Genetic Analysis and Fingerprint Construction for Isatis indigotica Fort. Using SSR Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction
2.3. Genotyping with SSR Markers
2.4. Data Analysis
3. Results
3.1. Genetic Diversity
3.2. Genetic Differentiation
3.3. Unique Alleles
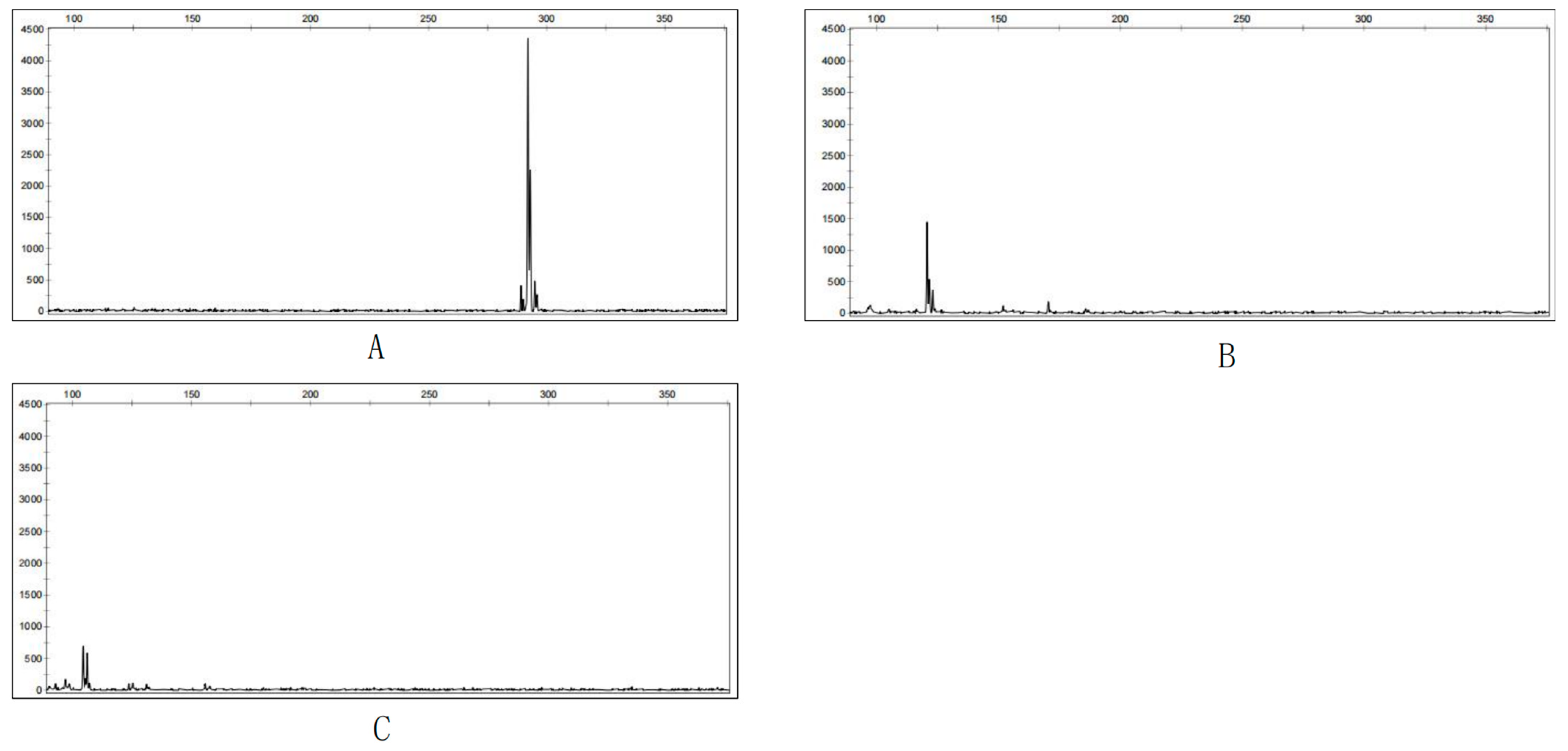
3.4. Fingerprinting of Cultivars
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, C.; Fan, N.; Xu, C.; Shao, S.; Shi, G.; Zhou, Y.; Wei, Y.; Wu, L.; Wang, B.; Shi, J.; et al. A synthetic derivative of bioactive constituents from Isatis indigotica ameliorates hypersensitivity and arthritis by inhibiting JAK2-STAT3 pathway in mice. Int. Immunopharmacol. 2023, 124, 110884. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lin, T.; Zhang, H.; Su, Y. Immune responses to foot-and-mouth disease DNA vaccines can be enhanced by coinjection with the Isatis indigotica extract. Intervirology 2005, 48, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Ma, Y.; He, Y.; Yang, H.; Chen, Y.; Wang, L.; Huang, D.; Qiu, S.; Tao, X.; Chen, W. A Network Pharmacology- Based Investigation to the Pharmacodynamic Material Basis and Mechanisms of the Anti-Inflammatory and Anti-Viral Effect of Isatis indigotica. Drug Des. Dev. Ther. 2021, 15, 3193–3206. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Gao, D.; Jeong, W.S.; Kim, C.T.; Cho, C.W.; Kim, H.M.; Kang, J.S. Anti-Wrinkle Effect of Isatis indigotica Leaf Extract: Evaluation of Antioxidant, Anti-Inflammation, and Clinical Activity. Antioxidants 2021, 10, 1339. [Google Scholar] [CrossRef]
- Shi, X.; Geng, J.; Feng, J.; Yang, Y.; Ma, X.; Chen, W.; Xiao, Y. Identification and investigation of a novel NADP+-dependent secoisolariciresinol dehydrogenase from Isatis indigotica. Front. Plant Sci. 2022, 13, 1035121. [Google Scholar] [CrossRef]
- Yang, S.; Wang, Z. The complete chloroplast genome sequence of the medicinal and economic plant woad Isatis indigotica (Brassicaceae). Mitochondrial DNA Part B Resour. 2017, 2, 514–515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.-K.; Tan, Y.-P.; Xu, H.-H.; Sun, S.-F.; Xiao, J.-C.; Xu, Z.-Y.; Wang, C.-C.; Zhu, H.-X.; Yang, J.; Li, D.-Y.; et al. Identification and expression analysis of whole gene family of Isatis indigotica 4-coumarate: CoA ligase. Zhongguo Zhong Yao Za Zhi Zhongguo Zhongyao Zazhi China J. Chin. Mater. Med. 2024, 49, 361–369. [Google Scholar]
- Shi, Y.-H.; Xie, Z.-Y.; Wang, R.; Huang, S.-J.; Li, Y.-M.; Wang, Z.-T. Quantitative and Chemical Fingerprint Analysis for the Quality Evaluation of Isatis indigotica based on Ultra-Performance Liquid Chromatography with Photodiode Array Detector Combined with Chemometric Methods. Int. J. Mol. Sci. 2012, 13, 9035–9050. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Hong, Y.; Koh, H.L. Chemical fingerprinting of Isatis indigotica root by RP-HPLC and hierarchical clustering analysis. J. Pharm. Biomed. Anal. 2005, 38, 514–520. [Google Scholar] [CrossRef]
- Cho, J.; Sa, K.J.; Park, H.; Heo, T.H.; Lee, S.; Lee, J.K. Association analysis of leaf aromatic substances in cultivated and weedy types of Perilla crop using SSR markers. Heliyon 2024, 10, e34995. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, P.; Tang, W.; Zheng, J.; Fan, F.; Jiang, X.; Li, Z.; Fang, Y. Simultaneous determination of subspecies and geographic origins of 110 rice cultivars by microsatellite markers. Food Chem. 2024, 445, 138657. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Bhargava, A. In-silico identification of simple sequence repeat (SSR) markers and phylogenetic analysis from chloroplast genomes of the genus Bambusa. Gene Rep. 2024, 37, 102048. [Google Scholar] [CrossRef]
- Saxena, P.; Malik, L.; Pattanaik, S.; Gudipalli, P. Genetic diversity analysis of a germplasm collection of red sanders (Pterocarpus santalinus L.f.) using cross-species SSR, ISSR and RAPD markers. S. Afr. J. Bot. 2024, 174, 856–869. [Google Scholar] [CrossRef]
- Wu, F.; Cai, G.; Xi, P.; Guo, Y.; Xu, M.; Li, A. Genetic Diversity Analysis and Fingerprint Construction for 87 Passionfruit (Passiflora spp.) Germplasm Accessions on the Basis of SSR Fluorescence Markers. Int. J. Mol. Sci. 2024, 25, 10815. [Google Scholar] [CrossRef]
- Laosatit, K.; Amkul, K.; Chankaew, S.; Somta, P. Molecular genetic diversity of winged bean gene pool in Thailand assessed by SSR markers. Hortic. Plant J. 2022, 8, 81–88. [Google Scholar] [CrossRef]
- Riangwong, K.; Wanchana, S.; Aesomnuk, W.; Saensuk, C.; Nubankoh, P.; Ruanjaichon, V.; Kraithong, T.; Toojinda, T.; Vanavichit, A.; Arikit, S. Mining and validation of novel genotyping-by-sequencing (GBS)-based simple sequence repeats (SSRs) and their application for the estimation of the genetic diversity and population structure of coconuts (Cocos nucifera L.) Thailand. Hortic. Res. 2020, 7, 156. [Google Scholar] [CrossRef]
- Xue, Y.; Liu, R.; Xue, J.; Wang, S.; Zhang, X. Genetic diversity and relatedness analysis of nine wild species of tree peony based on simple sequence repeats markers. Hortic. Plant J. 2021, 7, 579–588. [Google Scholar] [CrossRef]
- Lai, R.; Xia, Y.; Li, R.; Yuan, Q.; Zhao, W.; Siddique, K.H.M.; Guo, P. Identifying SSR/InDel loci related to tobacco bacterial wilt resistance using association mapping. Heliyon 2024, 10, e38939. [Google Scholar] [CrossRef] [PubMed]
- Mukta, S.; Bappy, M.N.I.; Bhuiyan, J.; Zohora, F.T.; Afrin, D. Assessment of genetic diversity in Bangladeshi rice (Oryza sativa L.) varieties utilizing SSR markers. Gene Rep. 2024, 37, 102051. [Google Scholar]
- Tiruneh, A.A.; Geletu, K.T.; Yao, N.K.; Weldegiorgis, K.D. The genetic diversity and population structure of wild and cultivated Avena species in Ethiopia using a SSR markers. Heliyon 2024, 10, e38942. [Google Scholar] [CrossRef]
- Lawson, M.J.; Zhang, L. Distinct patterns of SSR distribution in the Arabidopsis thaliana and rice genomes. Genome Biol. 2006, 7, R14. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Marcel, T.C.; Ramsay, L.; Russell, J.; Roeder, M.S.; Stein, N.; Waugh, R.; Langridge, P.; Niks, R.E.; Graner, A. A high density barley microsatellite consensus map with 775 SSR loci. Theor. Appl. Genet. 2007, 114, 1091–1103. [Google Scholar] [CrossRef]
- Luo, C.; Chen, D.; Cheng, X.; Liu, H.; Li, Y.; Huang, C. SSR Analysis of Genetic Relationship and Classification in Chrysanthemum Germplasm Collection. Hortic. Plant J. 2018, 4, 73–82. [Google Scholar] [CrossRef]
- Arlotta, C.; Toscano, V.; Genovese, C.; Calderaro, P.; Puglia, G.D.; Raccuia, S.A. Nutraceutical Content and Genetic Diversity Share a Common Pattern in New Pomegranate Genotypes. Molecules 2022, 27, 389. [Google Scholar] [CrossRef]
- Ouni, R.; Zborowska, A.; Sehic, J.; Choulak, S.; Hormaza, J.I.; Garkava-Gustavsson, L.; Mars, M. Genetic Diversity and Structure of Tunisian Local Pear Germplasm as Revealed by SSR Markers. Hortic. Plant J. 2020, 6, 61–70. [Google Scholar] [CrossRef]
- Karkhaneh, A.; Salari, H.; Cheghamirza, K.; Zarei, L. Agronomic and Molecular Identification of Drought-Tolerant Bread Wheat Varieties in Iran. J. Plant Growth Regul. 2025, 1–12. [Google Scholar] [CrossRef]
- Romano, A.; Masi, P.; Aversano, R.; Carucci, F.; Palomba, S.; Carputo, D. Microstructure and tuber properties of potato varieties with different genetic profiles. Food Chem. 2018, 239, 789–796. [Google Scholar] [CrossRef]
- Patel, R.; Memon, J.; Kumar, S.; Patel, D.A.; Sakure, A.A.; Patel, M.B.; Das, A.; Karjagi, C.G.; Patel, S.; Patel, U.; et al. Genetic Diversity and Population Structure of Maize (Zea mays L.) Inbred Lines in Association with Phenotypic and Grain Qualitative Traits Using SSR Genotyping. Plants 2024, 13, 823. [Google Scholar]
- Rezk, A.A.; Mohamed, H.I.; El-Beltagi, H.S. Genetic variability and diversity analysis in Oryza sativa L. genotypes using quantitative traits and SSR markers. Saudi J. Biol. Sci. 2024, 31, 103944. [Google Scholar] [CrossRef]
- Jang, S.J.; Sa, K.J.; Fu, Z.Y.; Lee, J.K. Association mapping analysis for cultivated and weedy types of Perilla crop collected from South Korea using morphological characteristics and SSR markers. Heliyon 2024, 10, e26720. [Google Scholar] [CrossRef] [PubMed]
- Sharma, L.; Kumar, R.; Tare, K.; Lamichaney, S. Assessment of genetic diversity and population structure analysis in Himalayan garlic genotypes from Sikkim using SSR markers. Mol. Biol. Rep. 2025, 52, 17. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Tian, Y.; Yu, F.; Qian, J.S.; Wang, F.J.; Li, X.; Li, T.Y.; Zhang, X.F.; Huang, D.Z.; Zhao, X.J. Association analysis of the molecular characteristics and floral traits of Iris x germanica. Czech J. Genet. Plant Breed. 2025, 1–12. [Google Scholar] [CrossRef]
- Ye, Y.; Tan, J.; Lin, J.; Zhang, Y.; Zhu, G.; Nie, C.; Huang, L.; Zhou, Y.; Xu, Y. Genome-wide identification of SSR markers for Curcuma alismatifolia Gagnep., and their potential for wider application in this genus. J. Appl. Res. Med. Aromat. Plants 2024, 42, 100572. [Google Scholar] [CrossRef]
- Gao, C.; Chen, C.; Liu, N.; Liu, F.; Su, X.; Liu, C.; Huang, Q. Genetic Diversity and Association Analysis of Traits Related to Water-Use Efficiency and Nitrogen-Use Efficiency of Populus deltoides Based on SSR Markers. Int. J. Mol. Sci. 2024, 25, 11515. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.; Li, Y.; Deng, F.; Yang, H.; Li, J.; Chen, J.; Sun, J.; Li, G.; Fernando, W.G.D.; Gao, H. Molecular Insights into the Reproductive Patterns and Genetic Structure of Wheat Stripe Rust in Ili, Xinjiang. Int. J. Mol. Sci. 2024, 25, 12357. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wang, H.X.; Yang, F.Q.; Xi, J.F.; Zou, Z.W.; Xia, B. Genetic diversity of Isatis indigotica germplasm resources based on ISSR-PCR technique. J. Nanchang Univ. (Nat. Sci.) 2019, 43, 577–582. [Google Scholar]
- Luo, B.; Jiang, J.X.; Liu, C.X.; Wei, J.; Wang, X.; Cao, Z.F.; Zhang, C.J.; Liu, N.A.; Lv, Y.Y.; Wei, Z.W.; et al. Characterization of the KNOX Gene Family in Regulating Alfalfa Compound Leaf Development. J. Plant Growth Regul. 2025, 1–16. [Google Scholar] [CrossRef]
- Gupta, P.; Udupa, S.M.; Gupta, D.S.; Kumar, J.; Kumar, S. Population structure analysis and determination of neurotoxin content in a set of grass pea (Lathyrus sativus L.) accessions of Bangladesh origin. Crop J. 2018, 6, 435–442. [Google Scholar]
- Zhang, L.; Cai, R.; Yuan, M.; Tao, A.; Xu, J.; Lin, L.; Fang, P.; Qi, J. Genetic diversity and DNA fingerprinting in jute (Corchorus spp.) based on SSR Markers. Crop J. 2015, 3, 416–422. [Google Scholar] [CrossRef]
- Zhao, J.J.; Sun, L.L.; Pi, Z.; Li, S.N.; Wu, Z.D. Genetic Diversity Analysis of 89 Monogerm Maintainer Lines of Sugar Beet. Sugar Tech. 2025, 1–15. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.D. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Devi, A.; Kumari, N.; Masand, M.; Sharma, B.; Majeed, A.; Rahim, M.S.; Singh, S.; Mohapatra, K.P.; Sharma, R.K. Microsatellite marker resource creation, genetic diversity assessment and core prediction in Valeriana jatamansi Jones. J. Appl. Res. Med. Aromat. Plants 2025, 45, 100616. [Google Scholar] [CrossRef]
- Su, Y.; Fu, J.; Xie, H.; Huang, Z.; Li, Y.; Luo, Y.; Zhou, X.; Li, Y.; Li, J.; Sun, Y.; et al. SSR markers development and their application in genetic diversity of burdock (Arctium lappa L.) germplasm. BMC Plant Biol. 2025, 25, 196. [Google Scholar] [CrossRef] [PubMed]
- Baite, M.S.; Biswal, S.; Pandit, E.; Pradhan, S.K.; Barik, S.R. Molecular screening of germplasm lines for false smut resistance in rice. Physiol. Mol. Plant Pathol. 2025, 136, 102571. [Google Scholar] [CrossRef]
- Feng, J.; Huang, D.; Yang, Y.; Chen, J.; Qiu, S.; Lv, Z.; Ma, X.; Li, Y.; Li, R.; Xiao, Y.; et al. Isatis indigotica: From (ethno) botany, biochemistry to synthetic biology. Mol. Hortic. 2021, 1, 17. [Google Scholar] [CrossRef]
- Chen, Q.; Lan, H.-Y.; Peng, W.; Rahman, K.; Liu, Q.-C.; Luan, X.; Zhang, H. Isatis indigotica: A review of phytochemistry, pharmacological activities and clinical applications. J. Pharm. Pharmacol. 2021, 73, 1137–1150. [Google Scholar] [CrossRef] [PubMed]
- Sai Timmarao, K.; Ponnam, N.; Lakshmanareddy, D.C.; Krishna Reddy, M.; Venkataravanappa, V.; Roshini, P.; Shaik, M.; Manoj, B.P.; Madhavi Reddy, K. Molecular mapping and development of SSR markers associated with Chilli leaf curl virus resistance in chilli (Capsicum annuum L.). Genomics 2025, 117, 111015. [Google Scholar] [CrossRef] [PubMed]
- Morlova, E.D.; Mladin, D. Evaluation of the unavailability of the primary circuit of Triga SSR reactor, importance factors and risk criteria for its components. Kerntechnik 2024, 89, 632–643. [Google Scholar] [CrossRef]







| Number | Cultivar | Origin | Leave Shape | Number | Cultivar | Origin | Leave Shape |
|---|---|---|---|---|---|---|---|
| 1 | HLJDXALSa | Heilongjiang, Daxinganling | Small | 15 | HBHSL | HeBei, HengShui | Large |
| 2 | JSSQS | JiangSu, SuQian | Small | 16 | HBBDS | HeBei, BaoDing | Small |
| 3 | YNHHSa | YunNan, HongHe | Small | 17 | GSZYSBL | GanSu, ZhangYe | Large |
| 4 | SDWFS | Shandong, Weifang | Small | 18 | GSZYSBS | GanSu, ZhangYe | Small |
| 5 | HNXYL | HeNan, XinYang | Large | 19 | HLJDQL | HeiLongJiang, DaQing | Large |
| 6 | JSHAS | Jiangsu, Huaian | Small | 20 | HLJDQS | HeiLongJiang, DaQing | Small |
| 7 | GSZYYLS | Gansu, Minle | Small | 21 | SXXZL | ShanXi, XinZhou | Large |
| 8 | GSLZS | Gansu, Lanzhou | Small | 22 | SXXZS | ShanXi, XinZhou | Small |
| 9 | AHBZS | Anhui, Bozhou | Small | 23 | GDXNL | GuangDong, XingNing | Large |
| 10 | YNHHSb | YunNan, HongHe | Small | 24 | GSDXLa | GanSu, DingXi | Large |
| 11 | HLJDBS | Heilongjiang, Duerbote | Small | 25 | GSDXLb | GanSu, DingXi | Large |
| 12 | SXYCS | ShanXi, YunCheng | Small | 26 | GSZYYLS2 | Gansu, Minle | Small |
| 13 | SXLLS | ShanXi, LvLiang | Small | 27 | GSZYYLS3 | Gansu, Minle | Small |
| 14 | HLJDXALSb | Heilongjiang, Daxinganling | Small |
| Locus | Repeat Motif | Forward Primer (5′~3′) | Reverse Primer (5′~3′) | Size | Tm | Fluorescent Labeling |
|---|---|---|---|---|---|---|
| BLG-P1 | (GA)10 | TCTCTTGATTCTTTTTGACGGA | TCGTTTCCTGTTCCCTTTTG | 109 | 51 | FAM |
| BLG-P2 | (TC)10 | GTGTTTGTGTTTCCCCCATC | GAAAAACGGTGCCACAATCT | 121 | 53 | FAM |
| BLG-P3 | (GA)12 | CGAATTTACCACGAACCGAT | GAAAACGGTGGCATGTCTCT | 136 | 53 | FAM |
| BLG-P4 | (TC)10 | CAAGCACAAGTGGTCCAAAA | GCTTGGTTTTCAACATGAGG | 151 | 52 | FAM |
| BLG-P5 | (TG)12 | AGAAGGCTGCACCAAGTGTT | GAGGAAGGATCCAAATGCAA | 157 | 54 | FAM |
| BLG-P6 | (GA)11 | CTTCCCATTTAGCGAACCAA | CTTCCGGTTCGATTTTTCAA | 165 | 51 | HEX |
| BLG-P7 | (GTT)9 | TCGTTCGGTTATGACGGCTCTT | CGTAAGGTCCAATGGCGAATAT | 176 | 55 | HEX |
| BLG-P8 | (ACCAAT)7 | CTCCAAGACCATCTTCCCAA | TGGGAAAAAGACAGGCAATC | 180 | 53 | HEX |
| BLG-P9 | (AG)11 | ACTCTCAGGGCAGCGACAGAAA | TCTCCCACCACCACCACAAATA | 192 | 58 | HEX |
| BLG-P10 | (TC)13 | TTCGATTATTGGGCGAAGTT | TAGCCACACCGAGATCAAGA | 193 | 53 | HEX |
| BLG-P11 | (TC)10 | TAAACCGTCGCAACAGAGAC | ACCTGCCATTGCCTAACAAG | 201 | 55 | ROX |
| BLG-P12 | (TC)15 | ATTTCGGTGCATTGCTTTCT | TAACTTCTTCGGTCTTGCCG | 204 | 52 | ROX |
| BLG-P13 | (AT)16 | CACCATTAATAGGAATGTGGCA | TTTAATGCATGGTTGGCATC | 210 | 51 | ROX |
| BLG-P14 | (TG)12 | TGGAGCAAGAAGAGAGGTTAGG | TTTGAAGCTCTGCAGGGAAAGT | 212 | 55 | ROX |
| BLG-P15 | (AG)15 | TGAGCATGCGAATCAAACTC | CGAATTGGGGAGATATTGGA | 235 | 52 | ROX |
| BLG-P16 | (AG)11 | GACATTTCCACCAGCAAGGT | AAGTGCTAGTTGGAAGCCGA | 248 | 55 | NED |
| BLG-P17 | (AG)12 | CAAACCACCACCGGACCACTAT | GCCTCTCCATCCTCGTCGTATT | 253 | 58 | NED |
| BLG-P18 | (CA)15 | TCCCCTTCTTTCTTCTATTGC | TCTCCGCCATAGATTTCTGC | 257 | 53 | NED |
| BLG-P19 | (GA)13 | TATGTAGCCATCCCTGCCTC | ATGGCGTCAATGACATACCA | 274 | 53 | NED |
| BLG-P20 | (TC)11 | TGGGAAGGAAGAAGAAGCAA | TGACGACAACGACTTCAACA | 279 | 52 | NED |
| Locus | Sample Size | Na | Ne | Ho | He | I | PIC |
|---|---|---|---|---|---|---|---|
| BLG-P1 | (GA)10 | 5 | 1.20 | 0.86 | 0.14 | 0.42 | 0.16 |
| BLG-P2 | (TC)10 | 4 | 1.52 | 0.66 | 0.34 | 0.63 | 0.30 |
| BLG-P3 | (GA)12 | 7 | 2.53 | 0.34 | 0.66 | 1.21 | 0.55 |
| BLG-P4 | (TC)10 | 6 | 2.60 | 0.38 | 0.62 | 1.17 | 0.55 |
| BLG-P5 | (TG)12 | 7 | 3.73 | 0.143 | 0.86 | 1.50 | 0.69 |
| BLG-P6 | (GA)11 | 3 | 2.06 | 0.36 | 0.64 | 0.77 | 0.40 |
| BLG-P7 | (GTT)9 | 5 | 2.85 | 0.57 | 0.43 | 1.23 | 0.59 |
| BLG-P8 | (ACCAAT)7 | 5 | 2.65 | 0.52 | 0.48 | 1.13 | 0.56 |
| BLG-P9 | (AG)11 | 3 | 1.82 | 0.68 | 0.32 | 0.76 | 0.39 |
| BLG-P10 | (TC)13 | 5 | 1.40 | 0.71 | 0.29 | 0.64 | 0.27 |
| BLG-P11 | (TC)10 | 4 | 2.13 | 0.64 | 0.36 | 0.99 | 0.48 |
| BLG-P12 | (TC)15 | 10 | 5.39 | 0.29 | 0.71 | 1.87 | 0.79 |
| BLG-P13 | (AT)16 | 11 | 7 | 0.25 | 0.75 | 2.11 | 0.84 |
| BLG-P14 | (TG)12 | 2 | 1.67 | 0.74 | 0.26 | 0.59 | 0.32 |
| BLG-P15 | (AG)15 | 5 | 3.97 | 0.26 | 0.74 | 1.47 | 0.71 |
| BLG-P16 | (AG)11 | 5 | 1.25 | 0.86 | 0.14 | 0.48 | 0.19 |
| BLG-P17 | (AG)12 | 4 | 1.69 | 0.50 | 0.50 | 0.79 | 0.37 |
| BLG-P18 | (CA)15 | 4 | 2.55 | 0.52 | 0.48 | 1.10 | 0.55 |
| BLG-P19 | (GA)13 | 7 | 3.53 | 0.69 | 0.31 | 1.55 | 0.69 |
| BLG-P20 | (TC)11 | 7 | 3.58 | 0.38 | 0.62 | 1.47 | 0.68 |
| Mean | 5 | 2.33 | 0.52 | 0.48 | 0.98 | 0.465 |
| Locus | Unique Allele Lengths in bp (Locus) | Number of Unique Alleles |
|---|---|---|
| HLJDXALSa | 202 (BLGP12); 288 (BLGP16) | 2 |
| JSSQS | 165 (BLGP6) | 1 |
| YNHHSa | 212 (BLGP12) | 1 |
| HNXYL | 179 (BLGP4) | 1 |
| JSHAS | 248 (BLGP13); 121 (BLGP2) | 2 |
| GSLZS | 316 (BLGP19) | 1 |
| YNHHSb | 119 (BLGP1) | 1 |
| HLJDBS | 298 (BLGP16) | 1 |
| SXYCS | 260 (BLGP20) | 1 |
| HBBDS | 210 (BLGP12); 234 (BLGP13); 296 (BLGP19) | 3 |
| GSZYSBS | 117,124 (BLGP1) | 1 |
| HLJDQS | 128 (BLGP3) | 1 |
| SXXZL | 196 (BLGP12); 252 (BLGP13) | 2 |
| SXXZS | 284 (BLGP19) | 1 |
| GSDXLa | 113 (BLGP2); 153 (BLGP4); 179 (BLGP8); 240 (BLGP20) | 4 |
| GSZYYLS2 | 161 (BLGP5) | 1 |
| GSZYYLS3 | 140 (BLGP3) | 1 |
| BNIFDC | 177 (BLGP5); 191 (BLGP7); 201 (BLGP10); 209 (BLGP10); 294 (BLGP16) | 5 |
| Total | 29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, X.; Xu, H.; Dong, Y.; Cui, H.; Sun, M.; Wang, H.; Liu, Y.; Meng, L.; Zheng, C. Genetic Analysis and Fingerprint Construction for Isatis indigotica Fort. Using SSR Markers. Curr. Issues Mol. Biol. 2025, 47, 146. https://doi.org/10.3390/cimb47030146
Xing X, Xu H, Dong Y, Cui H, Sun M, Wang H, Liu Y, Meng L, Zheng C. Genetic Analysis and Fingerprint Construction for Isatis indigotica Fort. Using SSR Markers. Current Issues in Molecular Biology. 2025; 47(3):146. https://doi.org/10.3390/cimb47030146
Chicago/Turabian StyleXing, Xiangyu, Haijun Xu, Yan Dong, Hanwen Cui, Mingrui Sun, Hong Wang, Yang Liu, Li Meng, and Chunying Zheng. 2025. "Genetic Analysis and Fingerprint Construction for Isatis indigotica Fort. Using SSR Markers" Current Issues in Molecular Biology 47, no. 3: 146. https://doi.org/10.3390/cimb47030146
APA StyleXing, X., Xu, H., Dong, Y., Cui, H., Sun, M., Wang, H., Liu, Y., Meng, L., & Zheng, C. (2025). Genetic Analysis and Fingerprint Construction for Isatis indigotica Fort. Using SSR Markers. Current Issues in Molecular Biology, 47(3), 146. https://doi.org/10.3390/cimb47030146





