Research Progress on the KMT2A-AFF3 Fusion Gene in Childhood Acute Lymphoblastic Leukemia: Mechanisms, Clinical Implications, and Therapeutic Strategies
Abstract
1. Introduction
2. Molecular Characterization
2.1. Structure and Function of the KMT2A Gene
2.2. Structure and Function of the AFF3 Gene
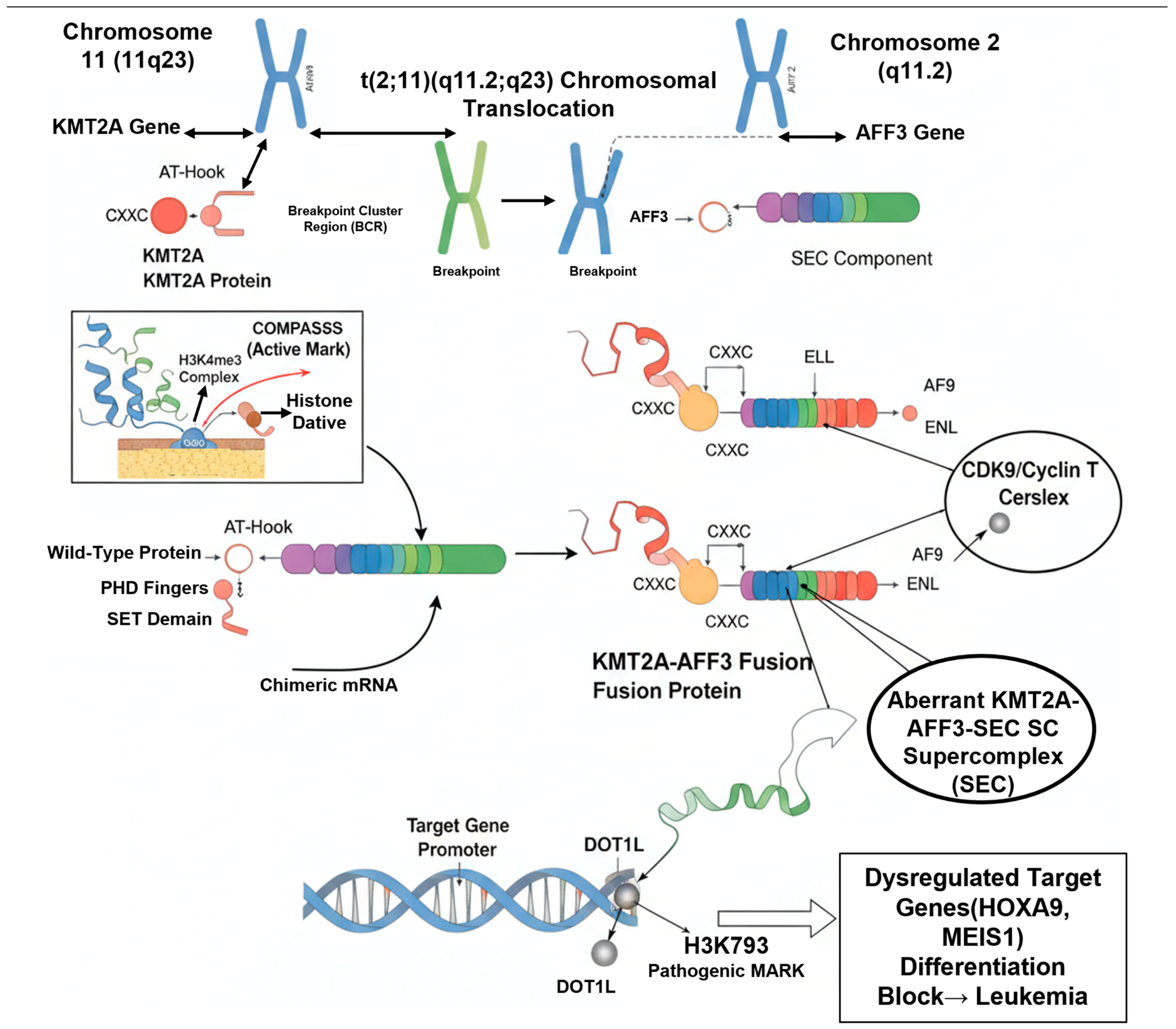
2.3. Formation of the Fusion Gene: t(2;11)(q11.2;q23)
2.4. Domain Structure and Biochemical Properties of the KMT2A-AFF3 Fusion Protein
3. Pathogenic Mechanisms
3.1. Aberrant Transcriptional Complex Formation
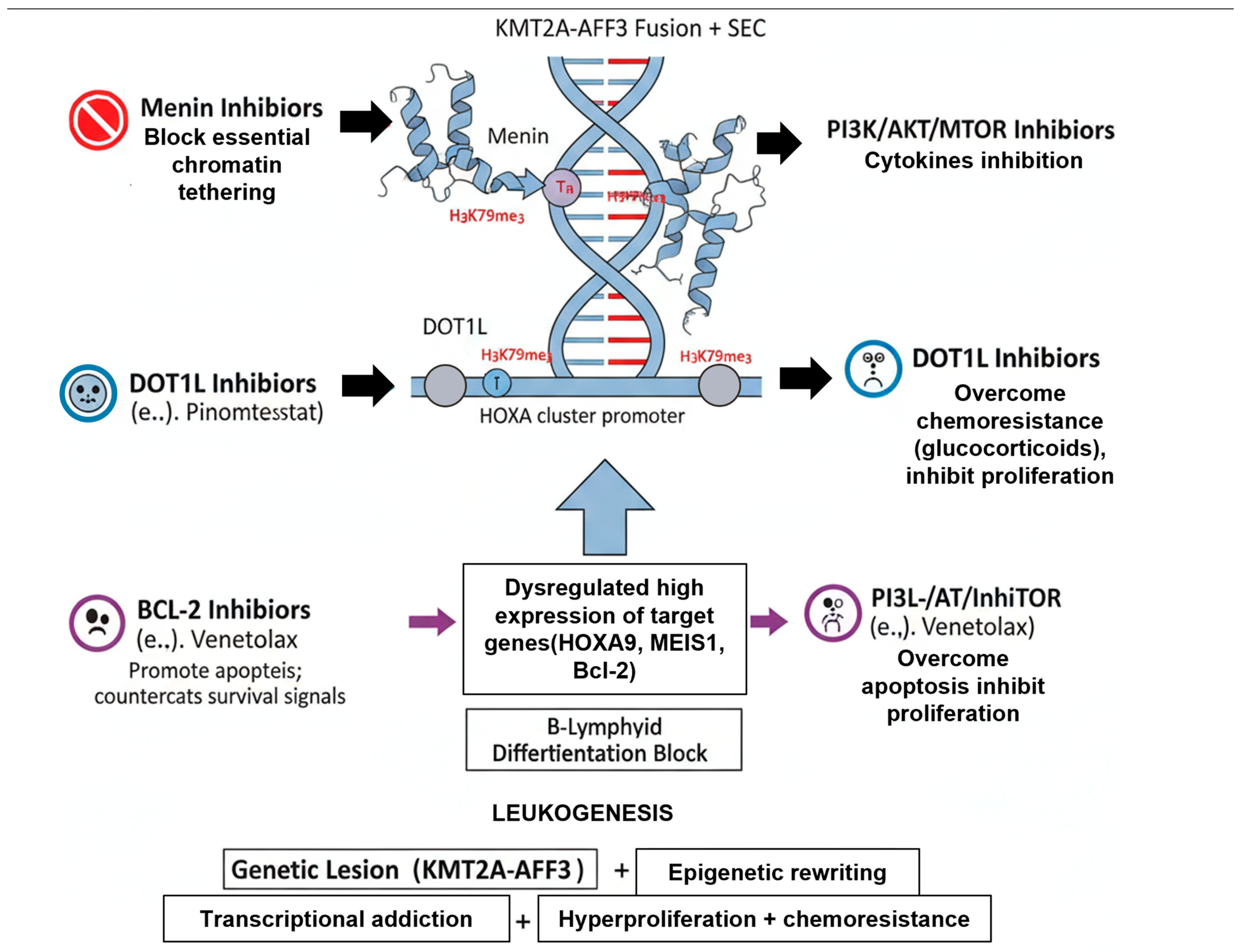
3.2. Epigenetic Reprogramming: The Role of DOT1L and Menin
3.3. Downstream Effects: Target Gene Dysregulation and Differentiation Arrest
3.4. Activation of Downstream Signaling Pathways
4. Clinical and Diagnostic Features
4.1. Epidemiology and Demographics
4.2. Clinical Presentation and Laboratory Findings
4.2.1. Hyperleukocytosis
4.2.2. Central Nervous System (CNS) Involvement
4.2.3. Organomegaly
4.2.4. Poor Initial Treatment Response
4.3. Immunophenotype
4.4. Cytogenetics and Molecular Diagnostics
4.4.1. Conventional Cytogenetics (Karyotyping)
4.4.2. Fluorescence In Situ Hybridization (FISH)
4.4.3. Reverse-Transcription PCR (RT-PCR)
4.4.4. Next-Generation Sequencing (NGS)
4.4.5. Minimal Residual Disease (MRD) Monitoring
5. Prognosis and Risk Stratification
5.1. Prognostic Significance of KMT2A-AFF3
5.2. Role in Clinical Risk Stratification
5.3. Key Prognostic Factors and Challenges
6. Treatment Strategies
6.1. Intensive Chemotherapy Backbones
6.2. Allogeneic Hematopoietic Stem Cell Transplantation (HSCT)
6.3. Targeted Therapies: Exploiting Core Dependencies
6.4. Immunotherapies
7. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharman, J.P.; Egyed, M.; Jurczak, W.; Skarbnik, A.; Pagel, J.M.; Flinn, I.W.; Kamdar, M.; Munir, T.; Walewska, R.; Corbett, G.; et al. Acalabrutinib with or without obinutuzumab versus chlorambucil and obinutuzmab for treatment-naive chronic lymphocytic leukaemia (ELEVATE TN): A randomised, controlled, phase 3 trial. Lancet 2020, 395, 1278–1291, Erratum in Lancet 2020, 395, 1694. [Google Scholar] [CrossRef]
- Tam, C.S.; Brown, J.R.; Kahl, B.S.; Ghia, P.; Giannopoulos, K.; Jurczak, W.; Simkovic, M.; Shadman, M.; Osterborg, A.; Laurenti, L.; et al. Zanubrutinib versus bendamustine and rituximab in untreated chronic lymphocytic leukaemia and small lymphocytic lymphoma (SEQUOIA): A randomised, controlled, phase 3 trial. Lancet Oncol. 2022, 23, 1031–1043, Correction in Lancet Oncol. 2023, 24, e106. [Google Scholar] [CrossRef]
- Fischer, K.; Al-Sawaf, O.; Bahlo, J.; Fink, A.M.; Tandon, M.; Dixon, M.; Robrecht, S.; Warburton, S.; Humphrey, K.; Samoylova, O.; et al. Venetoclax and Obinutuzumab in Patients with CLL and Coexisting Conditions. N. Engl. J. Med. 2019, 380, 2225–2236. [Google Scholar] [CrossRef] [PubMed]
- Eichhorst, B.; Niemann, C.U.; Kater, A.P.; Furstenau, M.; von Tresckow, J.; Zhang, C.; Robrecht, S.; Gregor, M.; Juliusson, G.; Thornton, P.; et al. First-Line Venetoclax Combinations in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2023, 388, 1739–1754. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Dohner, H.; Hillmen, P.; Keating, M.; Montserrat, E.; Chiorazzi, N.; et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 2018, 131, 2745–2760. [Google Scholar] [CrossRef] [PubMed]
- Rios-Olais, F.A.; Hilal, T. Measurable Residual Disease in Chronic Lymphocytic Leukemia: Current Understanding and Evolving Role in Clinical Practice. Curr. Treat. Options Oncol. 2023, 24, 907–928. [Google Scholar] [CrossRef]
- Miller, L.J.; Leventaki, V.; Harker-Murray, P.D.; Drendel, H.M.; Bone, K.M. A complex KMT2A::AFF3 fusion resulting from a three-way chromosomal rearrangement in pediatric B lymphoblastic leukemia. Cancer Genet. 2022, 262–263, 43–46. [Google Scholar] [CrossRef]
- Stilgenbauer, S.; Leblond, V.; Foa, R.; Bottcher, S.; Ilhan, O.; Knauf, W.; Mikuskova, E.; Renner, C.; Tausch, E.; Woszczyk, D.; et al. Obinutuzumab plus bendamustine in previously untreated patients with CLL: A subgroup analysis of the GREEN study. Leukemia 2018, 32, 1778–1786. [Google Scholar] [CrossRef]
- Ogino, J.; Dou, Y. Histone methyltransferase KMT2A: Developmental regulation to oncogenic transformation. J. Biol. Chem. 2024, 300, 107791. [Google Scholar] [CrossRef]
- Krivtsov, A.V.; Armstrong, S.A. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 2007, 7, 823–833. [Google Scholar] [CrossRef]
- Lin, C.; Smith, E.R.; Takahashi, H.; Lai, K.C.; Martin-Brown, S.; Florens, L.; Washburn, M.P.; Conaway, J.W.; Conaway, R.C.; Shilatifard, A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 2010, 37, 429–437. [Google Scholar] [CrossRef]
- Smith, E.; Lin, C.; Shilatifard, A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011, 25, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wang, Y.; Wang, X.; He, X.; Yang, C.; Naseri, A.; Pederson, T.; Zheng, J.; Zhang, S.; Xiao, X.; et al. Simultaneous epigenetic perturbation and genome imaging reveal distinct roles of H3K9me3 in chromatin architecture and transcription. Genome Biol. 2020, 21, 296. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.; Polly, P.; Liu, T. The histone methyltransferase DOT1L: Regulatory functions and a cancer therapy target. Am. J. Cancer Res. 2015, 5, 2823–2837. [Google Scholar] [PubMed]
- Kingsley, M.C.; Xie, H.M.; Chen, B.R.; Riedel, S.S.; Pastuer, T.; Bollig, M.K.; Shank, T.; Libbrecht, C.; Stabler, S.P.; Deshpande, A.J.; et al. Specific patterns of H3K79 methylation influence genetic interaction of oncogenes in AML. Blood Adv. 2020, 4, 3109–3122. [Google Scholar] [CrossRef]
- Chen, Y.; Jones, K.L.; Anastassiadis, K.; Kranz, A.; Stewart, A.F.; Grembecka, J.; Meyerson, M.; Ernst, P. Distinct pathways affected by menin versus MLL1/MLL2 in MLL-rearranged acute myeloid leukemia. Exp. Hematol. 2019, 69, 37–42. [Google Scholar] [CrossRef]
- Kwon, M.C.; Thuring, J.W.; Querolle, O.; Dai, X.; Verhulst, T.; Pande, V.; Marien, A.; Goffin, D.; Wenge, D.V.; Yue, H.; et al. Preclinical efficacy of the potent, selective menin-KMT2A inhibitor JNJ-75276617 (bleximenib) in KMT2A- and NPM1-altered leukemias. Blood 2024, 144, 1206–1220. [Google Scholar] [CrossRef]
- Sharman, J.P.; Brander, D.M.; Mato, A.R.; Ghosh, N.; Schuster, S.J.; Kambhampati, S.; Burke, J.M.; Lansigan, F.; Schreeder, M.T.; Lunin, S.D.; et al. Ublituximab plus ibrutinib versus ibrutinib alone for patients with relapsed or refractory high-risk chronic lymphocytic leukaemia (GENUINE): A phase 3, multicentre, open-label, randomised trial. Lancet Haematol. 2021, 8, e254–e266, Correction in Lancet Haematol. 2021, 8, e249. [Google Scholar] [CrossRef]
- Benito, J.M.; Godfrey, L.; Kojima, K.; Hogdal, L.; Wunderlich, M.; Geng, H.; Marzo, I.; Harutyunyan, K.G.; Golfman, L.; North, P.; et al. MLL-Rearranged Acute Lymphoblastic Leukemias Activate BCL-2 through H3K79 Methylation and Are Sensitive to the BCL-2-Specific Antagonist ABT-199. Cell Rep. 2015, 13, 2715–2727. [Google Scholar] [CrossRef]
- Kater, A.P.; Levin, M.D.; Dubois, J.; Kersting, S.; Enggaard, L.; Veldhuis, G.J.; Mous, R.; Mellink, C.H.M.; van der Kevie-Kersemaekers, A.F.; Dobber, J.A.; et al. Minimal residual disease-guided stop and start of venetoclax plus ibrutinib for patients with relapsed or refractory chronic lymphocytic leukaemia (HOVON141/VISION): Primary analysis of an open-label, randomised, phase 2 trial. Lancet Oncol. 2022, 23, 818–828. [Google Scholar] [CrossRef]
- Chen, X.; Burkhardt, D.B.; Hartman, A.A.; Hu, X.; Eastman, A.E.; Sun, C.; Wang, X.; Zhong, M.; Krishnaswamy, S.; Guo, S. MLL-AF9 initiates transformation from fast-proliferating myeloid progenitors. Nat. Commun. 2019, 10, 5767, Correction in Nat. Commun. 2020, 11, 681. [Google Scholar] [CrossRef]
- Lewis, T.S.; Mccormick, R.S.; Zeng, W.; Miyamoto, J.B.; Kennedy, D.; Sievers, E.L.; Mcearchern, J.A.; Law, C.L. Abstract 1789: Auristatin-based antibody-drug conjugates are synergistic in combination with PI3K-AKT-mTOR pathway inhibitors in hematologic malignancies and carcinoma. Cancer Res. 2011, 71 (Suppl. S8), 1789. [Google Scholar] [CrossRef]
- Stam, R.; Boer, M.L.; Passier, M.; Sallan, S.; Armstrong, S.; Pieters, P. MLL Rearranged Infant Acute Lymphoblastic Leukemia Is Characterized by Silencing of the Putative Tumor Suppressor Gene FHIT. Blood 2004, 104, 525. [Google Scholar] [CrossRef]
- Isobe, T.; Takagi, M.; Sato-Otsubo, A.; Nishimura, A.; Nagae, G.; Yamagishi, C.; Tamura, M.; Tanaka, Y.; Asada, S.; Takeda, R.; et al. Multi-omics analysis defines highly refractory RAS burdened immature subgroup of infant acute lymphoblastic leukemia. Nat. Commun. 2022, 13, 4501. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Sun, Y.; Wang, J.; Song, W.; Xu, T.; Kapur, R.; Hess, J.L. The Role of STAT5 in HOXA9-Associated Leukemia. Blood 2016, 128, 3919. [Google Scholar] [CrossRef]
- Bartram, J.; Ancliff, P.; Vora, A. How I treat infant acute lymphoblastic leukemia. Blood 2025, 145, 35–42. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, J.J.; Seriu, T.; Panzer-Grumayer, E.R.; Biondi, A.; Pongers-Willemse, M.J.; Corral, L.; Stolz, F.; Schrappe, M.; Masera, G.; Kamps, W.A.; et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet 1998, 352, 1731–1738. [Google Scholar] [CrossRef]
- Yin, L.; Wan, L.; Zhang, Y.; Hua, S.; Shao, X. Recent Developments and Evolving Therapeutic Strategies in KMT2A-Rearranged Acute Leukemia. Cancer Med. 2024, 13, e70326. [Google Scholar] [CrossRef]
- Massoth, L.R.; Hung, Y.P.; Nardi, V.; Nielsen, G.P.; Hasserjian, R.P.; Louissaint, A., Jr.; Fisch, A.S.; Deshpande, V.; Zukerberg, L.R.; Lennerz, J.K.; et al. Pan-sarcoma genomic analysis of KMT2A rearrangements reveals distinct subtypes defined by YAP1-KMT2A-YAP1 and VIM-KMT2A fusions. Mod. Pathol. 2020, 33, 2307–2317. [Google Scholar] [CrossRef]
- Thompson, P.A.; Srivastava, J.; Peterson, C.; Strati, P.; Jorgensen, J.L.; Hether, T.; Keating, M.J.; O’Brien, S.M.; Ferrajoli, A.; Burger, J.A.; et al. Minimal residual disease undetectable by next-generation sequencing predicts improved outcome in CLL after chemoimmunotherapy. Blood 2019, 134, 1951–1959. [Google Scholar] [CrossRef]
- Matsuo, H.; Yoshida, K.; Fukumura, K.; Nakatani, K.; Noguchi, Y.; Takasaki, S.; Noura, M.; Shiozawa, Y.; Shiraishi, Y.; Chiba, K.; et al. Recurrent CCND3 mutations in MLL-rearranged acute myeloid leukemia. Blood Adv. 2018, 2, 2879–2889. [Google Scholar] [CrossRef] [PubMed]
- Schneider, P.; Crump, N.T.; Arentsen-Peters, S.; Smith, A.L.; Hagelaar, R.; Adriaanse, F.R.S.; Bos, R.S.; de Jong, A.; Nierkens, S.; Koopmans, B.; et al. Modelling acquired resistance to DOT1L inhibition exhibits the adaptive potential of KMT2A-rearranged acute lymphoblastic leukemia. Exp. Hematol. Oncol. 2023, 12, 81. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Karsa, M.; Ronca, E.; Bongers, A.; Kosciolek, A.; El-Ayoubi, A.; Revalde, J.L.; Seneviratne, J.A.; Cheung, B.B.; Cheung, L.C.; et al. The Combination of Curaxin CBL0137 and Histone Deacetylase Inhibitor Panobinostat Delays KMT2A-Rearranged Leukemia Progression. Front. Oncol. 2022, 12, 863329. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.N.; Hu, Y.X.; Gao, L.; Xiao, P.F.; Lu, J.; Wu, S.Y.; Wang, M.; Shao, X.J.; Zhou, C.Y.; Ling, J.; et al. The therapeutic efficacy of pediatric ALL patients with MLL gene rearrangement treated with CCLG-ALL2008 protocol. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6020–6029. [Google Scholar]
- Sun, D.; Du, X.; Su, P. Molecular evolution of transcription factors AF4/FMR2 family member (AFF) gene family and the role of lamprey AFF3 in cell proliferation. Dev. Genes Evol. 2024, 234, 45–53. [Google Scholar] [CrossRef]
- Bassani, S.; Chrast, J.; Ambrosini, G.; Voisin, N.; Schutz, F.; Brusco, A.; Sirchia, F.; Turban, L.; Schubert, S.; Abou Jamra, R.; et al. Variant-specific pathophysiological mechanisms of AFF3 differently influence transcriptome profiles. Genome Med. 2024, 16, 72. [Google Scholar] [CrossRef]
- Molica, S.; Giannarelli, D.; Montserrat, E. Minimal Residual Disease and Survival Outcomes in Patients With Chronic Lymphocytic Leukemia: A Systematic Review and Meta-analysis. Clin. Lymphoma Myeloma Leuk. 2019, 19, 423–430. [Google Scholar] [CrossRef]
- Issa, G.C.; Aldoss, I.; DiPersio, J.; Cuglievan, B.; Stone, R.; Arellano, M.; Thirman, M.J.; Patel, M.R.; Dickens, D.S.; Shenoy, S.; et al. The menin inhibitor revumenib in KMT2A-rearranged or NPM1-mutant leukaemia. Nature 2023, 615, 920–924. [Google Scholar] [CrossRef]
- Aldoss, I.; Issa, G.C.; Thirman, M.J.; Dipersio, J.; Arellano, M.; Blachly, J.S.; Mannis, G.; Perl, A.; Dickens, D.; Mcmahon, C.M. Revumenib Monotherapy in Patients with Relapsed/Refractory KMT2Ar Acute Leukemias: Efficacy and Safety Results from the Augment-101 Phase 1/2 Study. Blood 2023, 142 (Suppl. S1), 2907. [Google Scholar] [CrossRef]
- Wang, E.S.; Altman, J.K.; Pettit, K.; De Botton, S.; Walter, R.P.; Fenaux, P.; Burrows, F.; Tomkinson, B.E.; Martell, B.; Fathi, A.T. Preliminary Data on a Phase 1/2A First in Human Study of the Menin-KMT2A (MLL) Inhibitor KO-539 in Patients with Relapsed or Refractory Acute Myeloid Leukemia. Blood 2020, 136 (Suppl. S1), 7–8. [Google Scholar] [CrossRef]
- Falkenstein, C.D.; Niswander, L.M.; Kessler, L.; Tomkinson, B.; Burrows, F.; Tasian, S.K. Preclinical In Vivo Activity of the Menin Inhibitor Ziftomenib (KO-539) in Pediatric KMT2A-Rearranged Acute Lymphoblastic Leukemia. Blood 2022, 140 (Suppl. S1), 3491–3492. [Google Scholar] [CrossRef]
- Issa, G.C.; Cuglievan, B.; DiNardo, C.D.; Short, N.J.; McCall, D.; Gibson, A.; Nunez, C.; Garcia, M.B.; Roth, M.; Bidikian, A.; et al. Early Results of the Phase I/II Study Investigating the All-Oral Combination of the Menin Inhibitor Revumenib (SNDX-5613) with Decitabine/Cedazuridine (ASTX727) and Venetoclax in Acute Myeloid Leukemia (SAVE). Blood 2023, 142 (Suppl. S1), 58. [Google Scholar] [CrossRef]
- Masago, K.; Kuroda, H.; Sasaki, E.; Fujita, Y.; Fujita, S.; Horio, Y.; Endo, M.; Ishihara, H.; Hanai, N.; Matsushita, H. Novel gene fusions in human oropharyngeal carcinoma. Cancer Genet. 2024, 286–287, 29–34. [Google Scholar] [CrossRef]
- Jeanselme, P.; Tavitian, S.; Lapierre, L.; Vergez, F.; Rigolot, L.; Huynh, A.; Bertoli, S.; Delabesse, E.; Huguet, F. Long-term exposure and response to azacitidine for post-hematopoietic stem cell transplantation relapse of early T-cell precursor acute lymphoblastic leukemia: A case report and review of the literature. Leuk. Lymphoma 2024, 65, 2025–2030. [Google Scholar] [CrossRef]
- Rafeeinia, A.; Asadikaram, G.; Karimi-Darabi, M.; Moazed, V. High Levels of Organochlorines Are Associated with Induction of ABL1 Promoter Methylation in Children with Acute Lymphoblastic Leukemia. DNA Cell Biol. 2022, 41, 727–734. [Google Scholar] [CrossRef]
- Nickels, E.M.; Li, S.; Myint, S.S.; Arroyo, K.; Feng, Q.; Siegmund, K.D.; de Smith, A.J.; Wiemels, J.L. DNA methylation at birth in monozygotic twins discordant for pediatric acute lymphoblastic leukemia. Nat. Commun. 2022, 13, 6077. [Google Scholar] [CrossRef]
| Signaling Pathway | General Role in ALL | Specific Relevance to KMT2A-r/KMT2A-AFF3 |
|---|---|---|
| Wnt/β-catenin | Regulates self-renewal, proliferation, and cell fate. | Often hyperactivated in KMT2A-r/β-catenin stabilization is crucial for maintaining the “leukemia stem cell” (LSC) properties and contributes to leukemic cell survival [21]. |
| PI3K/AKT/mTOR | Central regulator of cell growth, survival, metabolism, and proliferation. | Frequently activated in ALL; in KMT2A-r, it is a major driver of resistance to chemotherapy, particularly glucocorticoids [22,23]. It also promotes the Warburg effect (aerobic glycolysis) and protein translation via mTORC1. |
| RAS/MAPK | Transduces signals from growth factor receptors to control cell proliferation and survival. | Co-mutations in RAS pathway genes (e.g., KRAS, NRAS, PTPN11) are very common “second hits” in KMT2A-r ALL, cooperating with the fusion gene to provide a potent proliferative signal [24]. |
| JAK/STAT | Mediates cytokine signaling, crucial for hematopoietic cell proliferation and differentiation. | JAK/STAT signaling, particularly via STAT5, is often activated by KMT2A-r-driven autocrine loops (e.g., upregulation of FLT3 or cytokine receptors), promoting cell cycle progression and survival [19,25] |
| Methodology | Target | Purpose/Clinical Utility | Limitations |
| Flow Cytometry | Cell surface/intracellular proteins (CD markers) | Diagnosis and Classification. Identifies B-lineage (CD19+), pro-B/pre-B stage, and hallmark CD10-negative phenotype. Detects aberrant myeloid markers. | Not specific to KMT2A-AFF3. Cannot distinguish from other KMT2A-r subtypes. |
| Conventional Cytogenetics | Chromosomes (G-banding) | Initial identification of the t(2;11)(q11.2;q23) translocation. | Low resolution. May miss cryptic or complex translocations [7]. Labor-intensive. |
| FISH | KMT2A gene locus (11q23) | Rapid Screening. Confirms a KMT2A rearrangement (“break-apart” signal) with high sensitivity. | Does not identify the AFF3 partner. Requires follow-up testing. |
| RT-PCR | KMT2A-AFF3 chimeric mRNA | Specific Confirmation. Confirms the exact fusion transcript KMT2A-AFF3. | Requires a priori suspicion of AFF3 as the partner to select the correct primers. |
| NGS (RNA-seq) | Total transcriptome (cDNA) | Gold Standard Identification. Unbiased detection of KMT2A-AFF3 and any other fusions. Identifies precise breakpoints. Simultaneously finds cooperating mutations (e.g., RAS pathway) [24]. | Higher cost and longer turnaround time than PCR. Requires bioinformatics expertise. |
| qPCR/ddPCR | KMT2A-AFF3 DNA/cDNA junction | MRD Monitoring. Ultra-sensitive quantification of residual leukemia cells during and after therapy to guide treatment decisions [27]. | Patient-specific assay must be designed and validated after diagnosis. Not a primary diagnostic tool. |
| Prognostic Factor | Favorable Indicator | Unfavorable Indicator (High-Risk) | Clinical Significance and Relevance to KMT2A-AFF3 |
|---|---|---|---|
| Primary Genetic Lesion | ETV6-RUNX1, High Hyper diploidy | Presence of KMT2A-AFF3 (or any KMT2A-r) | Defines the primary risk group. The KMT2A-AFF3 fusion is the initiating driver and assigns the patient to high-risk therapy from diagnosis [3,28]. |
| Age at Diagnosis | 1–9.99 years | Infancy (<1 year), especially <6 months | A critical independent risk factor. KMT2A-AFF3 often occurs in infants, compounding the genetic risk and leading to a very poor prognosis [26]. |
| WBC at Diagnosis | <50,000/µL (B-ALL) | >50,000/µL (Hyperleukocytosis) | Indicates high tumor burden. KMT2A-AFF3 patients frequently present with hyperleukocytosis, another independent high-risk factor. |
| Early Treatment Response (MRD) | Rapid clearance (e.g., <0.01% at End of Induction) | Persistent MRD (e.g., >0.1% at End of Induction) | The most powerful prognostic indicator. Assesses in vivo chemosensitivity. Persistent MRD in KMT2A-AFF3 ALL is a strong indication for HSCT [29,30]. |
| Cooperating Mutations | Absence of “second hits” | Presence of RAS pathway mutations (KRAS, NRAS, PTPN11) | Modifies disease biology. KMT2A-r ALL has a high frequency of cooperating RAS mutations, which enhance proliferation and chemoresistance [24]. |
| CNS Status | CNS-1 (No blasts in CSF) | CNS-2 or CNS-3 (Blasts in CSF) | Indicates disease spread. KMT2A-r leukemias, including KMT2A-AFF3, have a high propensity for CNS infiltration, requiring intensive CNS-directed therapy [3]. |
| Drug Name (Code) | Target | Trial Name/ID | Phase | Target Population | Key Reference(s) |
|---|---|---|---|---|---|
| Revumenib (SNDX-5613) | Menin-KMT2A interaction | AUGMENT-101 (NCT04065399) | Phase I/II (Pivotal) | Relapsed/Refractory (R/R) KMT2A-r or NPM1-mutant (NPM1m) acute leukemia (ALL & AML); Adult and Pediatric | Issa et al., 2023 [38]; Aldoss et al., 2023 [39] |
| Ziftomenib (KO-539) | Menin-KMT2A interaction | KOMET-001 (NCT04067336) | Phase I/II | R/R KMT2A-r or NPM1m acute myeloid leukemia (AML); includes KMT2A-r ALL sub-study | Wang et al., 2020 [40] Falkenstein et al., 2022 [41] |
| Revumenib + Chemo. (e.g., Venetoclax) | Menin-KMT2A interaction and BCL-2 | SAVE (NCT05360160) | Phase I/II | R/R AML or Myeloid Leukemia with KMT2A-r, NPM1m, or NUP98-r | Issa et al., 2023 [42] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Liang, J. Research Progress on the KMT2A-AFF3 Fusion Gene in Childhood Acute Lymphoblastic Leukemia: Mechanisms, Clinical Implications, and Therapeutic Strategies. Curr. Issues Mol. Biol. 2025, 47, 988. https://doi.org/10.3390/cimb47120988
Zhang Y, Liang J. Research Progress on the KMT2A-AFF3 Fusion Gene in Childhood Acute Lymphoblastic Leukemia: Mechanisms, Clinical Implications, and Therapeutic Strategies. Current Issues in Molecular Biology. 2025; 47(12):988. https://doi.org/10.3390/cimb47120988
Chicago/Turabian StyleZhang, Yawei, and Juan Liang. 2025. "Research Progress on the KMT2A-AFF3 Fusion Gene in Childhood Acute Lymphoblastic Leukemia: Mechanisms, Clinical Implications, and Therapeutic Strategies" Current Issues in Molecular Biology 47, no. 12: 988. https://doi.org/10.3390/cimb47120988
APA StyleZhang, Y., & Liang, J. (2025). Research Progress on the KMT2A-AFF3 Fusion Gene in Childhood Acute Lymphoblastic Leukemia: Mechanisms, Clinical Implications, and Therapeutic Strategies. Current Issues in Molecular Biology, 47(12), 988. https://doi.org/10.3390/cimb47120988





