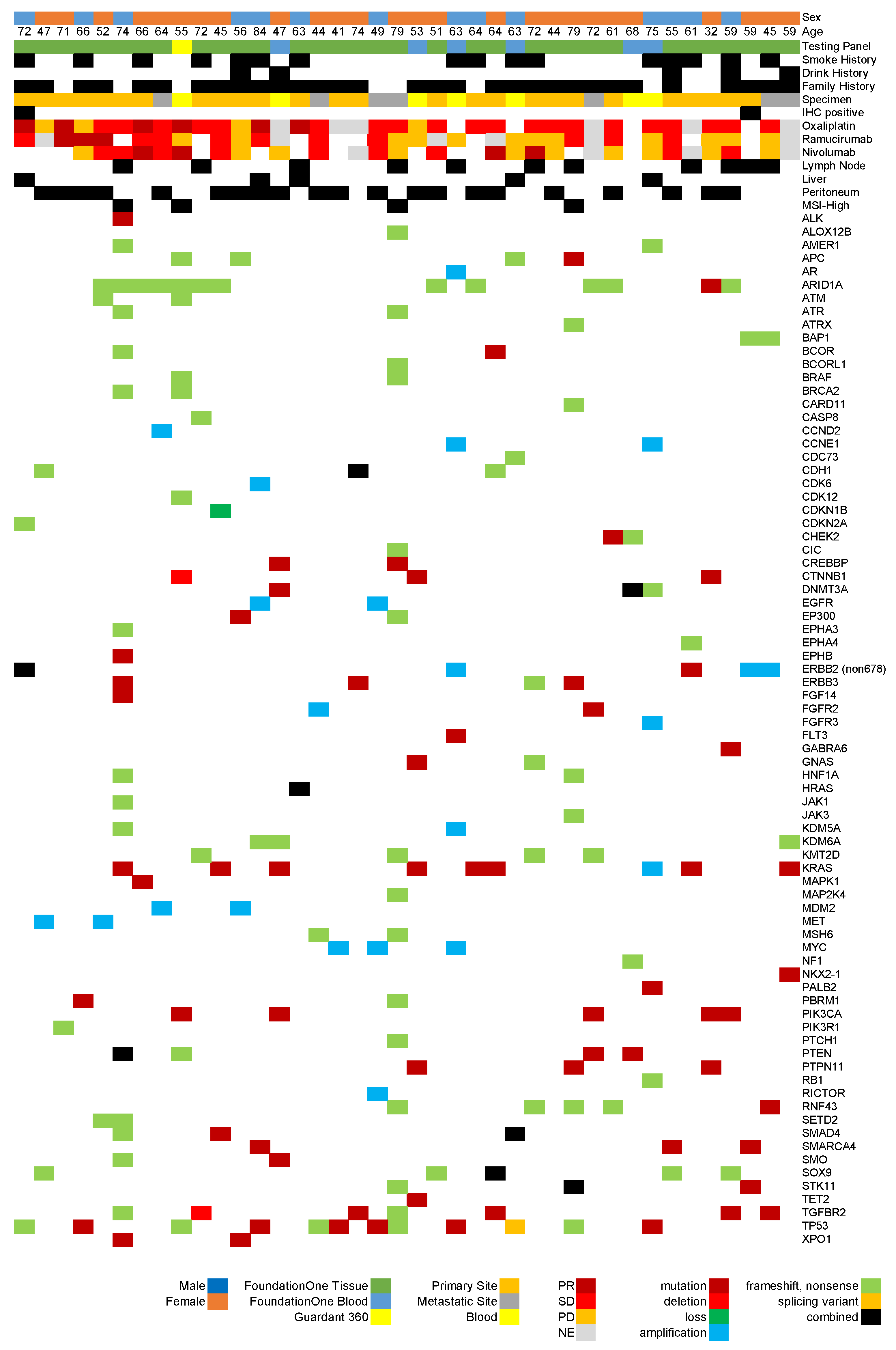
3. Results
3.1. Patient Characteristics
3.2. Overview of Genomic Testing Results
3.3. Assessment of Treatment Efficacy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ERBB2 | Epidermal growth factor receptor 2 |
| C-CAT | Center for Cancer Genomics and Advanced Therapeutics |
| GEJ | Gastroesophageal junction |
| TCGA | The Cancer Genome Atlas |
| ACRG | The Asian Cancer Research Group |
| MSI | Microsatellite instability |
| TMB | Tumor mutational burden |
| FFPE | Formalin-Fixed Paraffin-Embedded |
| N/A | Not applicable |
| RECIST | Response Evaluation Criteria in Solid Tumors |
| MSI-H | Microsatellite instability high |
| PR | Partial response |
| SD | Stable disease |
| PD | Progressive disease |
| NE | Not evaluated |
| CGP | Comprehensive genomic profiling |
| T-DXd | Trastuzumab deruxtecan |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.K.; Chen, L.T.; Ryu, M.H.; Oh, D.Y.; Oh, S.C.; Chung, H.C.; Lee, K.W.; Omori, T.; Shitara, K.; Sakuramoto, S.; et al. Nivolumab plus chemotherapy versus placebo plus chemotherapy in patients with MSI-negative, untreated, unresectable advanced or recurrent gastric or gastro-oesophageal junction cancer (ATTRACTION-4): A randomised, multicentre, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2022, 23, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Gravalos, C.; Jimeno, A. HER2 in gastric cancer: A new prognostic factor and a novel therapeutic target. Ann. Oncol. 2008, 19, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Niclauss, N.; Gütgemann, I.; Dohmen, J.; Kalff, J.C.; Lingohr, P. Novel Biomarkers of Gastric Adenocarcinoma: Current Research and Future Perspectives. Cancers 2021, 13, 5660. [Google Scholar] [CrossRef]
- Patel, T.H.; Cecchini, M. Targeted Therapies in Advanced Gastric Cancer. Curr. Treat. Options Oncol. 2020, 21, 70. [Google Scholar] [CrossRef]
- Salnikov, M.Y.; MacNeil, K.M.; Mymryk, J.S. The viral etiology of EBV-associated gastric cancers contributes to their unique pathology, clinical outcomes, treatment responses and immune landscape. Front. Immunol. 2024, 15, 1358511. [Google Scholar] [CrossRef]
- Furukawa, K.; Hatakeyama, K.; Terashima, M.; Nagashima, T.; Urakami, K.; Ohshima, K.; Notsu, A.; Sugino, T.; Yagi, T.; Fujiya, K.; et al. Molecular classification of gastric cancer predicts survival in patients undergoing radical gastrectomy based on project HOPE. Gastric Cancer 2022, 25, 138–148. [Google Scholar] [CrossRef]
- Pahuja, K.B.; Nguyen, T.T.; Jaiswal, B.S.; Prabhash, K.; Thaker, T.M.; Senger, K.; Chaudhuri, S.; Kljavin, N.M.; Antony, A.; Phalke, S.; et al. Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations. Cancer Cell 2018, 34, 792–806.e5. [Google Scholar] [CrossRef]
- Japanese Gastric Cancer Association. Japanese Gastric Cancer Treatment Guidelines 2021 (6th edition). Gastric Cancer 2023, 26, 1–25. [Google Scholar] [CrossRef]
- Koizumi, W.; Narahara, H.; Hara, T.; Takagane, A.; Akiya, T.; Takagi, M.; Miyashita, K.; Nishizaki, T.; Kobayashi, O.; Takiyama, W.; et al. S-1 plus cisplatin versus S-1 alone for first-line treatment of advanced gastric cancer (SPIRITS trial): A phase III trial. Lancet Oncol. 2008, 9, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Higuchi, K.; Nishikawa, K.; Gotoh, M.; Fuse, N.; Sugimoto, N.; Nishina, T.; Amagai, K.; Chin, K.; Niwa, Y.; et al. Phase III study comparing oxaliplatin plus S-1 with cisplatin plus S-1 in chemotherapy-naïve patients with advanced gastric cancer. Ann. Oncol. 2015, 26, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Mukai, Y.; Ueno, H. Establishment and implementation of Cancer Genomic Medicine in Japan. Cancer Sci. 2021, 112, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Kato, M.; Kohsaka, S.; Sudo, T.; Tamai, I.; Shiraishi, Y.; Okuma, Y.; Ogasawara, D.; Suzuki, T.; Yoshida, T.; et al. C-CAT: The National Datacenter for Cancer Genomic Medicine in Japan. Cancer Discov. 2022, 12, 2509–2515. [Google Scholar] [CrossRef]
- Suzuki, S.; Saito, Y. Genomic Analysis of Advanced Phyllodes Tumors Using Next-Generation Sequencing and Their Chemotherapy Response: A Retrospective Study Using the C-CAT Database. Medicina 2024, 60, 1898. [Google Scholar] [CrossRef]
- Suzuki, S.; Saito, Y.; Saito, K.; Yamada, Y.; Takahashi, K.; Kumanishi, R.; Fukui, T.; Yoshioka, T. Limited Efficacy of Anti-EGFR Monoclonal Antibodies in Colorectal Cancer Patients with Rare RAS Variants: Analysis of the C-CAT Database. Curr. Issues Mol. Biol. 2024, 46, 14476–14486. [Google Scholar] [CrossRef]
- Zhou, H.; Tan, S.; Li, H.; Lin, X. Expression and significance of EBV, ARID1A and PIK3CA in gastric carcinoma. Mol. Med. Rep. 2019, 19, 2125–2136. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhu, X.; Wei, X.; Tang, C.; Zhang, W. HER2-targeted therapies in gastric cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188549. [Google Scholar] [CrossRef]
- Pires, I.M.; Ward, T.H.; Dive, C. Oxaliplatin responses in colorectal cancer cells are modulated by CHK2 kinase inhibitors. Br. J. Pharmacol. 2010, 159, 1326–1338. [Google Scholar] [CrossRef]
- Sidorova, J. A game of substrates: Replication fork remodeling and its roles in genome stability and chemo-resistance. Cell Stress. 2017, 1, 115–133. [Google Scholar] [CrossRef]
- Bakkenist, C.J.; Lee, J.J.; Schmitz, J.C. ATM Is Required for the Repair of Oxaliplatin-Induced DNA Damage in Colorectal Cancer. Clin. Color. Cancer 2018, 17, 255–257. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Chang, J.Y. New Insights into Mechanisms of Cisplatin Resistance: From Tumor Cell to Microenvironment. Int. J. Mol. Sci. 2019, 20, 4136. [Google Scholar] [CrossRef] [PubMed]
- Sunami, K.; Naito, Y.; Komine, K.; Amano, T.; Ennishi, D.; Imai, M.; Kage, H.; Kanai, M.; Kenmotsu, H.; Koyama, T.; et al. Chronological improvement in precision oncology implementation in Japan. Cancer Sci. 2022, 113, 3995–4000. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Yamazaki, F.; Kubo, T.; Sunami, K.; Kumamoto, T.; Arakawa, A.; Sugiyama, M.; Watanabe, Y.; Nakajima, M.; Shirakawa, N.; et al. Pediatric Precision Medicine at the National Cancer Center Japan: Prospective Genomic Study of Pediatric Patients with Cancer as Part of the TOP-GEAR Project. JCO Precis. Oncol. 2023, 7, e2200266. [Google Scholar] [CrossRef]
- Shitara, K.; Bang, Y.J.; Iwasa, S.; Sugimoto, N.; Ryu, M.H.; Sakai, D.; Chung, H.C.; Kawakami, H.; Yabusaki, H.; Lee, J.; et al. Trastuzumab Deruxtecan in Previously Treated HER2-Positive Gastric Cancer. N. Engl. J. Med. 2020, 382, 2419–2430. [Google Scholar] [CrossRef]
- Conlon, N.T.; Kooijman, J.J.; van Gerwen, S.J.C.; Mulder, W.R.; Zaman, G.J.R.; Diala, I.; Eli, L.D.; Lalani, A.S.; Crown, J.; Collins, D.M. Comparative analysis of drug response and gene profiling of HER2-targeted tyrosine kinase inhibitors. Br. J. Cancer 2021, 124, 1249–1259. [Google Scholar] [CrossRef]
- Hu, Q.; Oki, E.; Yamada, T.; Kashiwada, T.; Sonoda, H.; Kataoka, M.; Kawanaka, H.; Tsuji, Y.; Makiyama, A.; Nakashima, Y.; et al. Genomic characterization between HER2-positive and negative gastric cancer patients in a prospective trial. Cancer Med. 2023, 12, 16649–16660. [Google Scholar] [CrossRef]
| Characteristic | FoundationOne CDx | FoundationOne Liquid CDx | NCC Oncopanel System | Guardant 360 CDx | GenMine TOP Cancer Panel |
|---|---|---|---|---|---|
| Specimen Type | FFPE tissue | Blood | FFPE tissue | Blood | FFPE tissue |
| Number of Genes | 324 | 324 | 114 | 74 | 723 |
| MSI Assessment | Yes | Yes | Yes * | Yes | No |
| Paired Analysis | No | No | Yes | No | Yes |
| TMB Evaluation | Yes | Yes | Yes | No | Yes |
| Minimal Neoplastic Content Required | 20% | N/A | 20% | N/A | 20% |
| Necessary DNA Input | 50 ng | 2 tubes | 50 ng | 2 tubes ** | 50 ng |
| Gastric or Gastroesophageal Junction Adenocarcinoma Cases (n = 3116) | |||
|---|---|---|---|
| Cancer Genomics Test | Treatment Response to Oxaliplatin * | ||
| FoundationOne CDx | 2262 | Complete Response | 28 |
| FoundationOne Liquid CDx | 394 | Partial Response | 813 |
| NCC Oncopanel System | 305 | Stable Disease | 815 |
| GenMineTM TOP Cancer Panel | 96 | Progressive Disease | 549 |
| Guardant360 CDx | 59 | Not Evaluated | 379 |
| Sex | Treatment Response to Ramucirumab * | ||
| Male | 5104 | Complete Response | 10 |
| Female | 1012 | Partial Response | 387 |
| Age Group (years) | Stable Disease | 828 | |
| 70–79 | Progressive Disease | 553 | |
| 60–69 | 992 | Not Evaluated | 545 |
| 50–59 | 908 | Treatment Response to Nivolumab * | |
| 40–49 | 549 | Complete Response | 18 |
| 30–29 | 339 | Partial Response | 451 |
| 80–89 | 146 | Stable Disease | 650 |
| 20–29 | 129 | Progressive Disease | 733 |
| 10–19 | 46 | Not Evaluated | 383 |
| 90– | 6 | ||
| Gastric or Gastroesophageal Junction Adenocarcinoma ERBB2 Mutation Cases (N = 130) | |||
|---|---|---|---|
| Cancer Genomics Test | Treatment Response to Oxaliplatin * | ||
| FoundationOne CDx | 103 | Complete Response | 2 |
| FoundationOne Liquid CDx | 19 | Partial Response | 29 |
| NCC Oncopanel System | 3 | Stable Disease | 33 |
| GenMineTM TOP Cancer Panel | 3 | Progressive Disease | 27 |
| Guardant360 CDx | 2 | Not Evaluated | 16 |
| Sex | Treatment Response to Ramucirumab * | ||
| Male | 60 | Complete Response | 0 |
| Female | 70 | Partial Response | 9 |
| Stable Disease | 32 | ||
| Age Group (years) | Progressive Disease | 553 | |
| 70–79 | 42 | Not Evaluated | 19 |
| 60–69 | 35 | Treatment Response to Nivolumab * | |
| 50–59 | 25 | Complete Response | 1 |
| 40–49 | 21 | Partial Response | 15 |
| 80–89 | 5 | Stable Disease | 23 |
| 30–39 | 2 | Progressive Disease | 40 |
| Not Evaluated | 16 | ||
| Gastric or Gastroesophageal Junction Adenocarcinoma ERBB2 E678Q Cases (N = 40) | |||
|---|---|---|---|
| Age Group (Years; Median 62) | Cancer Testing Panel | ||
| 60–69 | 11 | FoundationOne CDx | 33 |
| 70–79 | 10 | FoundationOne Liquid CDx | 6 |
| 50–59 | 9 | Guardant360 CDx | 1 |
| 40–49 | 8 | ||
| 30–39 | 1 | Treatment Response to Oxaliplatin * | |
| 80–89 | 1 | Complete Response | 0 |
| Partial Response | 5 | ||
| Sex | Stable Disease | 17 | |
| Female | 26 | Progressive Disease | 5 |
| Male | 14 | Not Evaluated | 8 |
| Smoking History | Treatment Response to Ramucirumab * | ||
| No | 23 | Complete Response | 0 |
| Yes | 16 | Partial Response | 2 |
| Unknown | 1 | Stable Disease | 10 |
| Progressive Disease | 10 | ||
| Drinking History | Not Evaluated | 7 | |
| No | 32 | ||
| Yes | 5 | Treatment Response to Nivolumab * | |
| Unknown | 3 | Complete Response | 0 |
| Partial Response | 5 | ||
| Metastatic Sites | Stable Disease | 10 | |
| Peritoneum | 21 | Progressive Disease | 10 |
| Lymph Node | 11 | Not Evaluated | 3 |
| Liver | 5 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, S.; Seino, M.; Sato, H.; Saito, Y.; Saito, K.; Yamada, Y.; Takahashi, K.; Kumanishi, R.; Fukui, T. Potential Resistance to Oxaliplatin-Based Regimens in Gastric Cancer Patients with ERBB2 R678Q Mutation: Evidence from a National Genomic Database. Curr. Issues Mol. Biol. 2025, 47, 430. https://doi.org/10.3390/cimb47060430
Suzuki S, Seino M, Sato H, Saito Y, Saito K, Yamada Y, Takahashi K, Kumanishi R, Fukui T. Potential Resistance to Oxaliplatin-Based Regimens in Gastric Cancer Patients with ERBB2 R678Q Mutation: Evidence from a National Genomic Database. Current Issues in Molecular Biology. 2025; 47(6):430. https://doi.org/10.3390/cimb47060430
Chicago/Turabian StyleSuzuki, Shuhei, Manabu Seino, Hidenori Sato, Yosuke Saito, Koki Saito, Yuta Yamada, Koshi Takahashi, Ryosuke Kumanishi, and Tadahisa Fukui. 2025. "Potential Resistance to Oxaliplatin-Based Regimens in Gastric Cancer Patients with ERBB2 R678Q Mutation: Evidence from a National Genomic Database" Current Issues in Molecular Biology 47, no. 6: 430. https://doi.org/10.3390/cimb47060430
APA StyleSuzuki, S., Seino, M., Sato, H., Saito, Y., Saito, K., Yamada, Y., Takahashi, K., Kumanishi, R., & Fukui, T. (2025). Potential Resistance to Oxaliplatin-Based Regimens in Gastric Cancer Patients with ERBB2 R678Q Mutation: Evidence from a National Genomic Database. Current Issues in Molecular Biology, 47(6), 430. https://doi.org/10.3390/cimb47060430






