Biodiversity and Evaluation of Genetic Resources of Some Coffee Trees Grown in Al-Baha, Saudi Arabia
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Morphological Measurements
2.2. DNA Extraction and ISSR Fingerprinting
2.3. Data Analysis
2.4. iMEC Analysis
3. Results
3.1. Morphological Variation Among Coffea arabica Cultivars
3.2. Diversity of Cultivars Based on Morphological Variations
3.3. ISSR and SCoT Fingerprinting Polymorphism in Coffea arabica Cultivars
3.4. Diversity of Coffea arabica Cultivars Based on ISSR Markers
3.5. Diversity of the Arabian Coffee Cultivars Based on SCoT Markers
3.6. Relationships of Arabian Coffee Cultivars Based on Morphological Variations Combined with ISSR and SCoT Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Legesse, A. Assessment of coffee (Coffea arabica L.) genetic erosion and genetic resources management in Ethiopia. Int. J. Agric. Ext. 2020, 7, 223–229. [Google Scholar] [CrossRef]
- Pohlan, H.A.J.; Janssens, M.J. Growth and production of coffee. In Soils, Plant Growth and Crop Production; Eolss Publishers: Oxford, UK, 2010; Volume 3, p. 101. [Google Scholar]
- Tounekti, T.; Mahdhi, M.; Al-Turki, T.A.; Khemira, H. Water relations and photo-protection mechanisms during drought stress in four coffee (Coffea arabica) cultivars from southwestern Saudi Arabia. South Afr. J. Bot. 2018, 117, 17–25. [Google Scholar] [CrossRef]
- Lewin, B.; Giovannucci, D.; Varangis, P. Coffee Markets: New Paradigms in Global Supply and Demand; Agriculture and Rural Development Discussion Paper 3; Agriculture and Rural Development Department, World Bank: Washington, DC, USA, 2004; pp. 1–150. [Google Scholar]
- Al-Turki, T.A. An initiative in exploration and management of plant genetic diversity in Saudi Arabia. In Managing Plant Genetic Diversity; Engels, J.M.M., Rao, V.R., Brown, A.H.D., Jackson, M.T., Eds.; CABI Publishing: Wallingford, UK, 2002; pp. 339–349. [Google Scholar]
- Tounekti, T.; Mahdhi, M.; Al-Turki, T.A.; Khemira, H. Genetic diversity analysis of coffee (Coffea arabica L.) germplasm accessions growing in the southwestern Saudi Arabia using quantitative traits. Nat. Resour. 2017, 8, 321–336. [Google Scholar]
- Badr, A. Molecular approaches in plant systematics and evolution. Taeckholmia 2008, 28, 127–167. [Google Scholar]
- Dida, G.; Bantte, K.; Disasa, T. Molecular characterization of Arabica coffee (Coffea arabica L.) germplasms and their contribution to biodiversity in Ethiopia. Plant Biotechnol. Rep. 2021, 15, 791–804. [Google Scholar] [CrossRef]
- Mannino, G.; Kunz, R.; Maffei, M.E. Discrimination of green coffee (Coffea arabica and Coffea canephora) of different geographical origin based on antioxidant activity, high-throughput metabolomics, and DNA RFLP fingerprinting. Antioxidants 2023, 12, 1135. [Google Scholar] [CrossRef] [PubMed]
- Khemira, H.; Mahdhi, M.; Tounekti, T.; Oteef, M.D.; Afzal, M.; Alfaifi, Z.; Sharma, M.; Alsolami, W.; Shargi, D. Diversity among Coffea arabica cultivars in southwestern Saudi Arabia as revealed by their morphometric features. Not. Bot. Horti Agrobot. Cluj-Napoca 2024, 52, 13452. [Google Scholar] [CrossRef]
- Al-Ghamedi, K.; Alaraidh, I.; Afzal, M.; Mahdhi, M.; Al-Faifi, Z.; Oteef, M.D.; Tounekti, T.; Alghamdi, S.S.; Khemira, H. Assessment of genetic diversity of local coffee cultivars in southwestern Saudi Arabia using SRAP markers. Agronomy 2023, 13, 302. [Google Scholar] [CrossRef]
- Migahid, A.M. Flora of Saudi Arabia; Riyadh University Publications: Riyadh, Saudi Arabia, 1996; Volume 1. [Google Scholar]
- Collenette, S. Wildflowers of Saudi Arabia; National Commission for Wildlife Conservation and Development (NCWCD): Riyadh, Saudi Arabia, 1999. [Google Scholar]
- Chaudhary, S.A. Flora of the Kingdom of Saudi Arabia; Ministry of Agriculture and Water: Riyadh, Saudi Arabia, 2001. [Google Scholar]
- Prakash, N.S.; Marques, D.V.; Varzea, V.; Silva, M.C.; Combes, M.; Lashermes, P. Introgression molecular analysis of a leaf rust resistance gene from Coffea liberica into C. arabica L. Theor. Appl. Genet. 2004, 109, 1311–1317. [Google Scholar] [CrossRef]
- Collard, B.C.; Mackill, D.J. Start codon targeted (SCoT) polymorphism: A simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol. Biol. Report. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Bornet, B.; Branchard, M. Nonanchored inter simple sequence repeat (ISSR) markers: Reproducible and specific tools for genome fingerprinting. Plant Mol. Biol. Rep. 2001, 19, 209–215. [Google Scholar] [CrossRef]
- Rohlf, F.J. Geometric morphometrics and phylogeny. In Morphology, Shape, and Phylogeny; MacLeod, N., Forey, P.L., Eds.; Taylor & Francis: London, UK, 2002; pp. 175–193. [Google Scholar]
- Sokal, R.R.; Michener, C.D. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 1958, 38, 1409–1438. [Google Scholar]
- Hammer, Ø. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Konopiński, M.K. Shannon diversity index: A call to replace the original Shannon’s formula with unbiased estimator in the population genetics studies. PeerJ 2020, 8, e9391. [Google Scholar] [CrossRef]
- Yunita, R.; Oktavioni, M.; Chaniago, I.; Syukriani, L.; Setiawan, M.A.; Jamsari, J. Analysis of genetic diversity of Arabica coffee (Coffea arabica L.) in Solok Regency by SRAP molecular markers. In IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2020; p. 012018. [Google Scholar]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. iMEC: Online marker efficiency calculator. Appl. Plant Sci. 2018, 6, e01159. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.Z.; Masoud, I.M.; Zedan, S.Z. Molecular characterization and genetic relationships of cultivated flax (Linum usitatissimum L.) genotypes using ISSR markers. Middle East J. Agric. Res. 2019, 8, 898–908. [Google Scholar]
- Hopley, T.; Byrne, M. Gene flow and genetic variation explain signatures of selection across a climate gradient in two riparian species. Genes 2019, 10, 579. [Google Scholar] [CrossRef]
- Chown, S.L.; Hodgins, K.A.; Griffin, P.C. Biological invasions, climate change, and genomics. In Crop Breeding; Springer: Berlin, Germany, 2016; pp. 59–114. [Google Scholar]
- Luo, Y.; Zhang, X.; Xu, J.; Zheng, Y.; Pu, S.; Duan, Z.; Li, Z.; Liu, G.; Chen, J.; Wang, Z. Phenotypic and molecular marker analysis uncovers the genetic diversity of the grass Stenotaphrum secundatum. BMC Genet. 2020, 21, 86. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef]
- Al-Murish, T.M.; Elshafei, A.A.; Al-Doss, A.A.; Barakat, M.N. Genetic diversity of coffee (Coffea arabica L.) in Yemen via SRAP, TRAP, and SSR markers. J. Food Agric. Environ. 2013, 11, 411–416. [Google Scholar]
- Kebede, M.; Bellachew, B. Phenotypic diversity in the Hararge coffee (Coffea arabica L.) germplasm for quantitative traits. East Afr. J. Sci. 2008, 2, 13–18. [Google Scholar] [CrossRef]
- Kumar, J.; Heikrujam, M.; Sharma, K.; Agrawal, V. SRAP and SSR marker-assisted genetic diversity, cultivar structure analysis, and sex identification in Jojoba (Simmondsia chinensis). Ind. Crops Prod. 2019, 133, 118–132. [Google Scholar] [CrossRef]
- Wintgens, J.N. Coffee Bean Quality Assessment; Springer: Berlin, Germany, 2009. [Google Scholar]
- Kathurima, C.; Ghosh, K.; Bandar, A.; Alwshigry, A.; Alojaimi, A.; Kimemia, J. Unveiling physical and sensory quality of Arabica coffee produced in the Kingdom of Saudi Arabia, Jazan Region. 2022. Available online: https://saudiarabia.un.org/en/download/118640/203992 (accessed on 9 January 2024).
- Andermann, T.; Antonelli, A.; Barrett, R.L.; Silvestro, D. Estimating alpha, beta, and gamma diversity through deep learning. Front. Plant Sci. 2022, 13, 839407. [Google Scholar] [CrossRef] [PubMed]
- Gana, A.S.; Shaba, S.Z.; Tsado, E.K. Principal component analysis of morphological traits in thirty-nine accessions of rice (Oryza sativa L.) grown in a rainfed lowland ecology of Nigeria. J. Plant Breed. Crop Sci. 2013, 5, 120–126. [Google Scholar]
- Revermann, R.; Finckh, M.; Stellmes, M.; Strohbach, B.J.; Frantz, D.; Oldeland, J. Linking land surface phenology and vegetation-plot databases to model terrestrial plant α-diversity of the Okavango Basin. Remote Sens. 2016, 8, 370. [Google Scholar] [CrossRef]
- Wickramasinghe, H.K.J.P. Nutritional interventions for dairy calves undergoing weaning and heat stresses. J. Anim. Sci. 2022, 100, skac247. [Google Scholar]
- Ovalle-Rivera, O.; Läderach, P.; Bunn, C.; Obersteiner, M.; Schroth, G. Projected shifts in Coffea arabica suitability among major global producing regions due to climate change. PLoS ONE 2015, 10, e0124155. [Google Scholar] [CrossRef]
- Montagnon, C.; Sheibani, F.; Benti, T.; Daniel, D.; Bote, A.D. Deciphering early movements and domestication of Coffea arabica through a comprehensive genetic diversity study covering Ethiopia and Yemen. Agronomy 2022, 12, 3203. [Google Scholar] [CrossRef]
- Montagnon, C.; Rossi, V.; Guercio, C.; Sheibani, F. Vernacular names and genetics of cultivated coffee (Coffea arabica) in Yemen. Agronomy 2022, 12, 1970. [Google Scholar] [CrossRef]
- Montagnon, C.; Mahyoub, A.; Solano, W.; Sheibani, F. Unveiling a unique genetic diversity of cultivated Coffea arabica L. in its main domestication center: Yemen. Genet. Resour. Crop Evol. 2021, 68, 2411–2422. [Google Scholar] [CrossRef]
- Eskes, A.B.; Mukred, A. Coffee Survey in PDR Yemen; FAO: Rome, Italy, 1990. [Google Scholar]
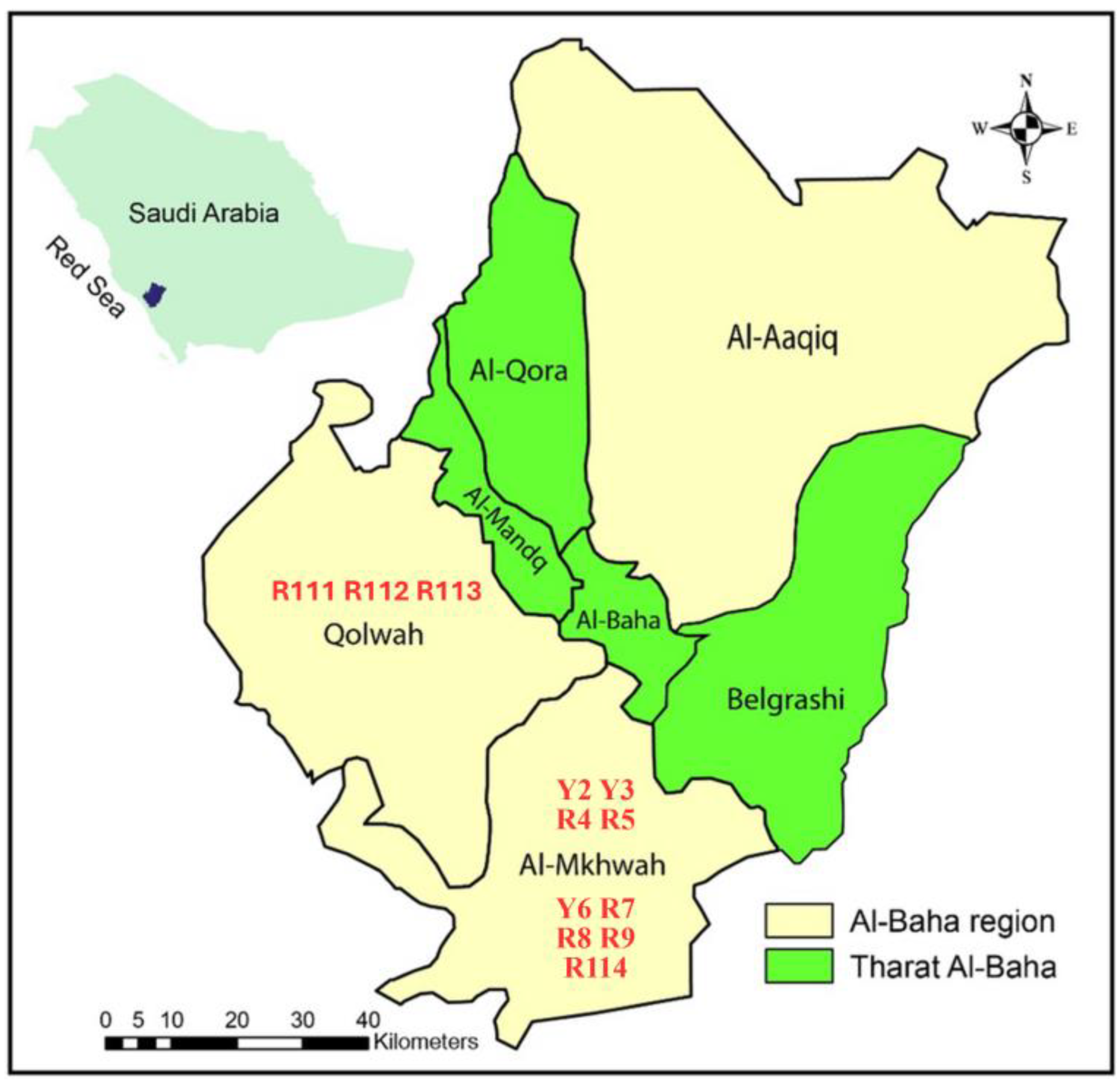


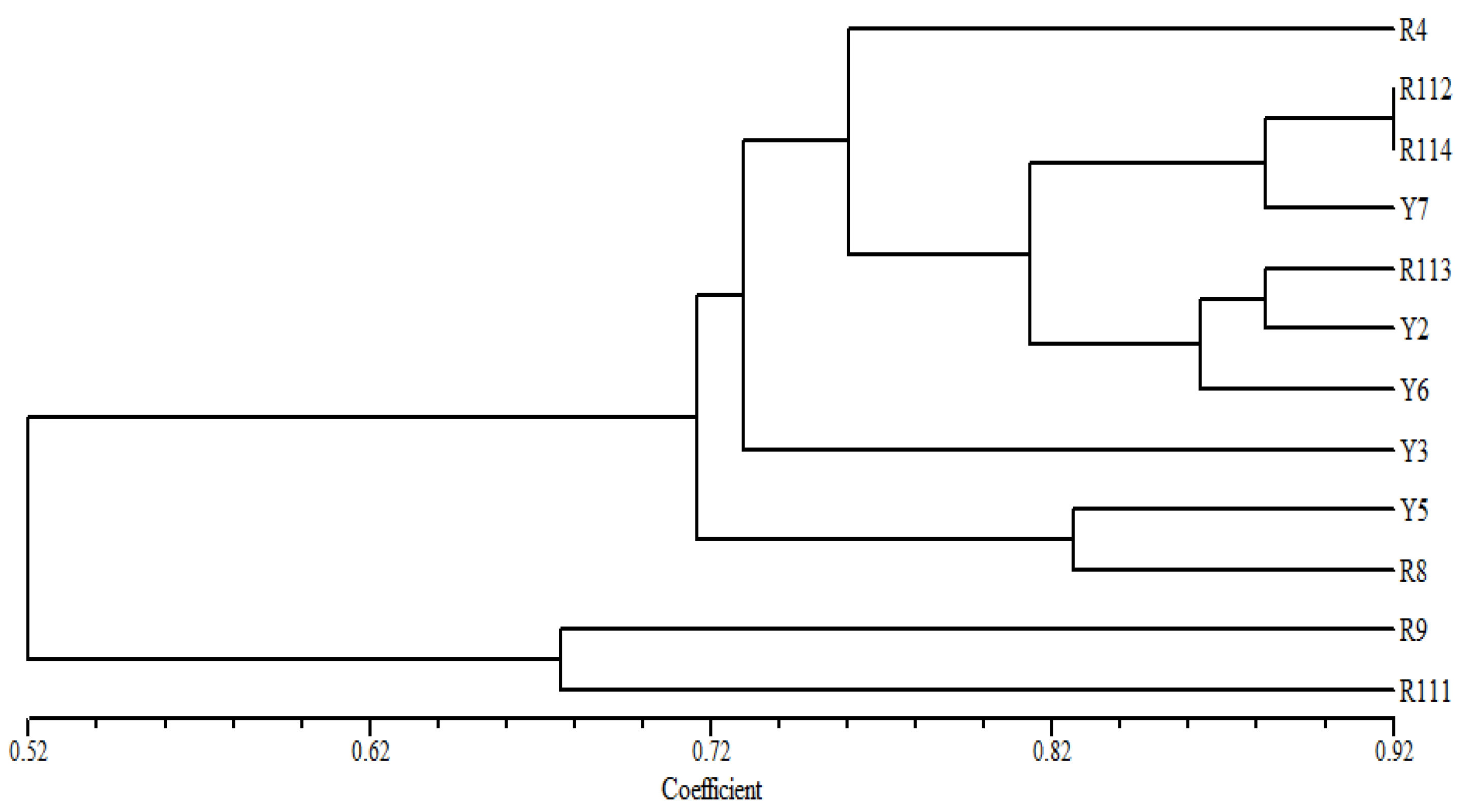
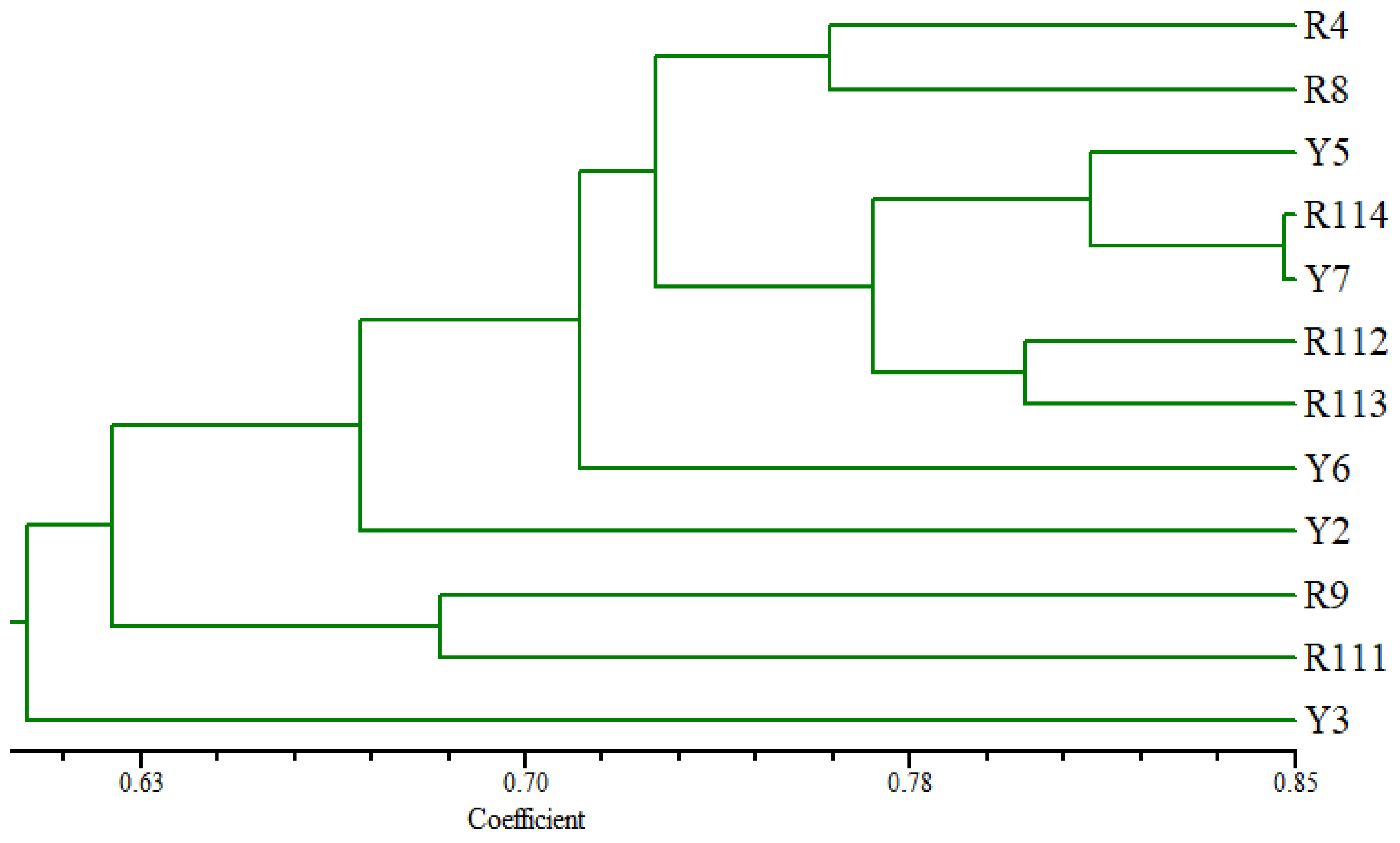

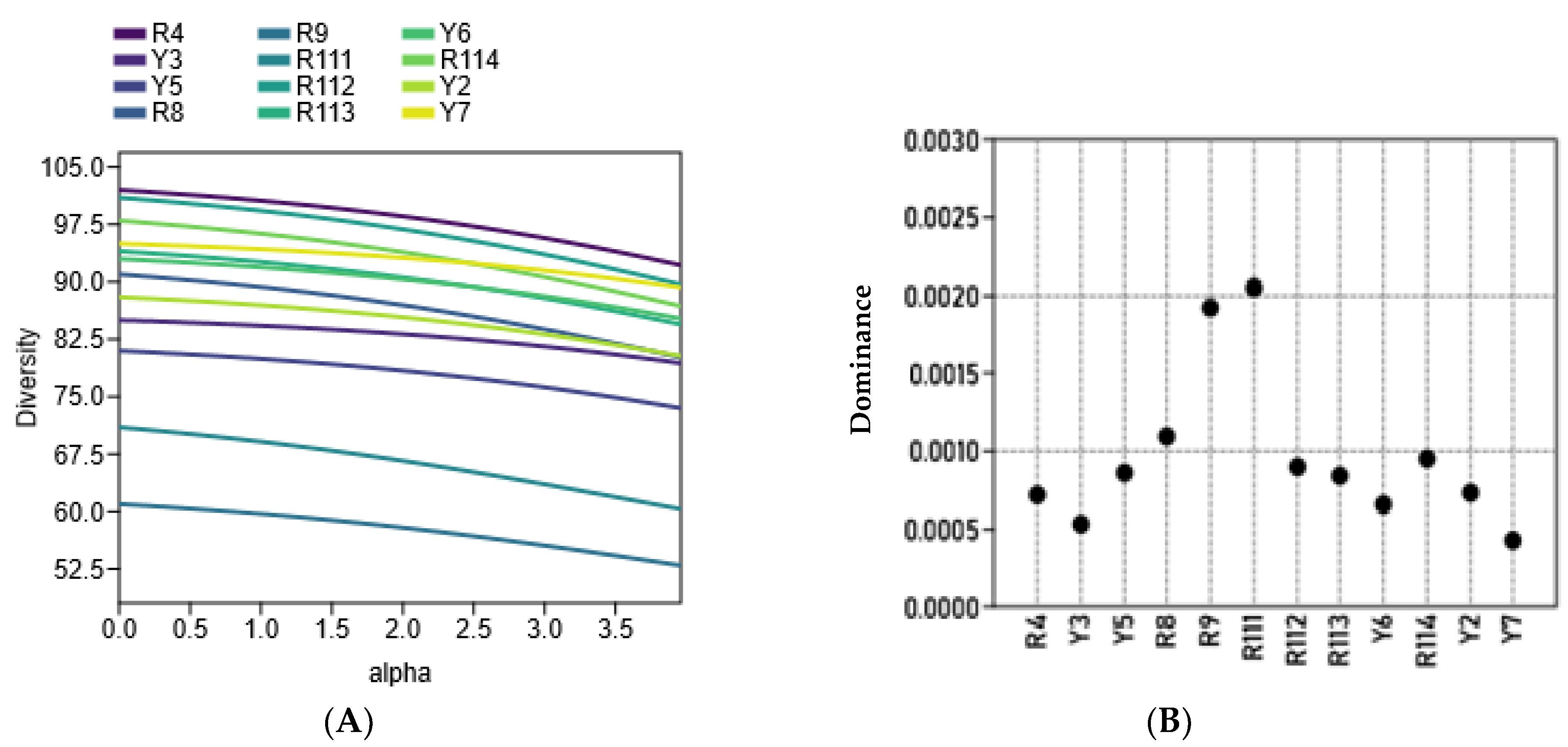
| No. | Cultivars Code | Area | GPS Location | Elevation (m) Asl | |
|---|---|---|---|---|---|
| 1 | R4 | Al-Mehkwa | 19°50′32.002″ N | 41°18′29.08″ E | 1685 |
| 2 | Y3 | Al-Mehkwa | 19°50′9.74″ N | 41°18′49.038″ E | 1684 |
| 3 | Y5 | Al-Mehkwa | 19°50′9.74″ N | 41°18′49.038″ E | 1684 |
| 4 | R8 | Al-Mehkwa | 19°50′32.002″ N | 41°18′29.08″ E | 1684 |
| 5 | R9 | Al-Mehkwa | 19°44′44.623″ N | 41°21′19.939″ E | 1104 |
| 6 | R111 | Qalwa | 19°55′40.242″ N | 41°9′15.18″ E | 1321 |
| 7 | R112 | Qalwa | 19°55′40.242″ N | 41°9′15.18″ E | 1321 |
| 8 | R113 | Qalwa | 19°55′41.242″ N | 41°9′18.18″ E | 1351 |
| 9 | Y6 | Al-Mehkwa | 19°50′9.74″ N | 41°18′49.038″ E | 1684 |
| 10 | R114 | Al-Mehkwa | 19°50′32.91″ N | 41°18′28.693″ E | 1107 |
| 11 | Y2 | Al-Mehkwa | 19°50′9.74″ N | 41°18′44.068″ E | 1684 |
| 12 | Y7 | Al-Mehkwa | 19°50′19.74″ N | 41°18′39.038″ E | 1684 |
| No. | Character | Arabian Coffee Cultivars | ||||||
|---|---|---|---|---|---|---|---|---|
| R4 | Y3 | Y5 | R8 | R9 | R111 | |||
| Qualitative Characters | 1 | Plant Shape | Pyramidal | Ellipsoid | Ellipsoid | Pyramidal | Ellipsoid | Pyramidal |
| 2 | Growth Habit | Tree | Shrub | Shrub | Tree | Shrub | Tree | |
| 3 | Young Leaf Anthocyanin Coloration | Week or absent | Strong | Strong | Strong | Strong | Strong | |
| 4 | Leaf Petiole Color | Green | Dark Brown | Dark Brown | Green | Dark Brown | Green | |
| 5 | Inflorescence Position | Axillary | Terminal | Terminal | Axillary | Terminal | Axillary | |
| 6 | Inflorescence on Old Wood | Absent | Present | Present | Absent | Present | Absent | |
| 7 | Fruit adherence to branch | Strong | Strong | Strong | Strong | Weak | Weak | |
| 8 | Cherry Color | Light red | Light red | Light red | Light red | Light red | Light red | |
| 9 | Leaf Shape | lanceolate | Lanceolate | lanceolate | lanceolate | lanceolate | Ovate | |
| 10 | Fruit Shape | Circular | Elliptic | Elliptic | Circular | Elliptic | Circular | |
| Quantitative Characters | 11 | Plant Height | Long | Long | Long | Intermediate | Short | Long |
| 12 | Canopy Diameter | Narrow | Wide | Wide | Wide | Intermediate | Wide | |
| 13 | Internode Length on Primary Branch | Intermediate | Short | Short | Short | Short | Short | |
| 14 | Leaf Length | Short | Short | Intermediate | Intermediate | Short | Short | |
| 15 | Leaf Width | Intermediate | Intermediate | Narrow | Intermediate | Intermediate | Narrow | |
| 16 | Leaf Margin Undulation | Intermediate | Strong | Intermediate | Week or absent | Intermediate | Strong | |
| 17 | Leaf Petiole Length cm | 4.5 ± 0.24 | 4.6 ± 0.32 | 6.7 ± 0.02 | 5.7 ± 0.44 | 4.8 ± 0.29 | 4.4 ± 0.22 | |
| 18 | No. of Flowers | 25 | 18 | 14 | 19 | 18 | 22 | |
| 19 | No. of Flower Fascicles | 11 | 8 | 7 | 10 | 8 | 11 | |
| 20 | Fruit Size cm | 1.5 ± 0.69 | 1.5 ± 0.60 | 1.5 ± 0.54 | 1.9 ± 0.93 | 1.8 ± 0.19 | 0.8 ± 0.32 | |
| 21 | Fruit Thickness | Intermediate | Intermediate | Intermediate | Thin | Intermediate | Intermediate | |
| 22 | Seed width | Intermediate | Intermediate | Intermediate | Intermediate | Broad | Intermediate | |
| 23 | 100 Fruit fresh weight | 163 ± 4.28 | 147 ± 5.24 | 112 ± 2.39 | 258 ± 9.73 | 256 ± 7.09 | 176 ± 4.51 | |
| 24 | 100 Fruit dry weight | 49.7 ± 1.74 | 44.6 ± 0.24 | 56 ± 4.33 | 45.2 ± 5.64 | 46.9 ± 3.23 | 40 ± 3.63 | |
| 25 | 100Seed dry weight | 27 ± 0.93 | 22 ± 0.24 | 24 ± 1.04 | 35 ± 1.35 | 38 ± 2.53 | 30 ± 6.37 | |
| Qualitative Characters | 1 | Plant Shape | Ellipsoid | Conical | Ellipsoid | Pyramidal | Ellipsoid | Conical |
| 2 | Growth Habit | Tree | Shrub | Shrub | Tree | Tree | Shrub | |
| 3 | Young Leaf Anthocyanin Coloration | Week or absent | Strong | Strong | Week or absent | Strong | Week or absent | |
| 4 | Leaf Petiole Color | Dark Brown | Green | Dark Brown | Green | Green | Dark Brown | |
| 5 | Inflorescence Position | Axillary | Terminal | Axillary | Terminal | Terminal | Axillary | |
| 6 | Inflorescence on Old Wood | Present | Absent | Present | Absent | Absent | Present | |
| 7 | Fruit adherence to branch | Weak | Strong | Strong | Strong | Strong | Weak | |
| 8 | Cherry Color | Light red | Light red | Light red | Light red | Light red | Orang | |
| 9 | Leaf Shape | Ovate | Elliptic | lanceolate | Ovate | Lanceolate | lanceolate | |
| 10 | Fruit Shape | Circular | Elliptic | Circular | Elliptic | Elliptic | Circular | |
| Quantitative Characters | 11 | Plant Height | Short | Short | Intermediate | Short | Long | Intermediate |
| 12 | Canopy Diameter | Wide | Narrow | Intermediate | Wide | Intermediate | Narrow | |
| 13 | Internode Length on Primary Branch | Long | Short | Short | Short | Short | Short | |
| 14 | Leaf Length | Intermediate | Long | Intermediate | Short | Intermediate | Short | |
| 15 | Leaf Width | Intermediate | Wide | Intermediate | Intermediate | Intermediate | Narrow | |
| 16 | Leaf Margin Undulation | Strong | Strong | Strong | Strong | Strong | Intermediate | |
| 17 | Leaf Petiole Length cm | 3.7 ± 1.29 | 4.1 ± 1.82 | 6.1 ± 1.22 | 4.3 ± 1.04 | 5.1 ± 1.39 | 4.9 ± 1.27 | |
| 18 | No. of Flowers/plant | 19 | 17 | 13 | 18 | 18 | 14 | |
| 19 | No. of Flower Fascicles | 10 | 8 | 6 | 7 | 8 | 6 | |
| 20 | Fruit Size cm | 1.5 ± 0.69 | 1.4 ± 0.60 | 1.4 ± 0.54 | 1.9 ± 0.93 | 1.8 ± 0.19 | 1.4 ± 0.32 | |
| 21 | Fruit Thickness | Thin | Intermediate | Intermediate | Thin | Intermediate | Thick | |
| 22 | Seed width | Intermediate | Intermediate | Broad | Intermediate | Intermediate | Narrow | |
| 23 | 100 Fruit fresh weight | 133 ± 6.73 | 166 ± 4.266 | 175 ± 4.22 | 176 ± 6.56 | 178 ± 3.83 | 180 ± 6.89 | |
| 24 | 100 Fruit dry weight | 44.9 ± 4.73 | 47 ± 2.53 | 49 ± 2.98 | 45.2 ± 3.39 | 48 ± 4.57 | 53 ± 4.34 | |
| 25 | 100 Seed dry weight | 34 ± 1.23 | 24 ± 2.25 | 24 ± 2.27 | 34 ± 1.83 | 26 ± 1.32 | 23 ± 1.23 | |
| No | Primer Code | Total Number of Bands | H | PIC | E | Rp | H. av | MI | D |
|---|---|---|---|---|---|---|---|---|---|
| 1 | HB-8 | 3 | 0.278 | 0.457 | 2.500 | 1.000 | 0.008 | 0.019 | 0.310 |
| 2 | HB-9 | 10 | 0.455 | 0.392 | 3.500 | 3.667 | 0.004 | 0.013 | 0.879 |
| 3 | HB-10 | 8 | 0.422 | 0.406 | 5.583 | 3.167 | 0.004 | 0.026 | 0.515 |
| 4 | HB-12 | 7 | 0.459 | 0.390 | 4.500 | 3.000 | 0.005 | 0.025 | 0.590 |
| 5 | HB-13 | 6 | 0.389 | 0.420 | 4.417 | 2.167 | 0.005 | 0.024 | 0.461 |
| 6 | HB-14 | 6 | 0.453 | 0.393 | 3.917 | 3.167 | 0.006 | 0.025 | 0.577 |
| 7 | 814 | 7 | 0.408 | 0.412 | 5.000 | 2.333 | 0.005 | 0.024 | 0.492 |
| 8 | 826 | 6 | 0.401 | 0.415 | 4.333 | 3.000 | 0.006 | 0.024 | 0.481 |
| 9 | ScoT3 | 8 | 0.437 | 0.400 | 2.583 | 4.500 | 0.005 | 0.012 | 0.898 |
| 10 | ScoT4 | 6 | 0.444 | 0.397 | 4.000 | 3.000 | 0.006 | 0.025 | 0.559 |
| 11 | ScoT7 | 10 | 0.495 | 0.373 | 4.500 | 6.000 | 0.004 | 0.019 | 0.800 |
| 12 | ScoT8 | 9 | 0.494 | 0.373 | 5.000 | 2.333 | 0.005 | 0.023 | 0.694 |
| 13 | ScoT9 | 6 | 0.375 | 0.425 | 4.500 | 2.000 | 0.005 | 0.023 | 0.440 |
| 14 | ScoT13 | 9 | 0.417 | 0.408 | 2.667 | 3.667 | 0.004 | 0.010 | 0.914 |
| 15 | ScoT14 | 12 | 0.490 | 0.375 | 5.167 | 5.000 | 0.003 | 0.018 | 0.816 |
| 16 | ScoT15 | 10 | 0.499 | 0.371 | 4.750 | 3.833 | 0.004 | 0.020 | 0.776 |
| 17 | ScoT16 | 9 | 0.479 | 0.381 | 5.417 | 3.500 | 0.004 | 0.024 | 0.640 |
| Average | 0.495 | 0.373 | 4.754 | 4.130 | 0.004 | 0.023 | 0.777 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omari Alzahrani, F.; Alshaharni, M.O.; El-Shaboury, G.A.; Badr, A. Biodiversity and Evaluation of Genetic Resources of Some Coffee Trees Grown in Al-Baha, Saudi Arabia. Curr. Issues Mol. Biol. 2025, 47, 136. https://doi.org/10.3390/cimb47030136
Omari Alzahrani F, Alshaharni MO, El-Shaboury GA, Badr A. Biodiversity and Evaluation of Genetic Resources of Some Coffee Trees Grown in Al-Baha, Saudi Arabia. Current Issues in Molecular Biology. 2025; 47(3):136. https://doi.org/10.3390/cimb47030136
Chicago/Turabian StyleOmari Alzahrani, Fatima, Mohammed Obeid Alshaharni, Gamal Awad El-Shaboury, and Abdelfattah Badr. 2025. "Biodiversity and Evaluation of Genetic Resources of Some Coffee Trees Grown in Al-Baha, Saudi Arabia" Current Issues in Molecular Biology 47, no. 3: 136. https://doi.org/10.3390/cimb47030136
APA StyleOmari Alzahrani, F., Alshaharni, M. O., El-Shaboury, G. A., & Badr, A. (2025). Biodiversity and Evaluation of Genetic Resources of Some Coffee Trees Grown in Al-Baha, Saudi Arabia. Current Issues in Molecular Biology, 47(3), 136. https://doi.org/10.3390/cimb47030136







