GWAS Reveals Stable Genetic Loci and Candidate Genes for Grain Protein Content in Wheat
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Phenotyping
2.3. Statistical Analyses
2.4. Association Mapping and Candidate Gene Search
2.5. Candidate Gene and Haplotype Analyses in 17 Chinese Wheat Cultivars Leveraging Chromosome-Level Genomes
3. Results
3.1. Phenotypic Variation
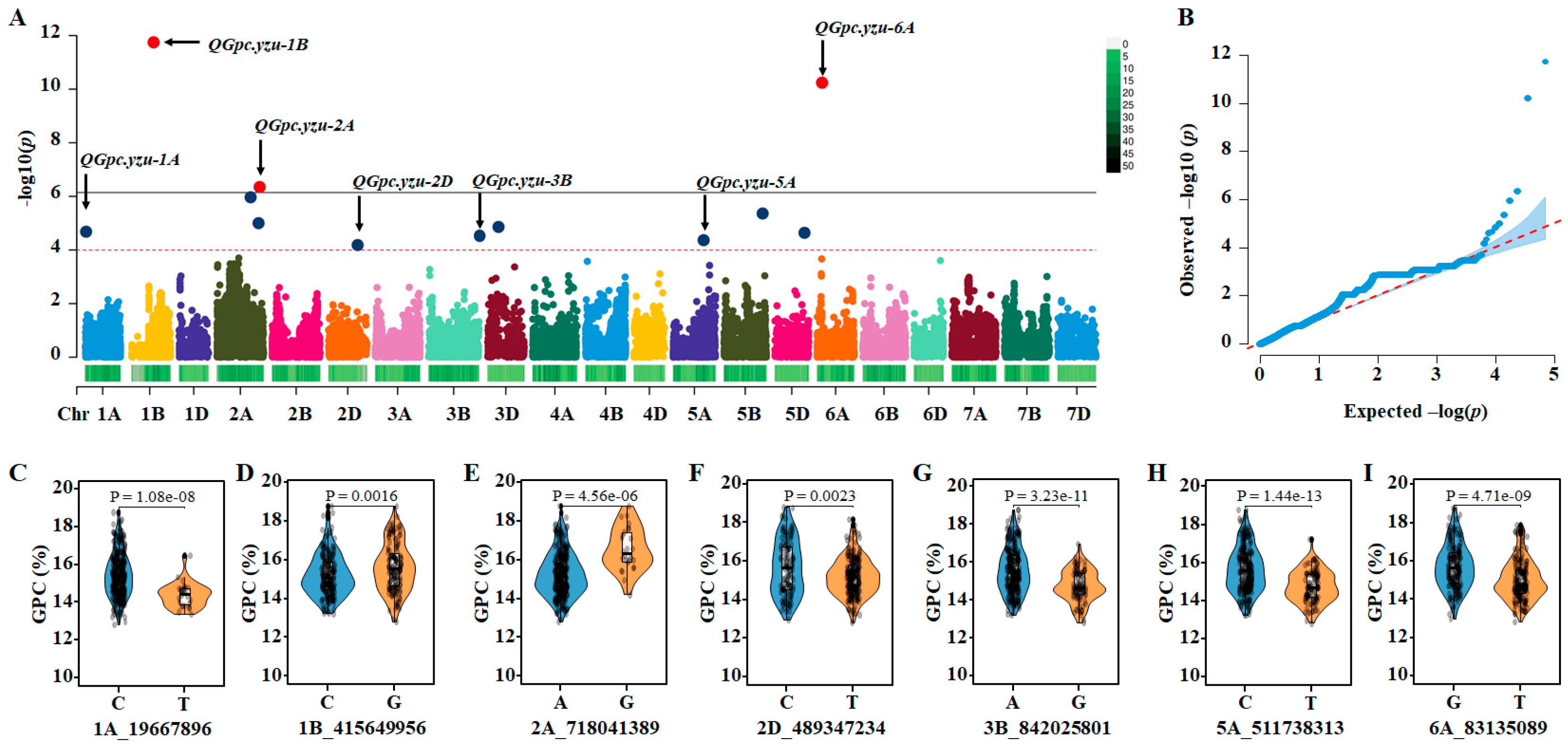
3.2. GWAS for Significant Quantitative Trait Nucleotides (QTNs)
3.3. Characterization of the Physical Regions of QGpc.yzu-2A Revealed Potential Candidate Genes for Grain Protein Content in Wheat
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Stein, N.; Choulet, F.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Battenfield, S.D.; Guzman, C.; Gaynor, R.C.; Singh, R.P.; Peña, R.J.; Dreisigacker, S.; Fritz, A.K.; Poland, J.A. Genomic selection for processing and end-use quality traits in the CIMMYT spring bread wheat breeding program. Plant Genome 2016, 9, 1–12. [Google Scholar] [CrossRef]
- Sukumaran, S.; Dreisigacker, S.; Lopes, M.; Chavez, P.; Reynolds, M.P. Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theor. Appl. Genet. 2015, 128, 353–363. [Google Scholar] [CrossRef]
- Groos, C.; Robert, N.; Bervas, E.; Charmet, G. Genetic analysis of grain protein-content, grain yield and thousand-kernel weight in bread wheat. Theor. Appl. Genet. 2003, 106, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Rapp, M.; Lein, V.; Lacoudre, F.; Lafferty, J.; Müller, E.; Vida, G.; Bozhanova, V.; Ibraliu, A.; Thorwarth, P.; Piepho, H.P.; et al. Simultaneous improvement of grain yield and protein content in durum wheat by different phenotypic indices and genomic selection. Theor. Appl. Genet. 2018, 131, 1315–1329. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Xia, X. From markers to genome-based breeding in wheat. Theor. Appl. Genet. 2019, 132, 767–784. [Google Scholar] [CrossRef] [PubMed]
- Kartseva, T.; Alqudah, A.M.; Aleksandrov, V.; Alomari, D.Z.; Doneva, D.; Arif, M.A.R.; Börner, A.; Misheva, S. Nutritional genomic approach for improving grain protein content in wheat. Foods 2023, 12, 1399. [Google Scholar] [CrossRef]
- Uauy, C.; Brevis, J.C.; Dubcovsky, J. The high grain protein content gene Gpc-B1 accelerates senescence and has pleiotropic effects on protein content in wheat. J. Exp. Bot. 2006, 57, 2785–2794. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef]
- Leonova, I.N.; Kiseleva, A.A.; Berezhnaya, A.A.; Stasyuk, A.I.; Likhenko, I.E.; Salina, E.A. Identification of QTLs for grain protein content in Russian spring wheat varieties. Plants 2022, 11, 437. [Google Scholar] [CrossRef]
- Kartseva, T.; Aleksandrov, V.; Alqudah, A.M.; Arif, M.A.R.; Kocheva, K.; Doneva, D.; Prokopova, K.; Börner, A.; Misheva, S. GWAS in a collection of bulgarian old and modern bread wheat accessions uncovers novel genomic loci for grain protein content and thousand kernel weight. Plants 2024, 13, 1084. [Google Scholar] [CrossRef]
- Basheir, S.; Hong, Y.; Lv, C.; Xu, H.; Zhu, J.; Guo, B.; Wang, F.; Xu, R. Identification of wheat germplasm resistance to late sowing. Agronomy 2023, 13, 1010. [Google Scholar] [CrossRef]
- Zhang, Z.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Jiao, C.; Xie, X.; Hao, C.; Chen, L.; Xie, Y.; Garg, V.; Zhao, L.; Wang, Z.; Zhang, Y.; Li, T.; et al. Pan-genome bridges wheat structural variations with habitat and breeding. Nature 2025, 637, 384–393. [Google Scholar] [CrossRef] [PubMed]
- The International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar] [CrossRef]
- Jukanti, A.; Heidlebaugh, N.; Parrott, D.; Fischer, I.A.; McInnerney, K.; Fischer, A.M. Comparative transcriptome profiling of near-isogenic barley (Hordeum vulgare) lines differing in the allelic state of a major grain protein content locus identifies genes with possible roles in leaf senescence and nitrogen reallocation. New Phytol. 2008, 177, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Tripet, B.P.; Mason, K.E.; Eilers, B.J.; Burns, J.; Powell, P.; Fischer, A.M.; Copié, V. Structural and biochemical analysis of the Hordeum vulgare L. HvGR-RBP1 protein, a glycine-rich RNA-binding protein involved in the regulation of barley plant development and stress response. Biochemistry 2014, 53, 7945–7960. [Google Scholar] [CrossRef]
- Lou, H.; Zhang, R.; Liu, Y.; Guo, D.; Zhai, S.; Chen, A.; Zhang, Y.; Xie, C.; You, M.; Peng, H.; et al. Genome-wide association study of six quality-related traits in common wheat (Triticum aestivum L.) under two sowing conditions. Theor. Appl. Genet. 2021, 134, 399–418. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Lou, H.; Zhai, S.; Zhang, R.; Liu, G.; Xu, W.; Yu, J.; Zhang, Y.; Ni, Z.; Sun, Q.; et al. TaNAM-6A is essential for nitrogen remobilisation and regulates grain protein content in wheat (Triticum aestivum L.). Plant Cell Environ. 2024, 47, 2310–2321. [Google Scholar] [CrossRef] [PubMed]
- Muqaddasi, Q.H.; Brassac, J.; Ebmeyer, E.; Kollers, S.; Korzun, V.; Argillier, O.; Stiewe, G.; Plieske, J.; Ganal, M.W.; Röder, M.S. Prospects of GWAS and predictive breeding for European winter wheat’s grain protein content, grain starch content, and grain hardness. Sci. Rep. 2020, 10, 12541. [Google Scholar] [CrossRef] [PubMed]
- Battenfield, S.D.; Sheridan, J.L.; Silva, L.D.C.E.; Miclaus, K.J.; Dreisigacker, S.; Wolfinger, R.D.; Peña, R.J.; Singh, R.P.; Jackson, E.W.; Fritz, A.K.; et al. Breeding-assisted genomics: Applying metaGWAS for milling and baking quality in CIMMYT wheat breeding program. PLoS ONE 2018, 13, e0204757. [Google Scholar] [CrossRef] [PubMed]
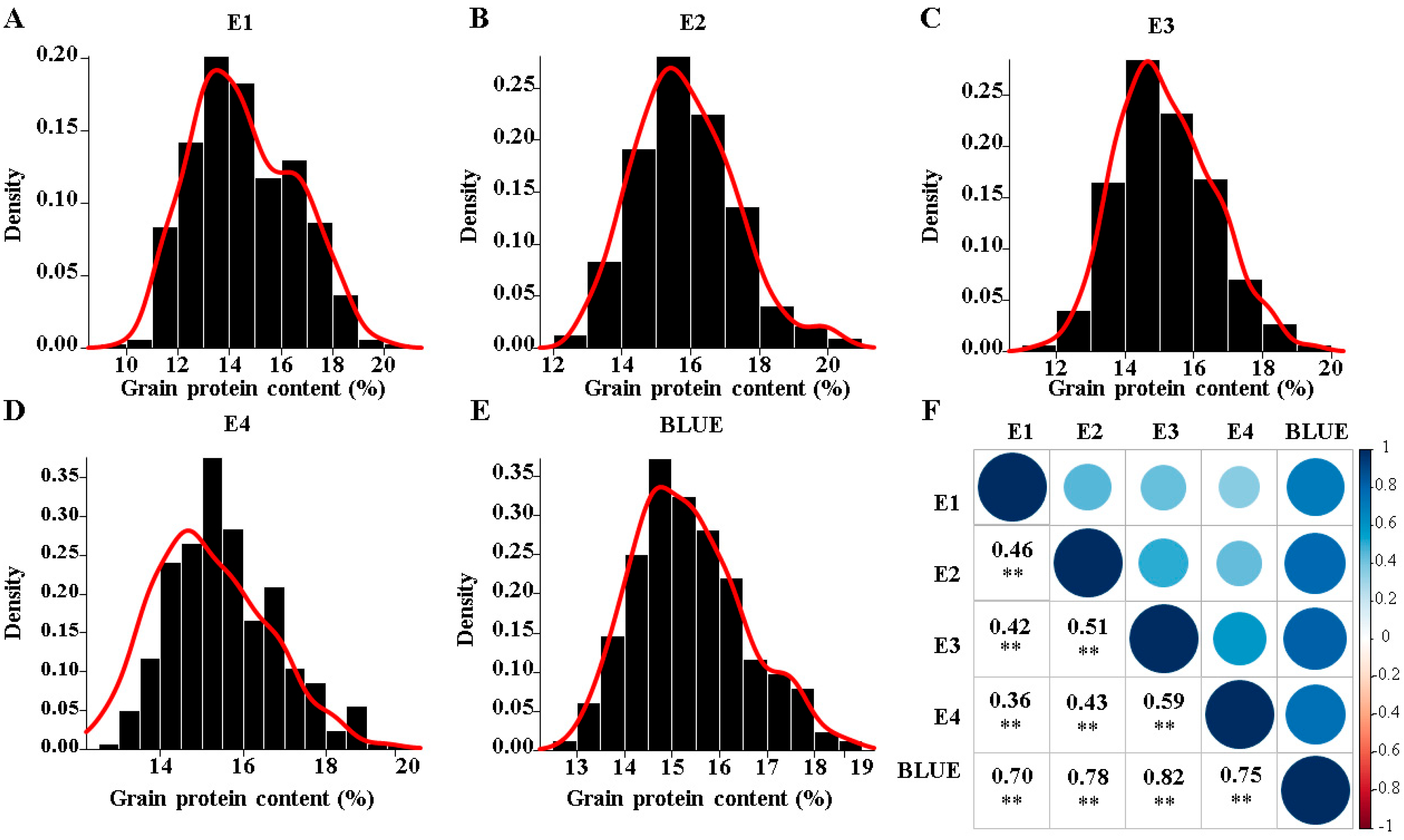

| Trait | Env. 1 | Mean * | Std. Dev. | Min. | Max. | CV% | h2 |
|---|---|---|---|---|---|---|---|
| GPC (%) | E1 | 14.56 a | 2.00 | 9.87 | 20.12 | 13.77 | 0.26 |
| E2 | 15.85 d | 1.47 | 12.81 | 20.54 | 9.26 | 0.49 | |
| E3 | 15.18 b | 1.39 | 11.46 | 19.53 | 9.18 | 0.20 | |
| E4 | 15.60 c | 1.28 | 12.99 | 19.89 | 8.23 | 0.33 | |
| BLUE | 15.37 | 1.17 | 12.82 | 18.79 | 7.58 | 0.52 |
| Source of Variation | SS | df | MS | F | p-Value | F Crit |
|---|---|---|---|---|---|---|
| Genotype (G) | 3291.177 | 326 | 10.096 | 8.936 | 0.000 | 1.149 |
| Environment (E) | 429.863 | 3 | 143.288 | 126.823 | 0.000 | 2.606 |
| G × E | 2189.864 | 968 | 2.262 | 2.002 | 0.000 | 1.113 |
| Total | 6238.638 | 1297 |
| QTLs | Chr. | Position (bp) 1 | QTNs | −log10 (P) | E |
|---|---|---|---|---|---|
| QGpc.yzu-1A | 1A | 19,667,896 | 1A_19667896 | 4.7 | BLUE |
| 4.3 | E3 | ||||
| QGpc.yzu-1B | 1B | 415,649,956 | 1B_415649956 | 11.7 | BLUE |
| 4.7 | E4 | ||||
| QGpc.yzu-2A | 2A | 718,041,389 | 2A_718041389 | 6.3 | BLUE |
| 4.3 | E3 | ||||
| 6.1 | E4 | ||||
| QGpc.yzu-2D | 2D | 489,347,234 | 2D_489347234 | 4.2 | BLUE |
| 4.4 | E3 | ||||
| QGpc.yzu-3B | 3B | 842,025,801 | 3B_842025801 | 4.4 | E3 |
| 4.7 | E4 | ||||
| QGpc.yzu-5A | 5A | 511,738,313 | 5A_511738313 | 4.4 | BLUE |
| 10.2 | E2 | ||||
| QGpc.yzu-6A | 6A | 83,135,089 | 6A_83135089 | 10.2 | BLUE |
| 4.2 | E4 |
| Gene ID 1 | Functional Annotation | H | SS | NS | Pi |
|---|---|---|---|---|---|
| TraesCS2A02G471300 | Benzyl alcohol O-benzoyltransferase | 3 | 0 | 6 | 0.00072 |
| TraesCS2A02G471500 | Homoserine kinase | 2 | 1 | 2 | 0.00055 |
| TraesCS2A02G471600 α | Phospholipase A2 | NA | NA | NA | NA |
| TraesCS2A02G472000 | IMPACT family member in pol 5′region | 2 | 1 | 2 | 0.00083 |
| TraesCS2A02G472700 | BTB/POZ and MATH domain-containing protein 2 | 2 | 3 | 2 | 0.00096 |
| TraesCS2A02G473000 | RNA-binding family protein | 2 | 0 | 0 | 0.00843 |
| TraesCS2A02G473100 | Polyadenylate-binding protein-interacting protein 4 | 3 | 0 | 0 | 0.01172 |
| TraesCS2A02G473200 α | AT hook motif DNA-binding family protein | NA | NA | NA | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Wang, R.; Tu, K.; Hong, Y.; Wang, F.; Zhu, J.; Lv, C.; Xu, R.; Guo, B. GWAS Reveals Stable Genetic Loci and Candidate Genes for Grain Protein Content in Wheat. Curr. Issues Mol. Biol. 2025, 47, 981. https://doi.org/10.3390/cimb47120981
Zhao Y, Wang R, Tu K, Hong Y, Wang F, Zhu J, Lv C, Xu R, Guo B. GWAS Reveals Stable Genetic Loci and Candidate Genes for Grain Protein Content in Wheat. Current Issues in Molecular Biology. 2025; 47(12):981. https://doi.org/10.3390/cimb47120981
Chicago/Turabian StyleZhao, Yuxuan, Renjie Wang, Keling Tu, Yi Hong, Feifei Wang, Juan Zhu, Chao Lv, Rugen Xu, and Baojian Guo. 2025. "GWAS Reveals Stable Genetic Loci and Candidate Genes for Grain Protein Content in Wheat" Current Issues in Molecular Biology 47, no. 12: 981. https://doi.org/10.3390/cimb47120981
APA StyleZhao, Y., Wang, R., Tu, K., Hong, Y., Wang, F., Zhu, J., Lv, C., Xu, R., & Guo, B. (2025). GWAS Reveals Stable Genetic Loci and Candidate Genes for Grain Protein Content in Wheat. Current Issues in Molecular Biology, 47(12), 981. https://doi.org/10.3390/cimb47120981








