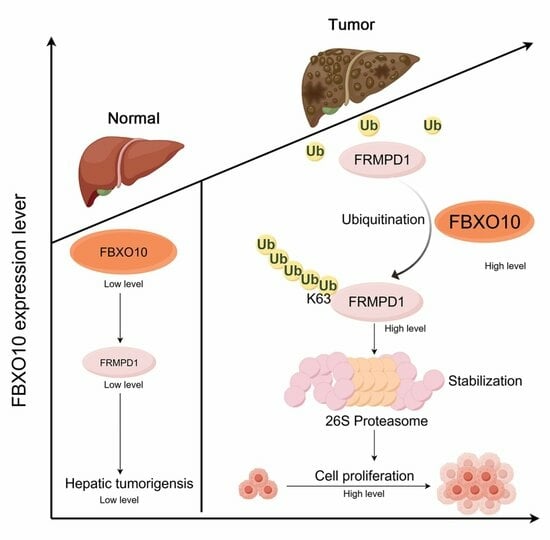
FBXO10 Drives Hepatocellular Carcinoma Proliferation via K63-Linked Ubiquitination and Stabilization of FRMPD1
Abstract
1. Introduction
2. Materials and Methods
2.1. TIMER Database Analysis
2.2. UALCAN Database Analysis
2.3. GEO Database Validation
2.4. Human Protein Atlas (HPA)
2.5. LinkedOmics Database Exploration
2.6. cBioPortal Genomic Profiling
2.7. Protein–Protein Interaction (PPI) Analysis
2.8. Cell Lines and Culture Conditions
2.9. Antibodies and Chemicals
2.10. Plasmids
2.11. Method Section Addition: Construction of Stable Cell Lines
2.12. Western Blot Analysis
2.13. Crystal Violet Proliferation Assay
2.14. Co-Immunoprecipitation (Co-IP) and Affinity Purification
2.15. Ubiquitination Analysis
2.16. Reverse Transcription-Quantitative PCR (RT-qPCR)
2.17. CCK-8 Cell Viability Assay
2.18. Illustrations
2.19. Statistical Analysis
3. Results
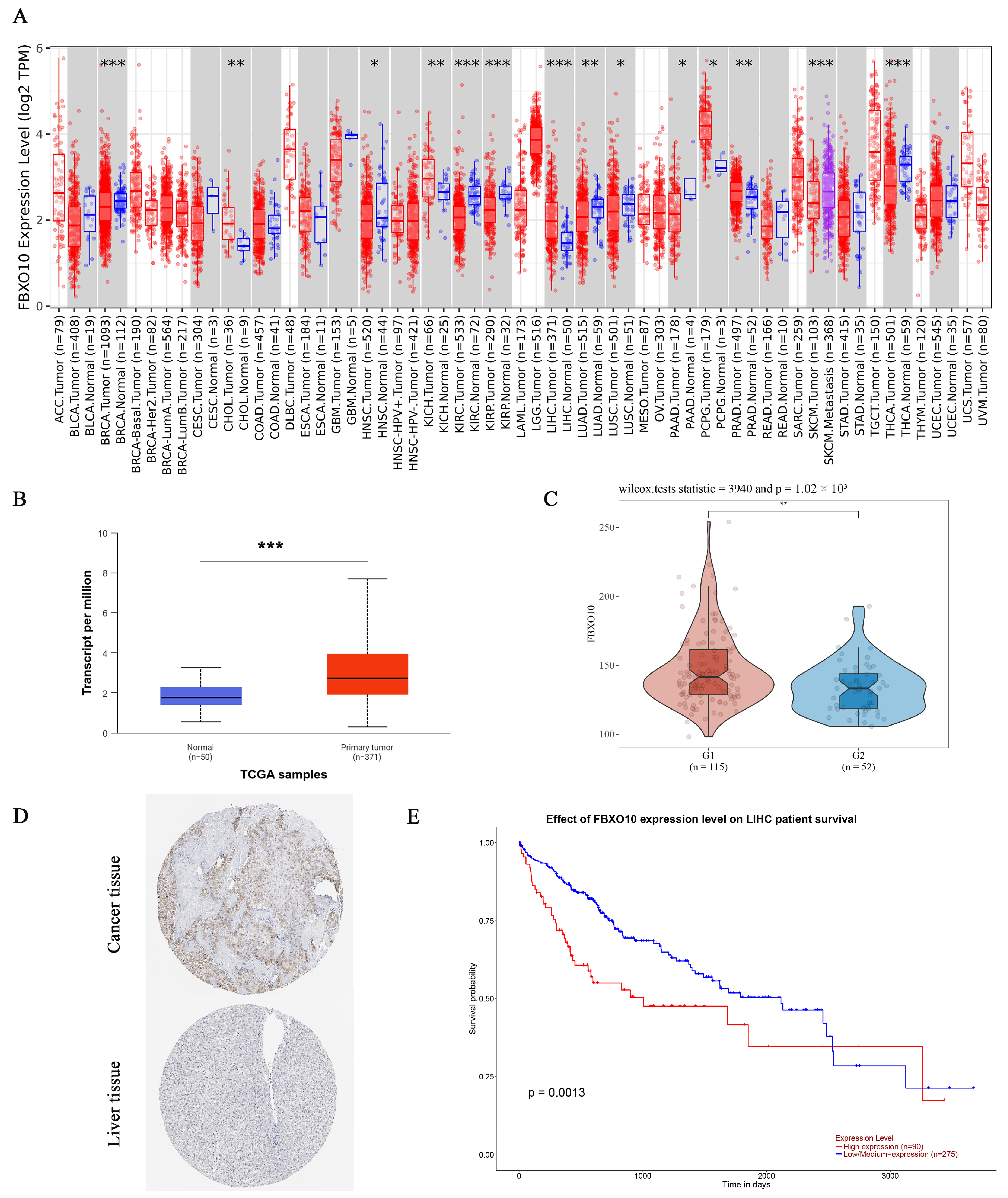
3.1. FBXO10 Exhibits Marked Upregulation and Is Strongly Correlated with Unfavorable Clinical Outcomes in Hepatocellular Carcinoma
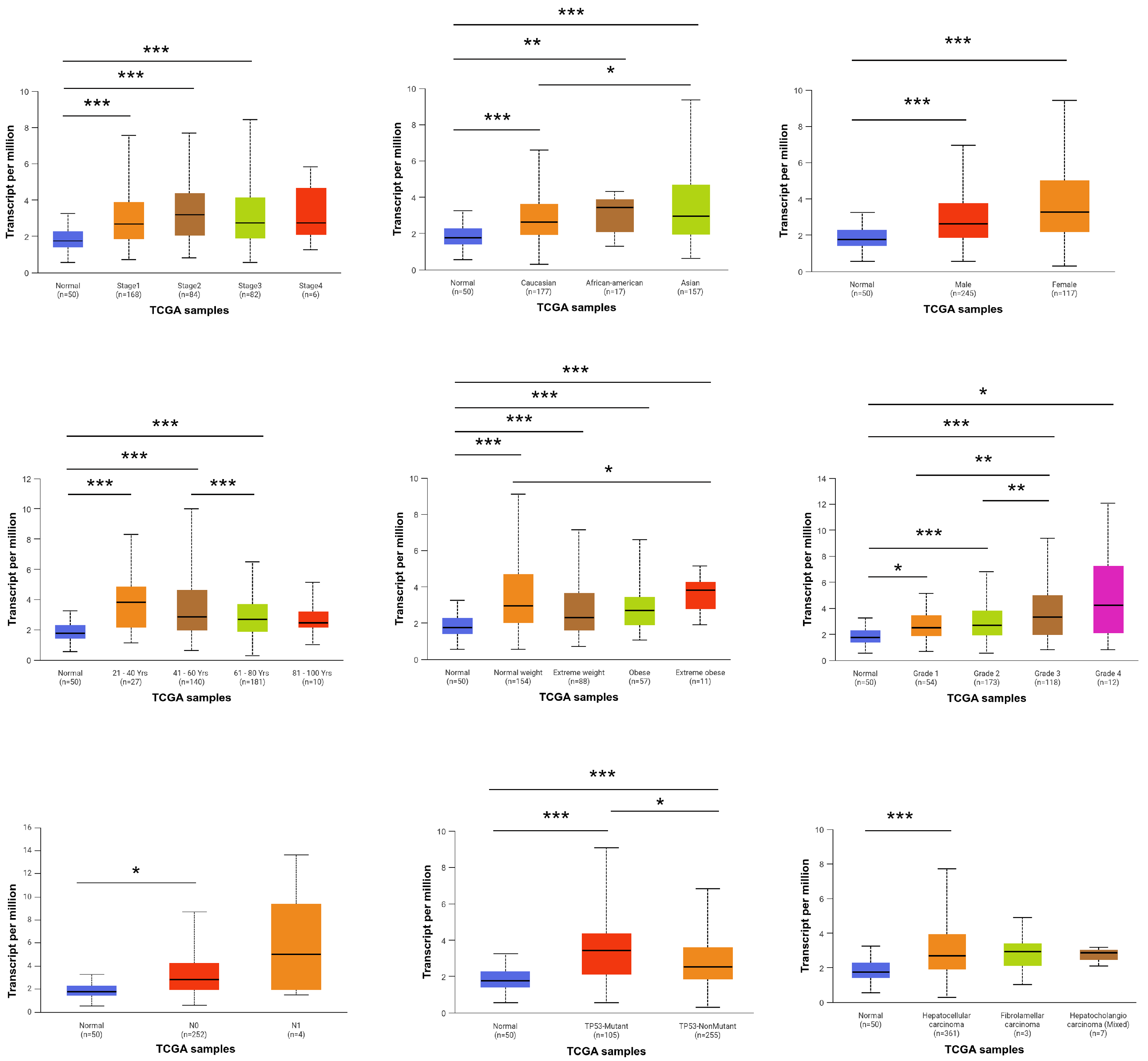
3.2. Association of FBXO10 Expression with Clinicopathological Features in HCC Patients
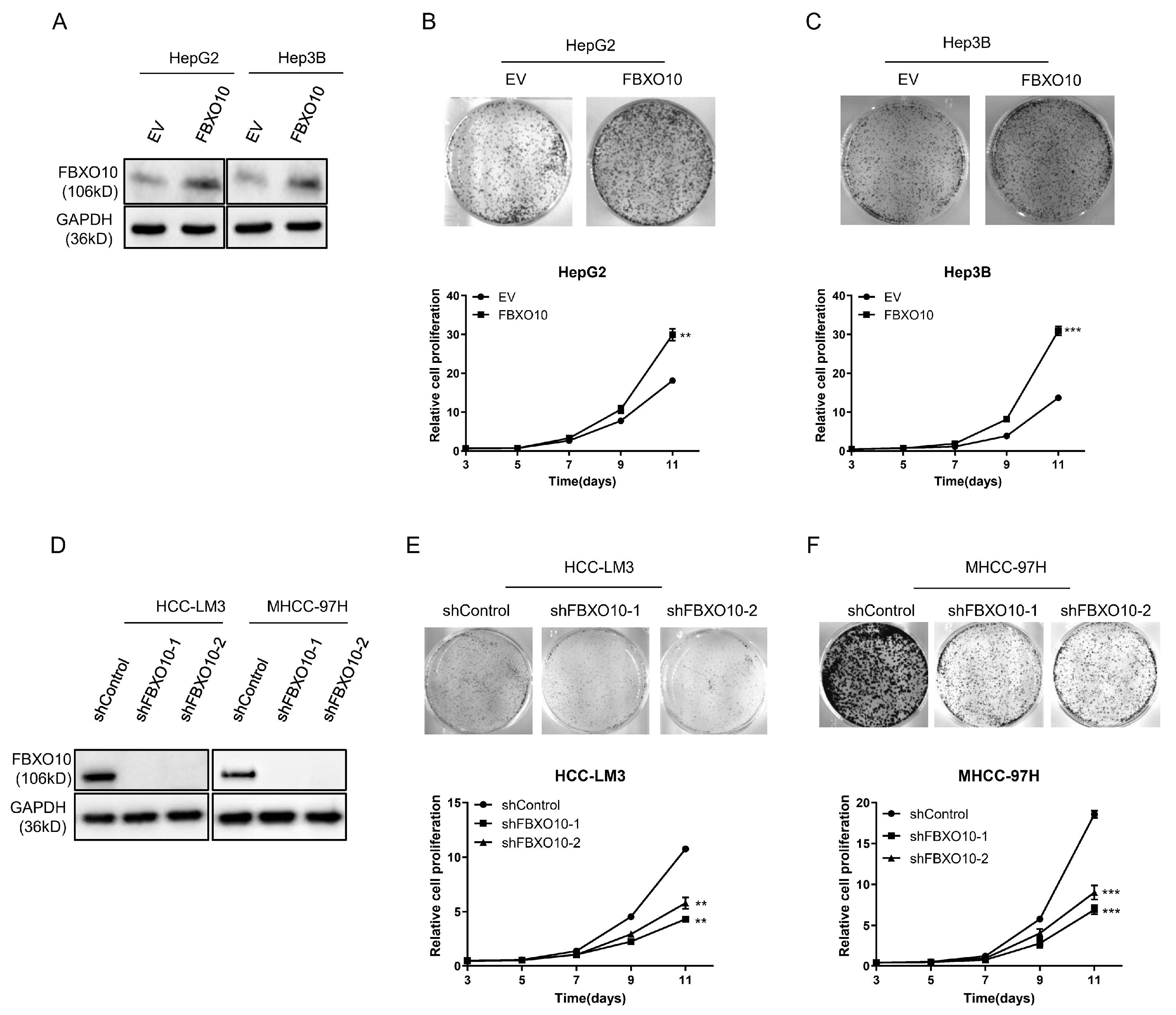
3.3. FBXO10 Exhibits Oncogenic Properties During Hepatocellular Carcinoma Proliferation
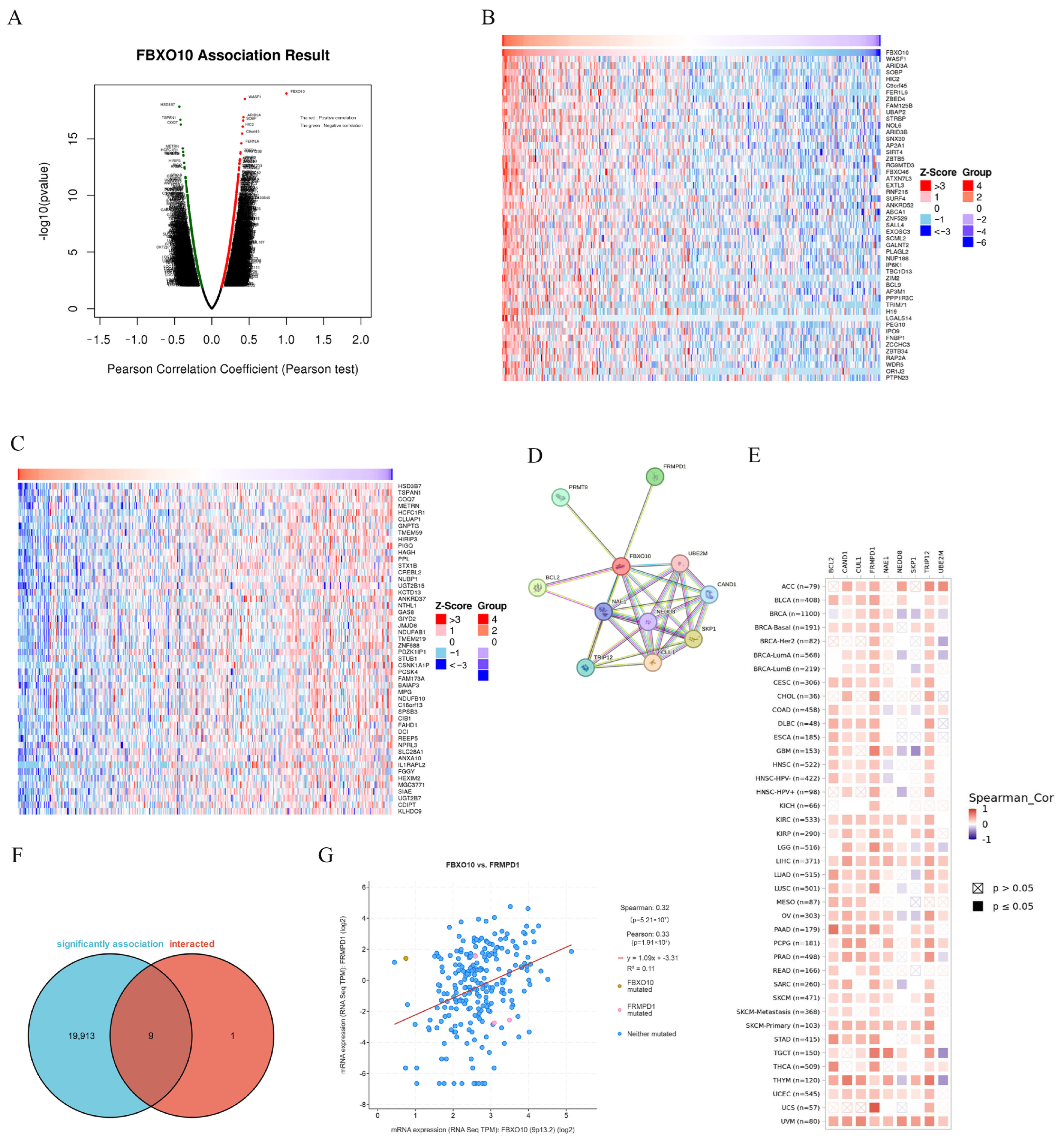
3.4. PPI Network Analysis of FBXO10-Related Genes
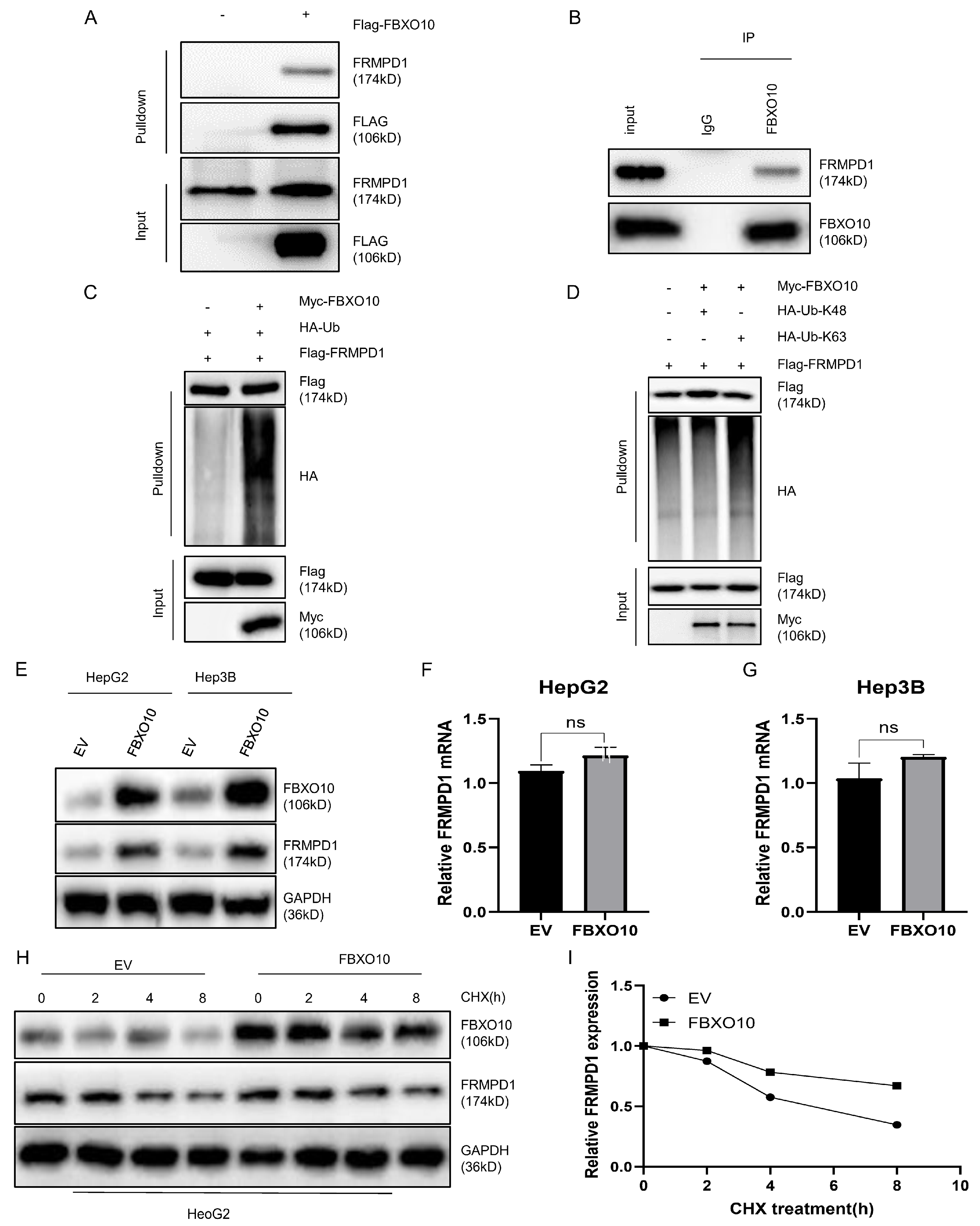
3.5. FBXO10 Facilitates K63-Linked Polyubiquitination of FRMPD1 to Stabilize Its Protein Levels
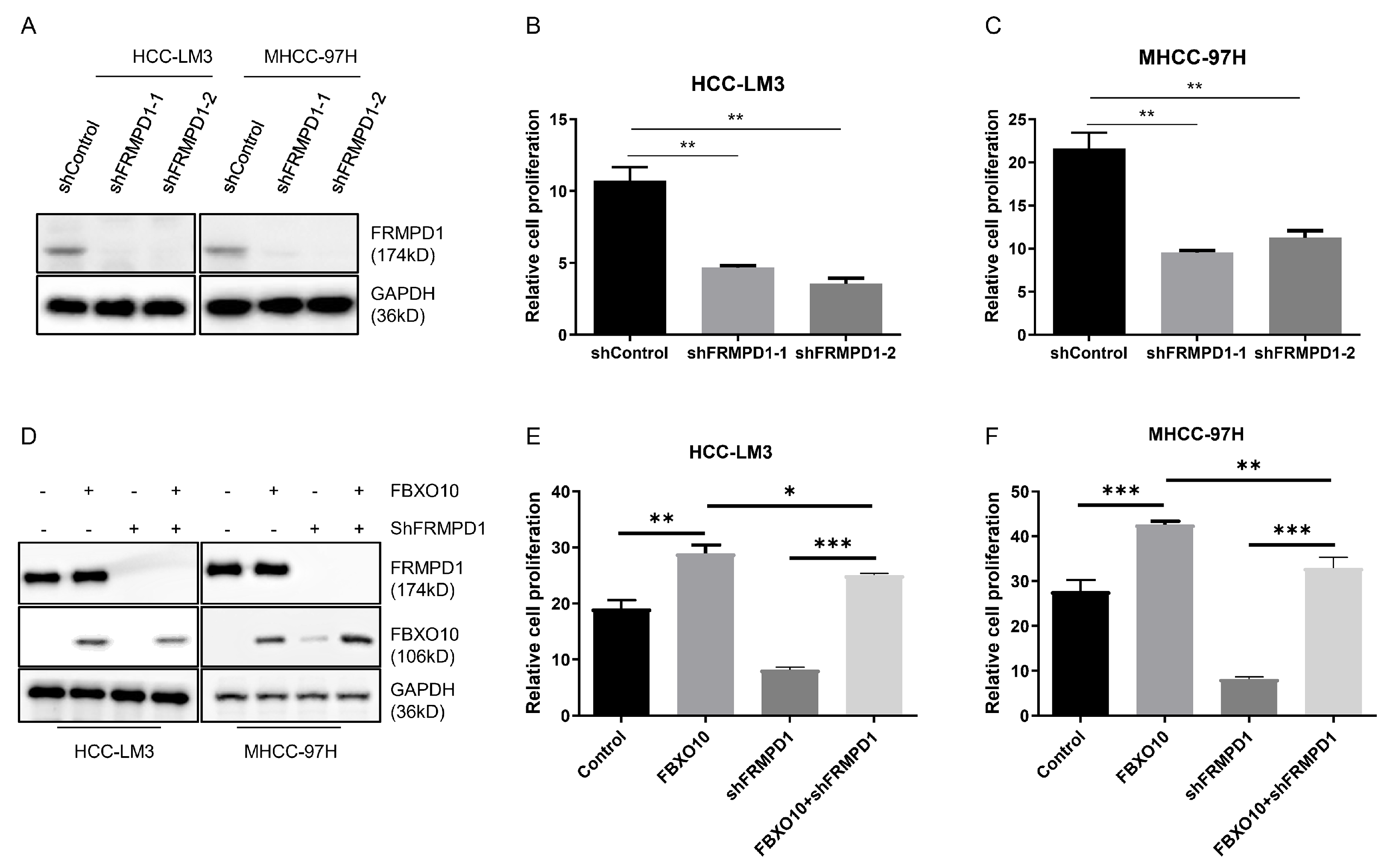
3.6. FBXO10 Enhances Hepatocellular Carcinoma Growth by Modulating FRMPD1
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HCC | Hepatocellular carcinoma |
| SCF | Skp1-Cullin-F-box |
| HGAL | Human germinal center-associated lymphoma |
| TIMER | Tumor Immune Estimation Resource |
| GEO | Gene Expression Omnibus |
| HPA | Human Protein Atlas |
| DEG | Differentially expressed gene |
| PPI | Protein–protein interaction |
| DMEM | Dulbecco’s Modified Eagle Medium |
| FBS | Fetal bovine serum |
| CHX | Cycloheximide |
| Co-IP | Co-immunoprecipitation |
| RT-qPCR | Reverse transcription-quantitative PCR |
| BCLA | Bladder urothelial carcinoma |
| BRCA | Breast invasive carcinoma |
| CESC | Cervical squamous cell carcinoma/endocervical adenocarcinoma |
| CHOL | Cholangiocarcinoma |
| COAD | Colon adenocarcinoma |
| ESCA | Esophageal carcinoma |
| HNSC | Head–neck squamous cell carcinoma |
| LIHC | Liver hepatocellular carcinoma |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| READ | Rectal adenocarcinoma |
| STAD | Stomach adenocarcinoma |
| UCEC | Uterine corpus endometrial carcinoma |
| KIRC | Kidney renal clear cell carcinoma |
| THCA | Thyroid carcinoma |
| UPS | Ubiquitin-proteasome system |
| PTM | Post-translational modification |
| DUB | Deubiquitinating enzyme |
| TPR | Tetratricopeptide repeat |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular Carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Pu, Z.; Ren, Z.; Xu, Q.; Wang, X.; Chen, J. Editorial: The role of tumor microenvironment in primary liver cancer therapeutic resistance. Front. Oncol. 2022, 12, 938557. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.-J.; Li, L.; Teng, Y.-X.; Zhang, Y.-X.; Liu, H.-T.; Huang, J.-L.; Liu, Z.-X.; Ma, L.; Zhong, J.-H. Treatments of Hepatocellular Carcinoma with Portal Vein Tumor Thrombus: Current Status and Controversy. J. Clin. Transl. Hepatol. 2021, 10, 147–158. [Google Scholar] [CrossRef]
- Li, Q.; Dong, Y.; Pan, Y.; Tang, H.; Li, D. Case Report: Clinical Responses to Tislelizumab as a First-Line Therapy for Primary Hepatocellular Carcinoma With B-Cell Indolent Lymphoma. Front. Immunol. 2021, 12, 634559. [Google Scholar] [CrossRef]
- Chan, T.; Wiltrout, R.H.; Weiss, J.M. Immunotherapeutic modulation of the suppressive liver and tumor microenvironments. Int. Immunopharmacol. 2011, 11, 879–889. [Google Scholar] [CrossRef]
- Tang, J.; Zhang, S.; Jiang, L.; Liu, J.; Xu, J.; Jiang, C.; Chen, Z.; Zhou, X.; Fuller, C.; Huang, J.; et al. Causal relationship between immune cells and hepatocellular carcinoma: A Mendelian randomisation study. J. Cancer 2024, 15, 4219–4231. [Google Scholar] [CrossRef]
- Su, K.; Wang, F.; Li, X.; Chi, H.; Zhang, J.; He, K.; Wang, Z.; Wen, L.; Song, Y.; Chen, J.; et al. Effect of external beam radiation therapy versus transcatheter arterial chemoembolization for non-diffuse hepatocellular carcinoma (≥5 cm): A multicenter experience over a ten-year period. Front. Immunol. 2023, 14, 1265959. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, L.; Ge, G.; Hu, K. Emerging Epigenetic-Based Nanotechnology for Cancer Therapy: Modulating the Tumor Microenvironment. Adv. Sci. 2023, 10, e2206169. [Google Scholar] [CrossRef]
- Donne, R.; Lujambio, A. The liver cancer immune microenvironment: Therapeutic implications for hepatocellular carcinoma. Hepatology 2023, 77, 1773–1796. [Google Scholar] [CrossRef]
- Su, L.; Luo, H.; Yan, Y.; Yang, Z.; Lu, J.; Xu, D.; Du, L.; Liu, J.; Yang, G.; Chi, H. Exploiting gender-based biomarkers and drug targets: Advancing personalized therapeutic strategies in hepatocellular carcinoma. Front. Pharmacol. 2024, 15, 1433540. [Google Scholar] [CrossRef] [PubMed]
- Skaar, J.R.; Pagan, J.K.; Pagano, M. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 2013, 14, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Tekcham, D.S.; Chen, D.; Liu, Y.; Ling, T.; Zhang, Y.; Chen, H.; Wang, W.; Otkur, W.; Qi, H.; Xia, T.; et al. F-box proteins and cancer: An update from functional and regulatory mechanism to therapeutic clinical prospects. Theranostics 2020, 10, 4150–4167. [Google Scholar] [CrossRef] [PubMed]
- Chiorazzi, M.; Rui, L.; Yang, Y.; Ceribelli, M.; Tishbi, N.; Maurer, C.W.; Ranuncolo, S.M.; Zhao, H.; Xu, W.; Chan, W.-C.C.; et al. Related F-box proteins control cell death in Caenorhabditis elegans and human lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 3943–3948. [Google Scholar] [CrossRef]
- Li, Y.; Bouchlaka, M.N.; Wolff, J.; Grindle, K.M.; Lu, L.; Qian, S.; Zhong, X.; Pflum, N.; Jobin, P.; Kahl, B.S.; et al. FBXO10 deficiency and BTK activation upregulate BCL2 expression in mantle cell lymphoma. Oncogene 2016, 35, 6223–6234. [Google Scholar] [CrossRef]
- Guo, F.; Luo, Y.; Jiang, X.; Lu, X.; Roberti, D.; Lossos, C.; Kunkalla, K.; Magistri, M.; Rui, L.; Verdun, R.; et al. Recent BCR stimulation induces a negative autoregulatory loop via FBXO10 mediated degradation of HGAL. Leukemia 2019, 34, 553–566. [Google Scholar] [CrossRef]
- Li, T.; JFan BWang NTraugh QChen JSLiu BLi, X.S.L.i.u. Timer: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.L.; Korch, C.T. Authentication of Human and Mouse Cell Lines by Short Tandem Repeat (Str) DNA Genotype Analysis. In Assay Guidance Manual; Markossian, S., Grossman, A., Arkin, M., Auld, D., Austin, C., Baell, J., Brimacombe, K., Chung, T.D.Y., Coussens, N.P., Dahlin, J.L., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative Pcr and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, P.; Inuzuka, H.; Wei, W. Roles of F-box proteins in cancer. Nat. Rev. Cancer 2014, 14, 233–247. [Google Scholar] [CrossRef]
- Zhang, Q.; Karnak, D.; Tan, M.; Lawrence, T.S.; Morgan, M.A.; Sun, Y. FBXW7 Facilitates Nonhomologous End-Joining via K63-Linked Polyubiquitylation of XRCC4. Mol. Cell 2016, 61, 419–433. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Mechanism of Lysine 48-Linked Ubiquitin-Chain Synthesis by the Cullin-RING Ubiquitin-Ligase Complex SCF-Cdc34. Cell 2005, 123, 1107–1120. [Google Scholar] [CrossRef]
- Kuang, P.; Tan, M.; Zhou, W.; Zhang, Q.; Sun, Y. SAG/RBX2 E3 ligase complexes with UBCH10 and UBE2S E2s to ubiquitylate β-TrCP1 via K11-linkage for degradation. Sci. Rep. 2016, 6, 37441. [Google Scholar] [CrossRef]
- Farinati, F.; Sergio, A.; Baldan, A.; Giacomin, A.; Di Nolfo, M.A.; Del Poggio, P.; Benvegnù, L.; Rapaccini, G.L.; Zoli, M.; Borzio, F.; et al. Early and very early hepatocellular carcinoma: When and how much do staging and choice of treatment really matter? A multi-center study. BMC Cancer 2009, 9, 33. [Google Scholar] [CrossRef]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in Globocan 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The Ubiquitin System. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Wang, W.; Yang, L.-Y.; Yang, Z.-L.; Peng, J.-X.; Yang, J.-Q. Elevated Expression of Autocrine Motility Factor Receptor Correlates with Overexpression of RhoC and Indicates Poor Prognosis in Hepatocellular Carcinoma. Dig. Dis. Sci. 2007, 52, 770–775. [Google Scholar] [CrossRef] [PubMed]
- Hoeller, D.; Dikic, I. Targeting the ubiquitin system in cancer therapy. Nature 2009, 458, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Yu, H.-Q.; Li, F.; Xiong, H.; Fang, L.; Zhang, J.; Bie, P.; Xie, C.-M. Elevated FBXL18 promotes RPS15A ubiquitination and SMAD3 activation to drive HCC. Hepatol. Commun. 2023, 7, e00198. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Zhao, Y.; Myung, J.K.; Yi, J.M.; Kim, M.-J.; Lee, S.-J. Suppression of breast cancer progression by FBXL16 via oxygen-independent regulation of HIF1α stability. Cell Rep. 2021, 37, 109996. [Google Scholar] [CrossRef]
- Rong, X.; Han, Q.; Lin, X.; Kremerskothen, J.; Wang, E. FRMPD1 activates the Hippo pathway via interaction with WWC3 to suppress the proliferation and invasiveness of lung cancer cells. Cancer Manag. Res. 2019, 11, 3395–3410. [Google Scholar] [CrossRef]
- Campla, C.K.; Mast, H.; Dong, L.; Lei, J.; Halford, S.; Sekaran, S.; Swaroop, A. Targeted deletion of an NRL- and CRX-regulated alternative promoter specifically silences FERM and PDZ domain containing 1 (Frmpd1) in rod photoreceptors. Hum. Mol. Genet. 2018, 28, 804–817. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Xu, B.; Kifayat, K.; Xie, Y.; Liu, X.; Dong, C.; Wang, L. FBXO10 Drives Hepatocellular Carcinoma Proliferation via K63-Linked Ubiquitination and Stabilization of FRMPD1. Curr. Issues Mol. Biol. 2025, 47, 391. https://doi.org/10.3390/cimb47060391
Liu W, Xu B, Kifayat K, Xie Y, Liu X, Dong C, Wang L. FBXO10 Drives Hepatocellular Carcinoma Proliferation via K63-Linked Ubiquitination and Stabilization of FRMPD1. Current Issues in Molecular Biology. 2025; 47(6):391. https://doi.org/10.3390/cimb47060391
Chicago/Turabian StyleLiu, Wuguang, Bin Xu, Kashif Kifayat, Yuhong Xie, Xiaolong Liu, Chengyong Dong, and Liming Wang. 2025. "FBXO10 Drives Hepatocellular Carcinoma Proliferation via K63-Linked Ubiquitination and Stabilization of FRMPD1" Current Issues in Molecular Biology 47, no. 6: 391. https://doi.org/10.3390/cimb47060391
APA StyleLiu, W., Xu, B., Kifayat, K., Xie, Y., Liu, X., Dong, C., & Wang, L. (2025). FBXO10 Drives Hepatocellular Carcinoma Proliferation via K63-Linked Ubiquitination and Stabilization of FRMPD1. Current Issues in Molecular Biology, 47(6), 391. https://doi.org/10.3390/cimb47060391