Exploring the Efficacy and Target Genes of Atractylodes Macrocephala Koidz Against Alzheimer’s Disease Based on Multi-Omics, Computational Chemistry, and Experimental Verification
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source
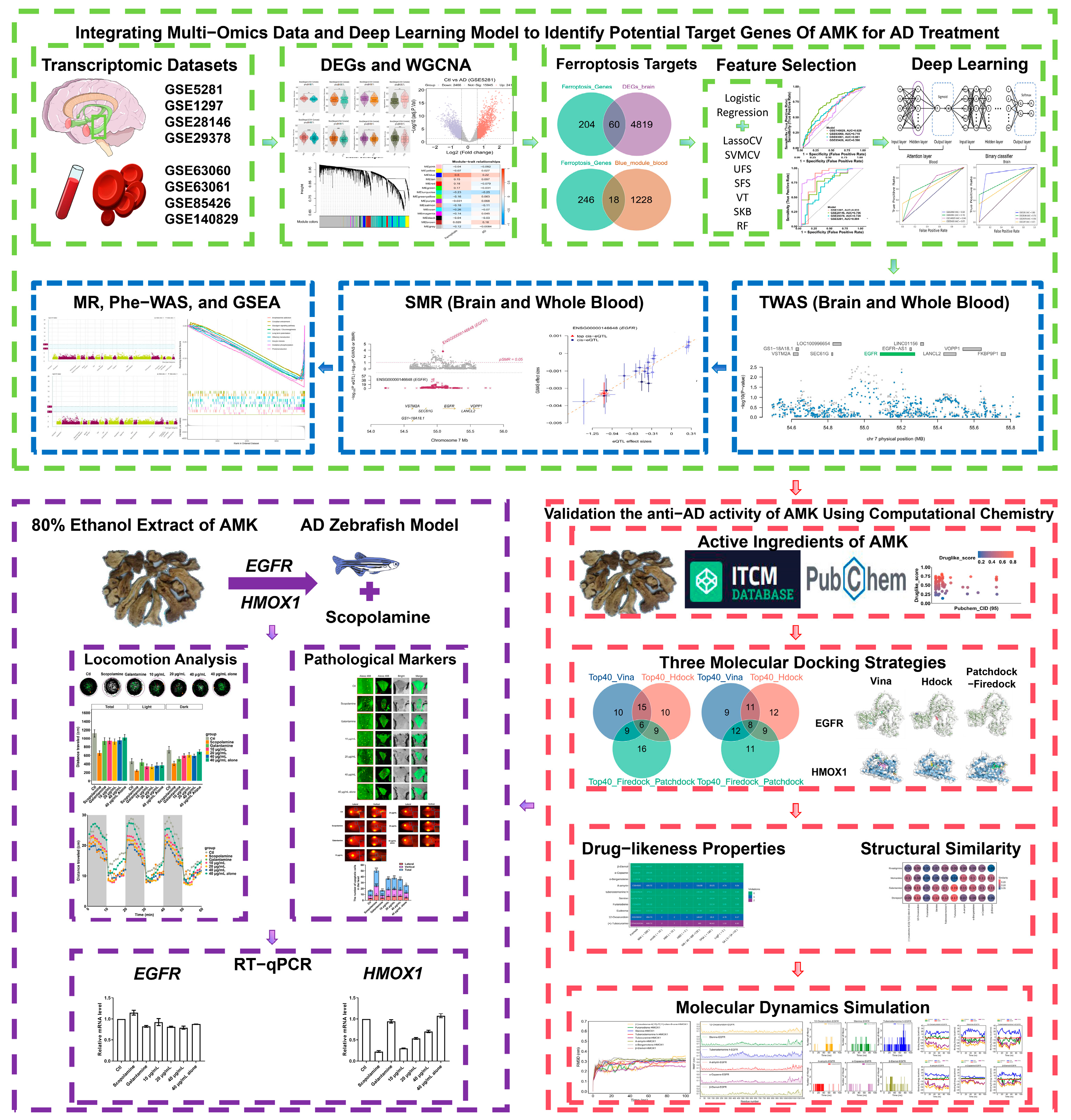
2.2. Identification of Potential Target Genes of AMK for AD Treatment Through the Integration of Multi-Omics and Logistic Regression with Seven Feature Selections and a Deep Learning Model
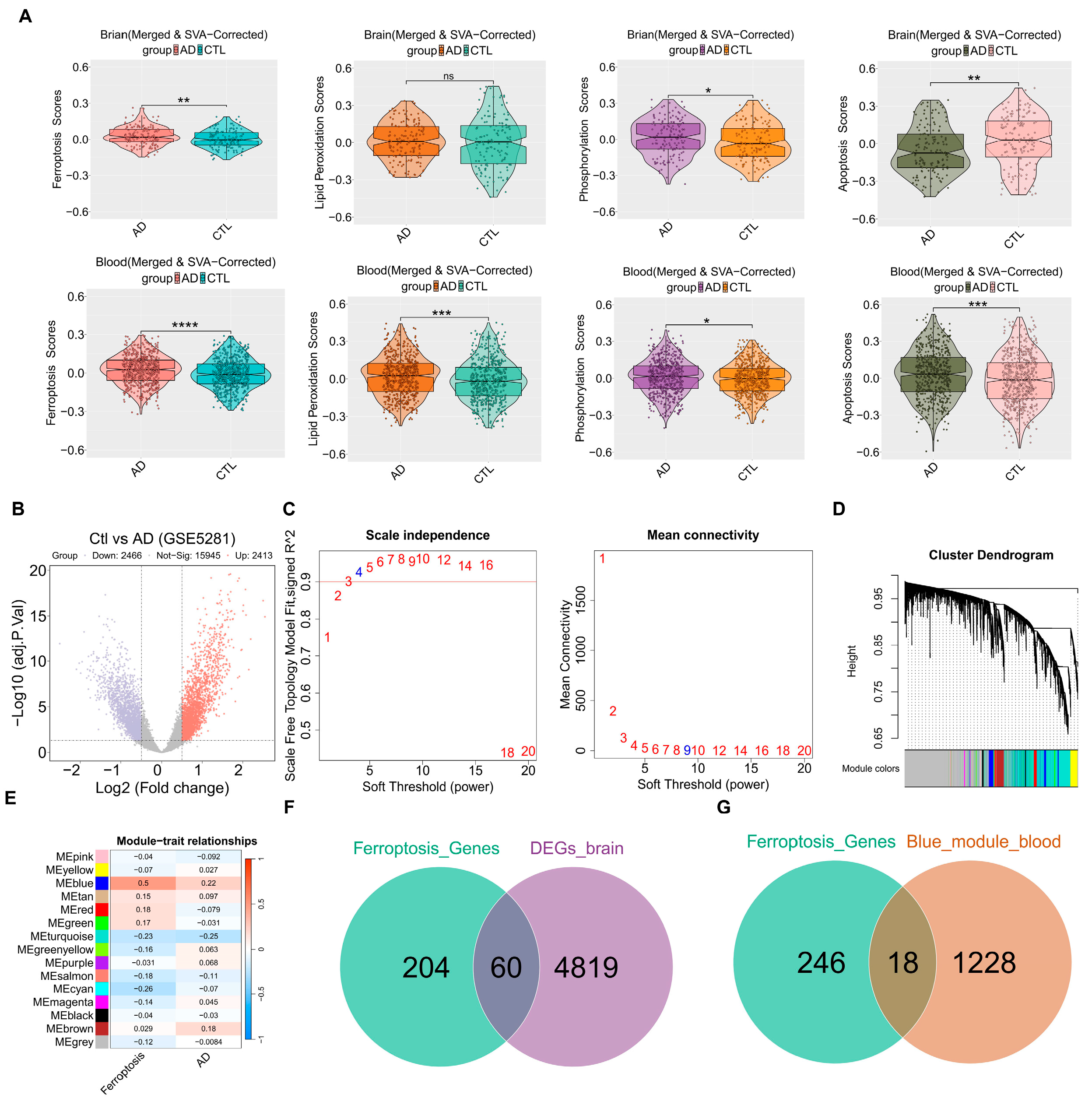
2.2.1. Gene Set Variation Analysis (GSVA)
2.2.2. Analysis of Differentially Expressed Genes (Brain and Hippocampus) and WGCNA (Blood)
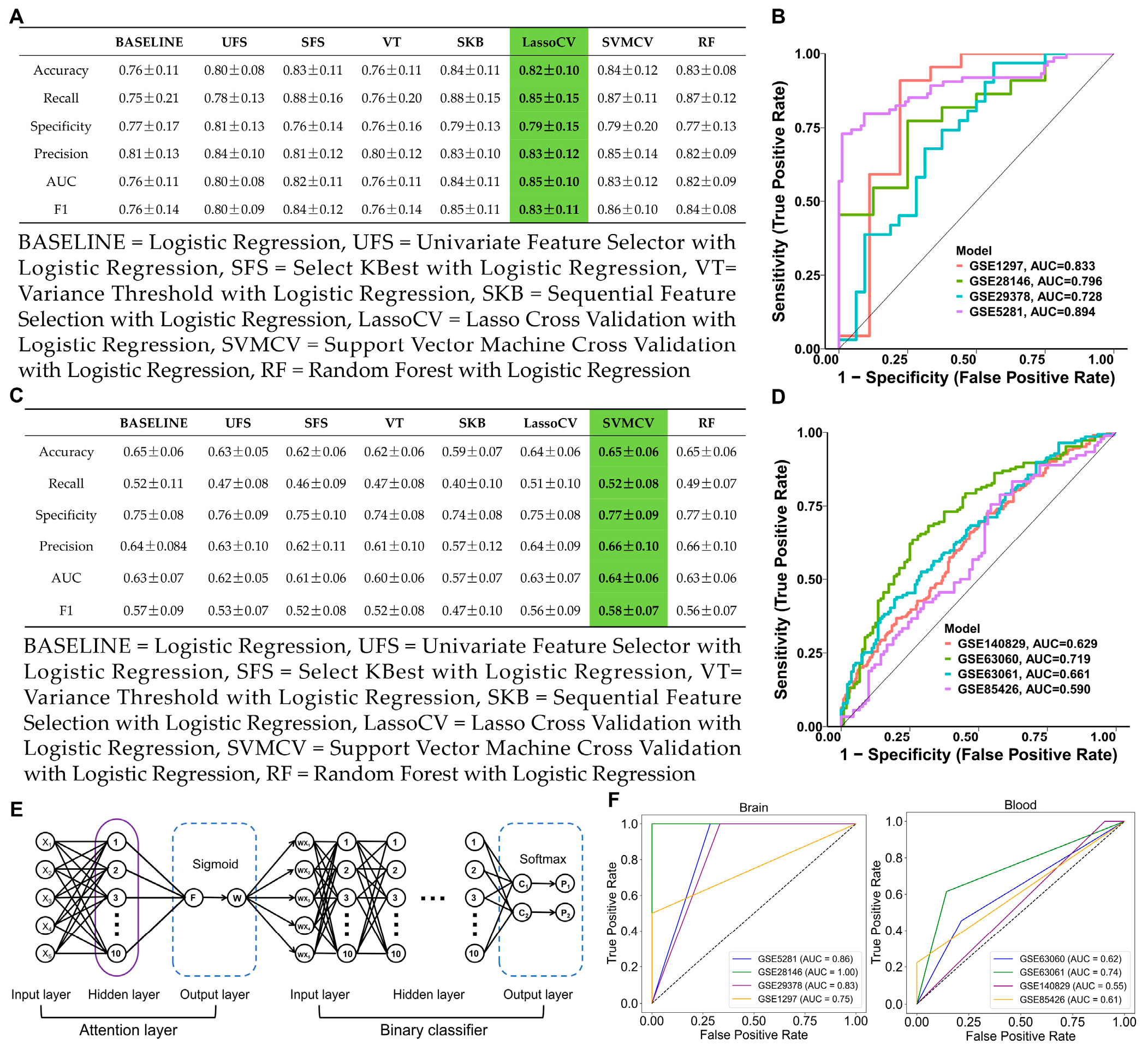
2.2.3. Machine Learning and Feature Selection
2.2.4. Deep Learning Model
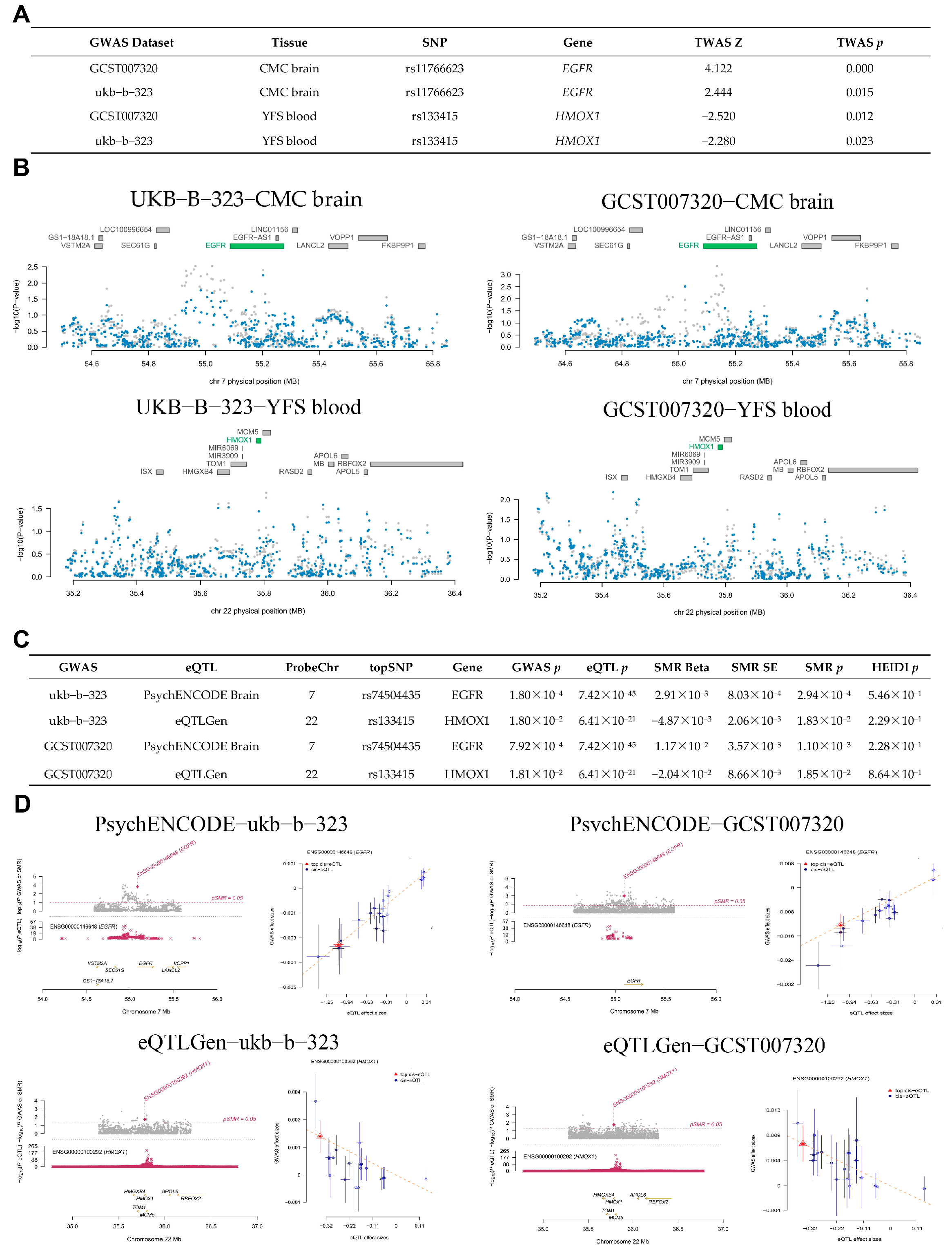
2.2.5. Transcriptome-Wide Association Study
2.2.6. Summary Data-Based Mendelian Randomization
2.2.7. Mendelian Randomization
2.2.8. Phenome-Wide Association Study (Phe-WAS)
2.2.9. Single-Gene Gene Set Enrichment Analysis (GSEA) Analysis
2.3. Validation of the Anti-AD Activity of AMK Using Computational Chemistry
2.3.1. Molecular Docking
2.3.2. Drug-likeness Properties and Structural Similarities
2.3.3. Molecular Dynamics (MD) Simulation
2.3.4. Binding-Free Energy Calculation
2.4. Validation of AMK’s Potential Use in Ameliorating AD-like Symptoms and Ferroptosis-Related Target Expression in a Zebrafish Model
2.4.1. Materials, Reagents and Animals
2.4.2. Scopolamine-Induced AD Model
2.4.3. AMK Preparation
2.4.4. AMK Treatment
2.4.5. Locomotion Analysis
2.4.6. Apoptosis Assessment
2.4.7. Detection of Aβ Deposition
2.4.8. RNA Extraction and Real-Time qPCR
2.5. Statistical Analysis
3. Results
3.1. The Ferroptosis-Related Targets from Brain and Blood
3.2. Feature Selection and Deep Learning Model Validation
3.3. Identification of Potential Drug Target Genes in Brain and Blood Using TWAS, SMR, and MR
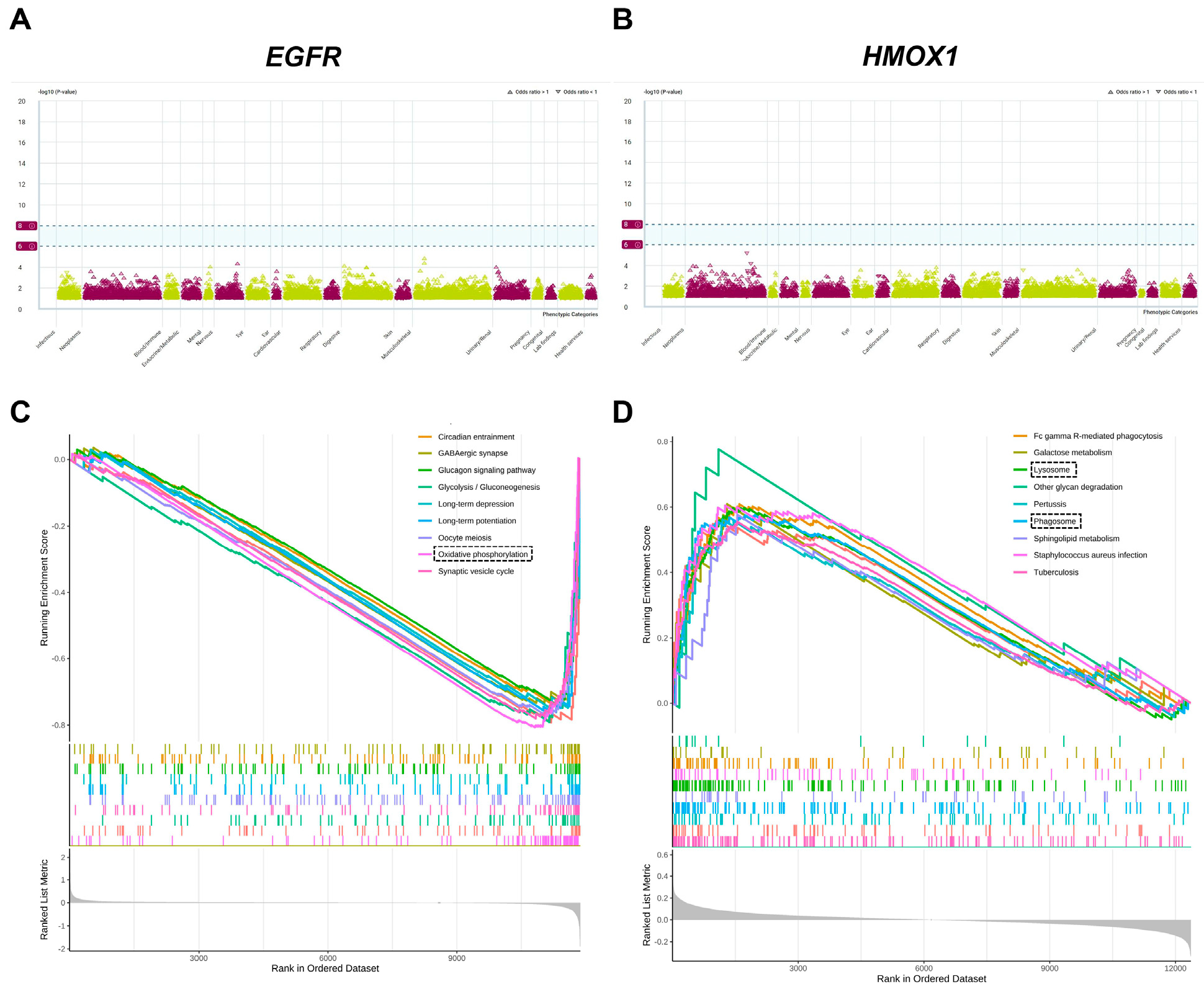
3.4. Phe-WAS and GSEA
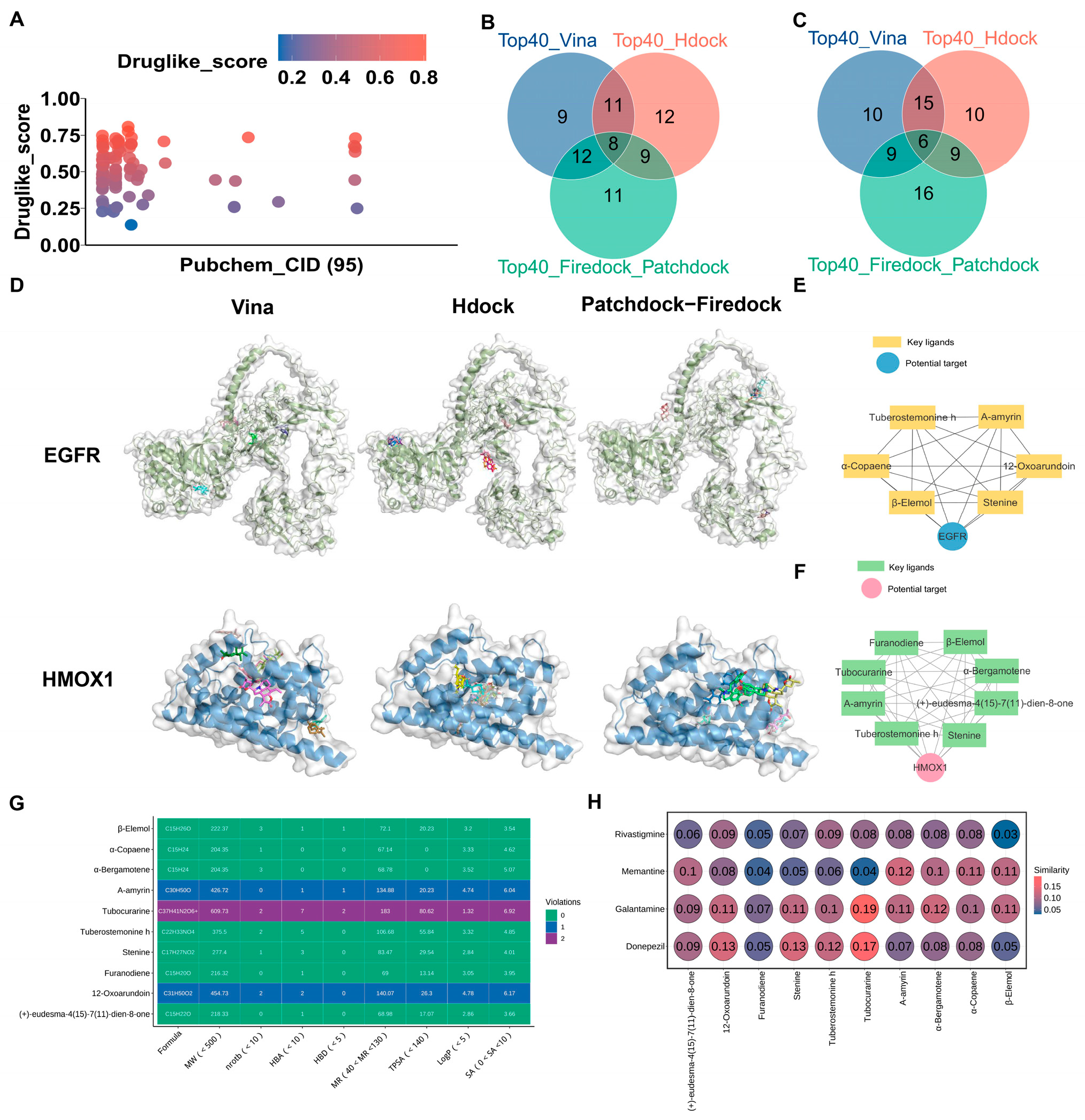
3.5. Three Molecular Docking Strategies, Drug-likeness Properties, and Structural Similarities
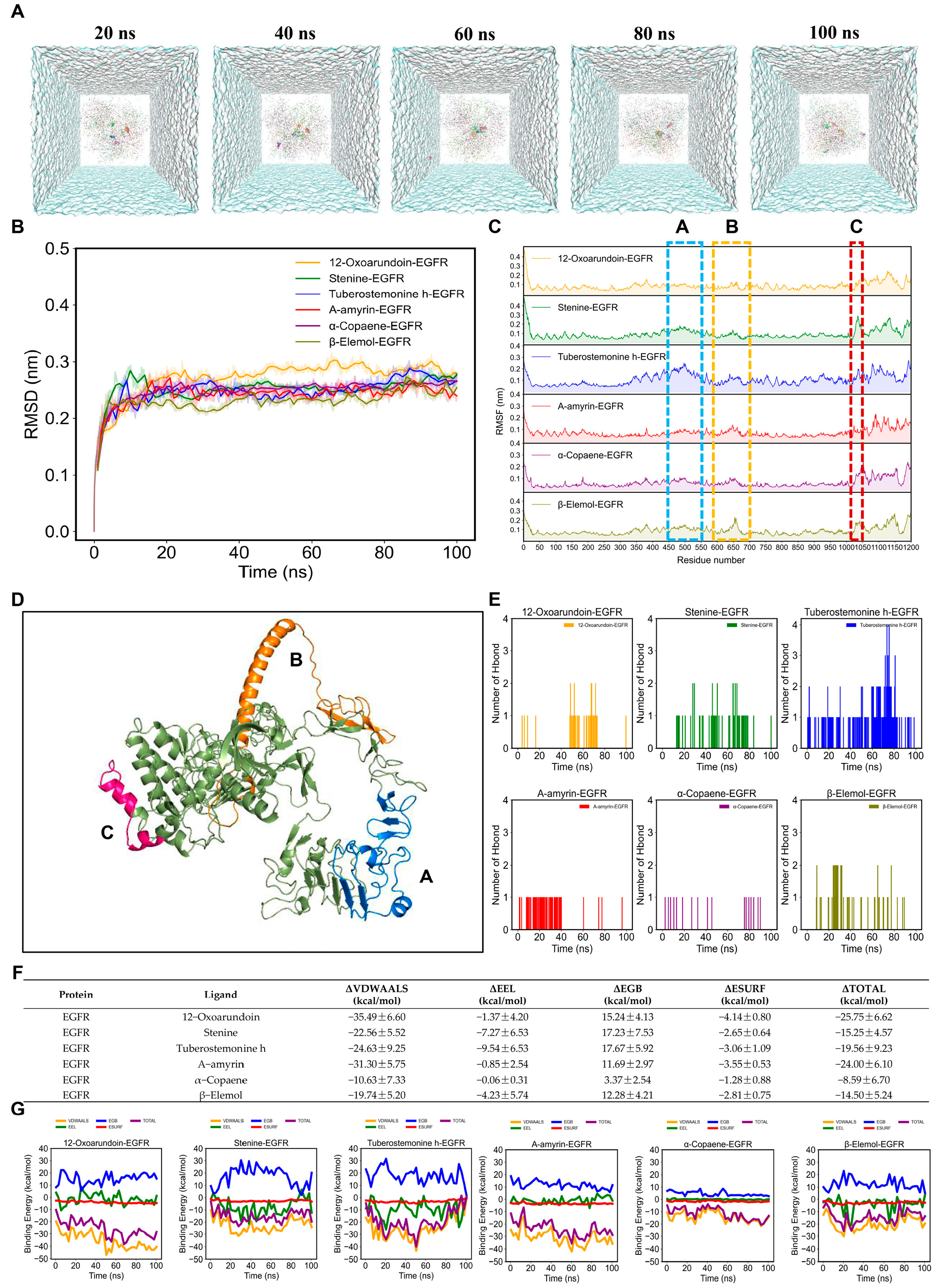
3.6. MD Simulation and MM-PBSA Energy Calculations
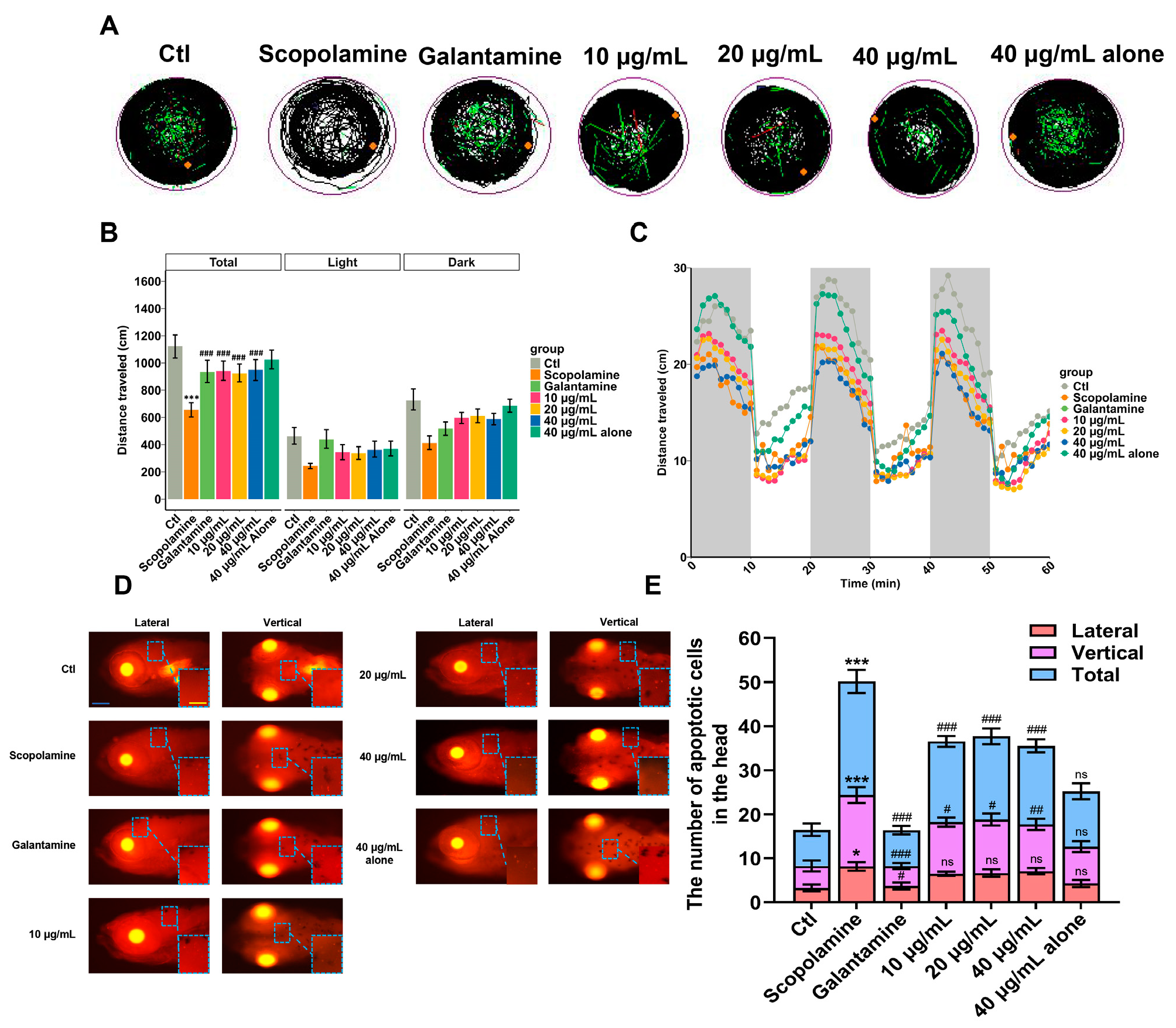
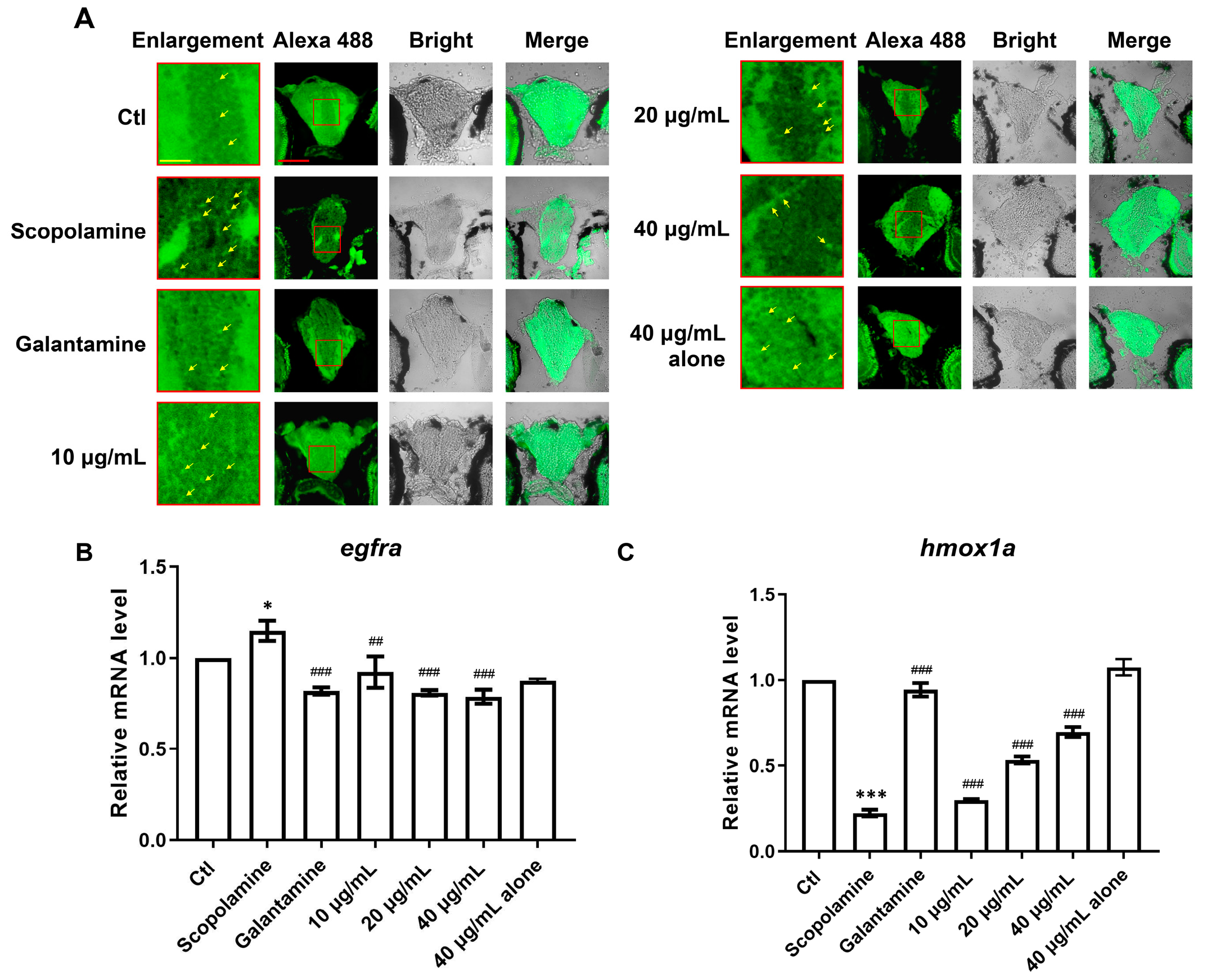
3.7. Zebrafish Model Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jicha, G.A.; Carr, S.A. Conceptual evolution in Alzheimer’s disease: Implications for understanding the clinical phenotype of progressive neurodegenerative disease. J. Alzheimer’s Dis. 2010, 19, 253–272. [Google Scholar] [CrossRef] [PubMed]
- Finder, V.H. Alzheimer’s disease: A general introduction and pathomechanism. J. Alzheimer’s Dis. 2010, 22, S5–S19. [Google Scholar] [CrossRef]
- Derry, P.J.; Hegde, M.L.; Jackson, G.R.; Kayed, R.; Tour, J.M.; Tsai, A.-L.; Kent, T.A. Revisiting the intersection of amyloid, pathologically modified tau and iron in Alzheimer’s disease from a ferroptosis perspective. Prog. Neurobiol. 2020, 184, 101716. [Google Scholar] [CrossRef]
- Sun, Z.-K.; Yang, H.-Q.; Chen, S.-D. Traditional Chinese medicine: A promising candidate for the treatment of Alzheimer’s disease. Transl. Neurodegener. 2013, 2, 6. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, X.-F.; Zhang, X.; Guo, D.-Y.; Duan, Y.-W.; Wang, Z.-C.; Pei, L.-S.; Ru, H.; Cheng, J.-X.; Shi, Y.-J. Evaluation of the therapeutic effect of traditional Chinese medicine on osteoarthritis: A systematic review and meta-analysis. Pain Res. Manag. 2020, 2020, 5712187. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Singh, G.; Bhushan, B.; Kumar, S.; Dhurandhar, Y.; Dixit, P. A Comprehensive Pharmacological Review of Atractylodes Macrocephala: Traditional Uses, Phytochemistry, Pharmacokinetics, And Therapeutic Potential. Pharmacol. Res. Mod. Chin. Med. 2024, 10, 100394. [Google Scholar] [CrossRef]
- Zhu, B.; Zhang, Q.-L.; Hua, J.-W.; Cheng, W.-L.; Qin, L.-P. The traditional uses, phytochemistry, and pharmacology of Atractylodes macrocephala Koidz.: A review. J. Ethnopharmacol. 2018, 226, 143–167. [Google Scholar] [CrossRef]
- He, Y.-J.; Cong, L.; Liang, S.-L.; Ma, X.; Tian, J.-N.; Li, H.; Wu, Y. Discovery and validation of Ferroptosis-related molecular patterns and immune characteristics in Alzheimer’s disease. Front. Aging Neurosci. 2022, 14, 1056312. [Google Scholar] [CrossRef]
- Sumirtanurdin, R.; Thalib, A.Y.; Cantona, K.; Abdulah, R. Effect of genetic polymorphisms on Alzheimer’s disease treatment outcomes: An update. Clin. Interv. Aging 2019, 14, 631–642. [Google Scholar] [CrossRef]
- Liu, J.; Yuan, S.; Niu, X.; Kelleher, R.; Sheridan, H. ESR1 dysfunction triggers neuroinflammation as a critical upstream causative factor of the Alzheimer’s disease process. Aging 2022, 14, 8595. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.-D.; Pang, P.; Zhou, X.-T.; Hu, F.; Xiong, W.; Chen, K.; Wang, J.; Wang, F.; Xie, D.; Hu, Y.-Z. Loss of ferroportin induces memory impairment by promoting ferroptosis in Alzheimer’s disease. Cell Death Differ. 2021, 28, 1548–1562. [Google Scholar] [CrossRef]
- Feng, H.; Stockwell, B.R. Unsolved mysteries: How does lipid peroxidation cause ferroptosis? PLoS Biol. 2018, 16, e2006203. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.; Reiter, R.J.; Manchester, L.C.; Yan, M.; El-Sawi, M.; Sainz, R.M.; Mayo, J.C.; Kohen, R.; Allegra, M.; Hardeland, R. Chemical and physical properties and potential mechanisms: Melatonin as a broad spectrum antioxidant and free radical scavenger. Curr. Top. Med. Chem. 2002, 2, 181–197. [Google Scholar] [CrossRef]
- Mohsen, H.; El-Dahshan, E.-S.A.; El-Horbaty, E.-S.M.; Salem, A.-B.M. Classification using deep learning neural networks for brain tumors. Future Comput. Inform. J. 2018, 3, 68–71. [Google Scholar] [CrossRef]
- Salloum, S.A.; Bettayeb, A.; Salloum, A.; Aburayya, A.; Khadragy, S.; Hamoudi, R.; Alfaisal, R. Novel machine learning based approach for analysing the adoption of metaverse in medical training: A UAE case study. Inform. Med. Unlocked 2023, 42, 101354. [Google Scholar] [CrossRef]
- Cui, S.; Li, L.; Zhang, Y.; Lu, J.; Wang, X.; Song, X.; Liu, J.; Li, K. Machine learning identifies metabolic signatures that predict the risk of recurrent angina in remitted patients after percutaneous coronary intervention: A multicenter prospective cohort study. Adv. Sci. 2021, 8, 2003893. [Google Scholar] [CrossRef] [PubMed]
- Suthaharan, S. Machine learning models and algorithms for big data classification. Integr. Ser. Inf. Syst. 2016, 36, 17–29. [Google Scholar]
- Wu, B.; Cox, M.P. Comparative genomics reveals a core gene toolbox for lifestyle transitions in Hypocreales fungi. Environ. Microbiol. 2021, 23, 3251–3264. [Google Scholar] [CrossRef] [PubMed]
- Wainberg, M.; Sinnott-Armstrong, N.; Mancuso, N.; Barbeira, A.N.; Knowles, D.A.; Golan, D.; Ermel, R.; Ruusalepp, A.; Quertermous, T.; Hao, K. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 2019, 51, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Pantsar, T.; Poso, A. Binding affinity via docking: Fact and fiction. Molecules 2018, 23, 1899. [Google Scholar] [CrossRef]
- Li, M.; Li, M.; Guo, J. Molecular mechanism of Ca2+ in the allosteric regulation of human parathyroid hormone receptor-1. J. Chem. Inf. Model. 2021, 62, 5110–5119. [Google Scholar] [CrossRef] [PubMed]
- Souza, P.C.; Thallmair, S.; Conflitti, P.; Ramírez-Palacios, C.; Alessandri, R.; Raniolo, S.; Limongelli, V.; Marrink, S.J. Protein–ligand binding with the coarse-grained Martini model. Nat. Commun. 2020, 11, 3714. [Google Scholar] [CrossRef] [PubMed]
- Walker, V.M.; Davey Smith, G.; Davies, N.M.; Martin, R.M. Mendelian randomization: A novel approach for the prediction of adverse drug events and drug repurposing opportunities. Int. J. Epidemiol. 2017, 46, 2078–2089. [Google Scholar] [CrossRef]
- Alonso, H.; Bliznyuk, A.A.; Gready, J.E. Combining docking and molecular dynamic simulations in drug design. Med. Res. Rev. 2006, 26, 531–568. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, J.; Bowler, E.; Cerezo, M.; Gil, L.; Hall, P.; Hastings, E.; Junkins, H.; McMahon, A.; Milano, A.; Morales, J. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017, 45, D896–D901. [Google Scholar] [CrossRef]
- Elsworth, B.; Lyon, M.; Alexander, T.; Liu, Y.; Matthews, P.; Hallett, J.; Bates, P.; Palmer, T.; Haberland, V.; Smith, G.D. The MRC IEU OpenGWAS data infrastructure. BioRxiv 2020. [Google Scholar] [CrossRef]
- van der Wijst, M.G.; de Vries, D.H.; Groot, H.E.; Trynka, G.; Hon, C.-C.; Bonder, M.-J.; Stegle, O.; Nawijn, M.; Idaghdour, Y.; van der Harst, P. The single-cell eQTLGen consortium. elife 2020, 9, e52155. [Google Scholar] [CrossRef]
- Akbarian, S.; Liu, C.; Knowles, J.A.; Vaccarino, F.M.; Farnham, P.J.; Crawford, G.E.; Jaffe, A.E.; Pinto, D.; Dracheva, S.; Geschwind, D.H. The psychencode project. Nat. Neurosci. 2015, 18, 1707–1712. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Yuan, X.; Du, Q.; Zhang, Z.; Shi, X.; Bao, J.; Ning, Y.; Peng, L. FerrDb V2: Update of the manually curated database of ferroptosis regulators and ferroptosis-disease associations. Nucleic Acids Res. 2023, 51, D571–D582. [Google Scholar] [CrossRef]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H. GeneCards Version 3: The human gene integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef] [PubMed]
- Labonte, A.C.; Kegerreis, B.; Geraci, N.S.; Bachali, P.; Madamanchi, S.; Robl, R.; Catalina, M.D.; Lipsky, P.E.; Grammer, A.C. Identification of alterations in macrophage activation associated with disease activity in systemic lupus erythematosus. PLoS ONE 2018, 13, e0208132. [Google Scholar] [CrossRef]
- Liu, S.; Wang, Z.; Zhu, R.; Wang, F.; Cheng, Y.; Liu, Y. Three differential expression analysis methods for RNA sequencing: Limma, EdgeR, DESeq2. JoVE (J. Vis. Exp.) 2021, 175, e62528. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Hackeling, G. Mastering Machine Learning with Scikit-Learn; Packt Publishing Ltd.: Birmingham, UK, 2017. [Google Scholar]
- Imambi, S.; Prakash, K.B.; Kanagachidambaresan, G.P. Programming with TensorFlow: Solution for Edge Computing Applications; Springer: Berlin/Heidelberg, Germany, 2021; pp. 87–104. [Google Scholar]
- Liu, S.; Zhong, H.; Zhu, J.; Wu, Y.; Deng, Y.; Wu, L. Regulome-wide association study identifies genetically driven accessible regions associated with pancreatic cancer risk. Int. J. Cancer 2024, 154, 670–678. [Google Scholar] [CrossRef]
- Wu, Y.; Zeng, J.; Zhang, F.; Zhu, Z.; Qi, T.; Zheng, Z.; Lloyd-Jones, L.R.; Marioni, R.E.; Martin, N.G.; Montgomery, G.W. Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat. Commun. 2018, 9, 918. [Google Scholar] [CrossRef]
- Shrine, N.; Izquierdo, A.G.; Chen, J.; Packer, R.; Hall, R.J.; Guyatt, A.L.; Batini, C.; Thompson, R.J.; Pavuluri, C.; Malik, V. Multi-ancestry genome-wide association analyses improve resolution of genes and pathways influencing lung function and chronic obstructive pulmonary disease risk. Nat. Genet. 2023, 55, 410–422. [Google Scholar] [CrossRef]
- Consortium, U. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.-Y. HDOCK: A web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef] [PubMed]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef] [PubMed]
- Mashiach, E.; Schneidman-Duhovny, D.; Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: A web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008, 36, W229–W232. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Godden, J.W.; Xue, L.; Bajorath, J. Combinatorial preferences affect molecular similarity/diversity calculations using binary fingerprints and Tanimoto coefficients. J. Chem. Inf. Comput. Sci. 2000, 40, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio Rerio); University of Oregon Press: Eugene, OR, USA, 1995. [Google Scholar]
- Li, D.; Cai, C.; Liao, Y.; Wu, Q.; Ke, H.; Guo, P.; Wang, Q.; Ding, B.; Fang, J.; Fang, S. Systems pharmacology approach uncovers the therapeutic mechanism of medicarpin against scopolamine-induced memory loss. Phytomedicine 2021, 91, 153662. [Google Scholar] [CrossRef] [PubMed]
- Belardo, C.; Boccella, S.; Perrone, M.; Fusco, A.; Morace, A.M.; Ricciardi, F.; Bonsale, R.; ELBini-Dhouib, I.; Guida, F.; Luongo, L. Scopolamine-Induced Memory Impairment in Mice: Effects of PEA-OXA on Memory Retrieval and Hippocampal LTP. Int. J. Mol. Sci. 2023, 24, 14399. [Google Scholar] [CrossRef]
- Burns, A.; Bernabei, R.; Bullock, R.; Jentoft, A.J.C.; Frölich, L.; Hock, C.; Raivio, M.; Triau, E.; Vandewoude, M.; Wimo, A. Safety and efficacy of galantamine (Reminyl) in severe Alzheimer’s disease (the SERAD study): A randomised, placebo-controlled, double-blind trial. Lancet Neurol. 2009, 8, 39–47. [Google Scholar] [CrossRef]
- Jin, M.; Ji, X.; Zhang, B.; Sheng, W.; Wang, R.; Liu, K. Synergistic effects of Pb and repeated heat pulse on developmental neurotoxicity in zebrafish. Ecotoxicol. Environ. Saf. 2019, 172, 460–470. [Google Scholar] [CrossRef]
- Jin, M.; Li, N.; Sheng, W.; Ji, X.; Liang, X.; Kong, B.; Yin, P.; Li, Y.; Zhang, X.; Liu, K. Toxicity of different zinc oxide nanomaterials and dose-dependent onset and development of Parkinson’s disease-like symptoms induced by zinc oxide nanorods. Environ. Int. 2021, 146, 106179. [Google Scholar] [CrossRef]
- Yang, F.-C.; Wang, C.; Zhu, J.; Gai, Q.-J.; Mao, M.; He, J.; Qin, Y.; Yao, X.-X.; Wang, Y.-X.; Lu, H.-M. Inhibitory effects of temozolomide on glioma cells is sensitized by RSL3-induced ferroptosis but negatively correlated with expression of ferritin heavy chain 1 and ferritin light chain. Lab. Investig. 2022, 102, 741–752. [Google Scholar] [CrossRef]
- Tang, D.; Chen, X.; Kang, R.; Kroemer, G. Ferroptosis: Molecular mechanisms and health implications. Cell Res. 2021, 31, 107–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Dhindsa, R.S.; Carss, K.; Harper, A.R.; Nag, A.; Tachmazidou, I.; Vitsios, D.; Deevi, S.V.; Mackay, A.; Muthas, D. Rare variant contribution to human disease in 281,104 UK Biobank exomes. Nature 2021, 597, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Bahcekapili, N.; Üzüm, G.; Gökkusu, C.; Kuru, A.; Ziylan, Y.Z. The relationship between erythropoietin pretreatment with blood–brain barrier and lipid peroxidation after ischemia/reperfusion in rats. Life Sci. 2007, 80, 1245–1251. [Google Scholar] [CrossRef] [PubMed]
- Negre-Salvayre, A.; Auge, N.; Ayala, V.; Basaga, H.; Boada, J.; Brenke, R.; Chapple, S.; Cohen, G.; Feher, J.; Grune, T. Pathological aspects of lipid peroxidation. Free Radic. Res. 2010, 44, 1125–1171. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Jiang, X.; Wu, M.; Cao, X.; Bao, W.; Zhu, L.-Q. Ferroptosis, a potential therapeutic target in Alzheimer’s disease. Front. Cell Dev. Biol. 2021, 9, 704298. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Sun, Y.-P.; Luo, Y.-M.; Peng, D.-H.; Li, X.; Yang, B.-Y.; Wang, Q.-H.; Kuang, H.-X. Biomarkers for the clinical diagnosis of Alzheimer’s disease: Metabolomics analysis of brain tissue and blood. Front. Pharmacol. 2021, 12, 700587. [Google Scholar] [CrossRef] [PubMed]
- Mufson, E.J.; Mahady, L.; Waters, D.; Counts, S.E.; Perez, S.E.; DeKosky, S.; Ginsberg, S.D.; Ikonomovic, M.D.; Scheff, S.; Binder, L. Hippocampal plasticity during the progression of Alzheimer’s disease. Neuroscience 2015, 309, 51–67. [Google Scholar] [CrossRef] [PubMed]
- Vickers, J.C.; Mitew, S.; Woodhouse, A.; Fernandez-Martos, C.M.; Kirkcaldie, M.T.; Canty, A.J.; McCormack, G.H.; King, A.E. Defining the earliest pathological changes of Alzheimer’s disease. Curr. Alzheimer Res. 2016, 13, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Moodley, K.; Chan, D. The hippocampus in neurodegenerative disease. Hippocampus Clin. Neurosci. 2014, 34, 95–108. [Google Scholar]
- Ain, Q.U.; Sarfraz, M.; Prasesti, G.K.; Dewi, T.I.; Kurniati, N.F. Confounders in identification and analysis of inflammatory biomarkers in cardiovascular diseases. Biomolecules 2021, 11, 1464. [Google Scholar] [CrossRef]
- Romano, R.; Bucci, C. Role of EGFR in the Nervous System. Cells 2020, 9, 1887. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Le Guen, Y.; Liu, L.; Lee, J.; Ma, S.; Yang, A.C.; Liu, X.; Rutledge, J.; Losada, P.M.; Song, B. Genome-wide analysis of common and rare variants via multiple knockoffs at biobank scale, with an application to Alzheimer disease genetics. Am. J. Hum. Genet. 2021, 108, 2336–2353. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.-J.; Jeong, Y.J.; Kim, J.; Hoe, H.-S. EGFR is a potential dual molecular target for cancer and Alzheimer’s disease. Front. Pharmacol. 2023, 14, 1238639. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fang, D. Comprehensive analysis of placental gene-expression profiles and identification of EGFR-mediated autophagy and ferroptosis suppression in intrahepatic cholestasis of pregnancy. Gene 2022, 834, 146594. [Google Scholar] [CrossRef] [PubMed]
- Jayaswamy, P.K.; Vijaykrishnaraj, M.; Patil, P.; Alexander, L.M.; Kellarai, A.; Shetty, P. Implicative role of epidermal growth factor receptor and its associated signaling partners in the pathogenesis of Alzheimer’s disease. Ageing Res. Rev. 2023, 83, 101791. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.Y.; Choi, B.-O.; Jeong, J.H.; Kong, K.A.; Hwang, J.; Ahn, J.-H. Amyloid beta-mediated hypomethylation of heme oxygenase 1 correlates with cognitive impairment in Alzheimer’s disease. PLoS ONE 2016, 11, e0153156. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-L.; Xu, C.-J. Astrocytes autophagy in aging and neurodegenerative disorders. Biomed. Pharmacother. 2020, 122, 109691. [Google Scholar] [CrossRef]
- Moreno-Navarrete, J.M.; Ortega, F.; Rodríguez, A.; Latorre, J.; Becerril, S.; Sabater-Masdeu, M.; Ricart, W.; Frühbeck, G.; Fernández-Real, J.M. HMOX1 as a marker of iron excess-induced adipose tissue dysfunction, affecting glucose uptake and respiratory capacity in human adipocytes. Diabetologia 2017, 60, 915–926. [Google Scholar] [CrossRef] [PubMed]
- Meyer, N.; Zielke, S.; Michaelis, J.B.; Linder, B.; Warnsmann, V.; Rakel, S.; Osiewacz, H.D.; Fulda, S.; Mittelbronn, M.; Münch, C. AT 101 induces early mitochondrial dysfunction and HMOX1 (heme oxygenase 1) to trigger mitophagic cell death in glioma cells. Autophagy 2018, 14, 1693–1709. [Google Scholar] [CrossRef]
- Dehm, S.M.; Bonham, K. SRC gene expression in human cancer: The role of transcriptional activation. Biochem. Cell Biol. 2004, 82, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; McMahon, M.; Chowdhry, S.; Dinkova-Kostova, A.T. Cancer chemoprevention mechanisms mediated through the Keap1–Nrf2 pathway. Antioxid. Redox Signal. 2010, 13, 1713–1748. [Google Scholar] [CrossRef]
- Peng, X.; Sun, B.; Tang, C.; Shi, C.; Xie, X.; Wang, X.; Jiang, D.; Li, S.; Jia, Y.; Wang, Y. HMOX1-LDHB interaction promotes ferroptosis by inducing mitochondrial dysfunction in foamy macrophages during advanced atherosclerosis. Dev. Cell 2024. [Google Scholar] [CrossRef]

| Exposure | Outcome | Method | p | OR | OR (95% LCI) | OR (95% UCI) |
|---|---|---|---|---|---|---|
| EGFR | GCST007320 | MR Egger | 1.28 × 10−5 | 1.014 | 1.009 | 1.019 |
| EGFR | GCST007320 | Inverse variance weighted | 1.07 × 10−25 | 1.014 | 1.011 | 1.016 |
| EGFR | GCST007320 | Weighted median | 4.31 × 10−13 | 1.013 | 1.01 | 1.017 |
| EGFR | ukb-b-323 | MR Egger | 1.20 × 10−4 | 1.003 | 1.001 | 1.004 |
| EGFR | ukb-b-323 | Inverse variance weighted | 2.56 × 10−22 | 1.003 | 1.002 | 1.003 |
| EGFR | ukb-b-323 | Weighted median | 1.81 × 10−12 | 1.003 | 1.002 | 1.004 |
| HMOX1 | GCST007320 | MR Egger | 3.53 × 10−8 | 0.974 | 0.966 | 0.982 |
| HMOX1 | GCST007320 | Weighted median | 6.52 × 10−7 | 0.987 | 0.982 | 0.992 |
| HMOX1 | GCST007320 | Inverse variance weighted | 3.07 × 10−6 | 0.992 | 0.988 | 0.995 |
| HMOX1 | GCST007320 | MR Egger | 3.53 × 10−8 | 0.974 | 0.966 | 0.982 |
| HMOX1 | GCST007320 | Weighted median | 4.12 × 10−7 | 0.987 | 0.982 | 0.992 |
| HMOX1 | GCST007320 | Inverse variance weighted | 3.07 × 10−6 | 0.992 | 0.988 | 0.995 |
| Exposure | Outcome | Method | Q | Q df | Q p |
|---|---|---|---|---|---|
| EGFR | GCST007320 | MR Egger | 16.172 | 27 | 0.950 |
| EGFR | GCST007320 | Inverse variance weighted | 16.174 | 28 | 0.963 |
| EGFR | ukb-b-323 | MR Egger | 22.455 | 27 | 0.714 |
| EGFR | ukb-b-323 | Inverse variance weighted | 22.643 | 28 | 0.751 |
| HMOX1 | GCST007320 | MR Egger | 14.444 | 24 | 0.936 |
| HMOX1 | GCST007320 | Inverse variance weighted | 24.520 | 25 | 0.490 |
| HMOX1 | ukb-b-323 | MR Egger | 37.865 | 59 | 0.985 |
| HMOX1 | ukb-b-323 | Inverse variance weighted | 45.509 | 60 | 0.917 |
| Exposure | Outcome | Egger Intercept | SE | p |
|---|---|---|---|---|
| EGFR | GCST007320 | 0.000 | 0.001 | 0.962 |
| EGFR | ukb-b-323 | 0.000 | 0.000 | 0.669 |
| HMOX1 | GCST007320 | 0.005 | 0.002 | 0.351 |
| HMOX1 | ukb-b-323 | 0.001 | 0.000 | 0.282 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Gao, X.; Tang, J.; Gao, L.; Cui, X.; Liu, K.; Zhang, X.; Jin, M. Exploring the Efficacy and Target Genes of Atractylodes Macrocephala Koidz Against Alzheimer’s Disease Based on Multi-Omics, Computational Chemistry, and Experimental Verification. Curr. Issues Mol. Biol. 2025, 47, 118. https://doi.org/10.3390/cimb47020118
Zheng Y, Gao X, Tang J, Gao L, Cui X, Liu K, Zhang X, Jin M. Exploring the Efficacy and Target Genes of Atractylodes Macrocephala Koidz Against Alzheimer’s Disease Based on Multi-Omics, Computational Chemistry, and Experimental Verification. Current Issues in Molecular Biology. 2025; 47(2):118. https://doi.org/10.3390/cimb47020118
Chicago/Turabian StyleZheng, Yuanteng, Xin Gao, Jiyang Tang, Li Gao, Xiaotong Cui, Kechun Liu, Xiujun Zhang, and Meng Jin. 2025. "Exploring the Efficacy and Target Genes of Atractylodes Macrocephala Koidz Against Alzheimer’s Disease Based on Multi-Omics, Computational Chemistry, and Experimental Verification" Current Issues in Molecular Biology 47, no. 2: 118. https://doi.org/10.3390/cimb47020118
APA StyleZheng, Y., Gao, X., Tang, J., Gao, L., Cui, X., Liu, K., Zhang, X., & Jin, M. (2025). Exploring the Efficacy and Target Genes of Atractylodes Macrocephala Koidz Against Alzheimer’s Disease Based on Multi-Omics, Computational Chemistry, and Experimental Verification. Current Issues in Molecular Biology, 47(2), 118. https://doi.org/10.3390/cimb47020118






