A Comprehensive Comparative Analysis and Phylogenetic Investigation of the Chloroplast Genome Sequences in Four Astragalus Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material Sampling, DNA Extraction and Sequencing
2.2. Chloroplast Genome Assembly and Annotation
2.3. Codon Usage Bias Analysis
2.4. Repeat Element and SSR Analysis
2.5. Comparative Genome and Sequence Divergence Analyses
2.6. Analysis of Synonymous (Ks) and Non-Synonymous (Ka) Substitution Rate
2.7. Phylogenetic Analysis
3. Results
3.1. Chloroplast Genome Features of Astragalus Species
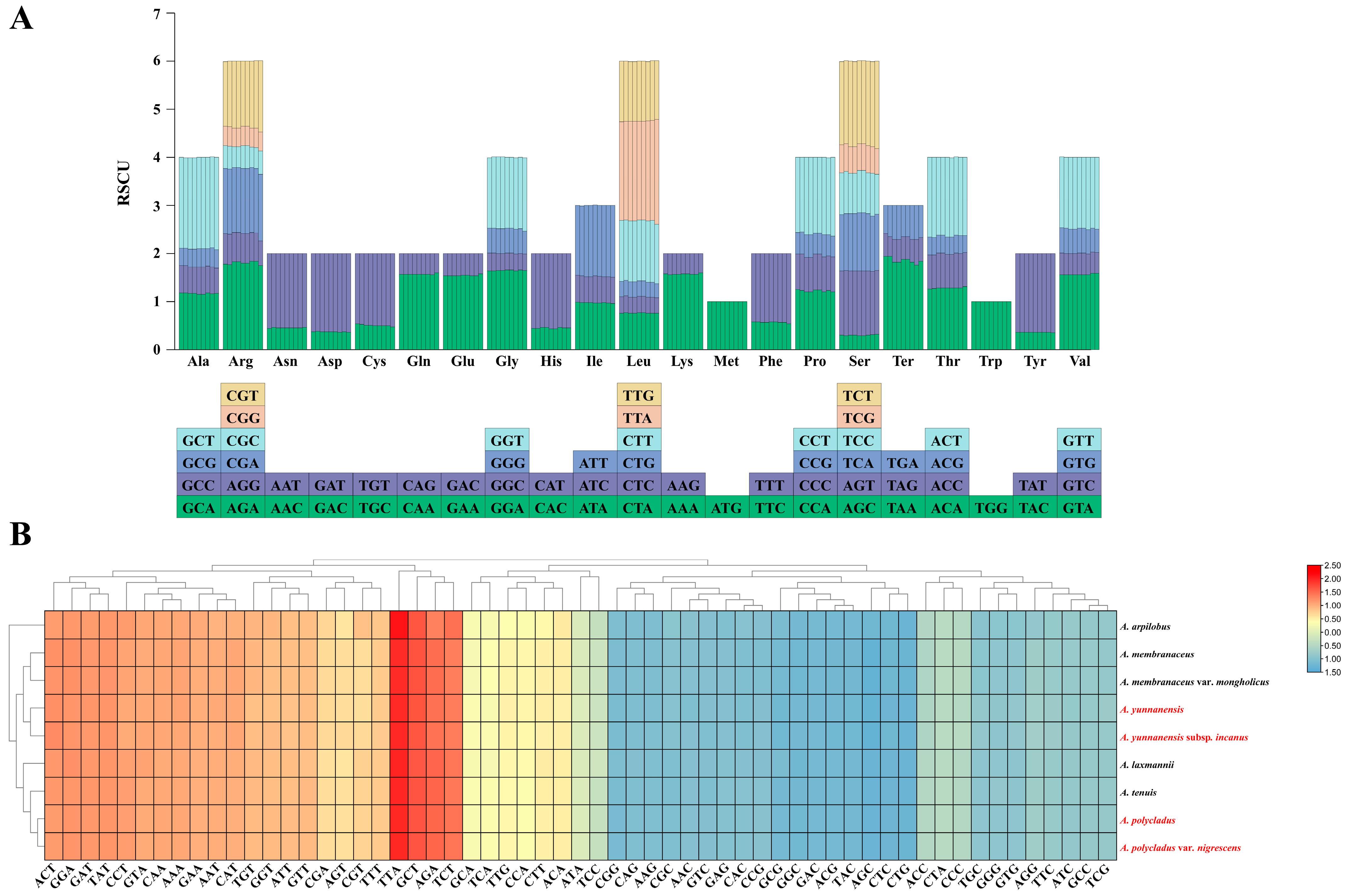
3.2. Codon Usage Analysis
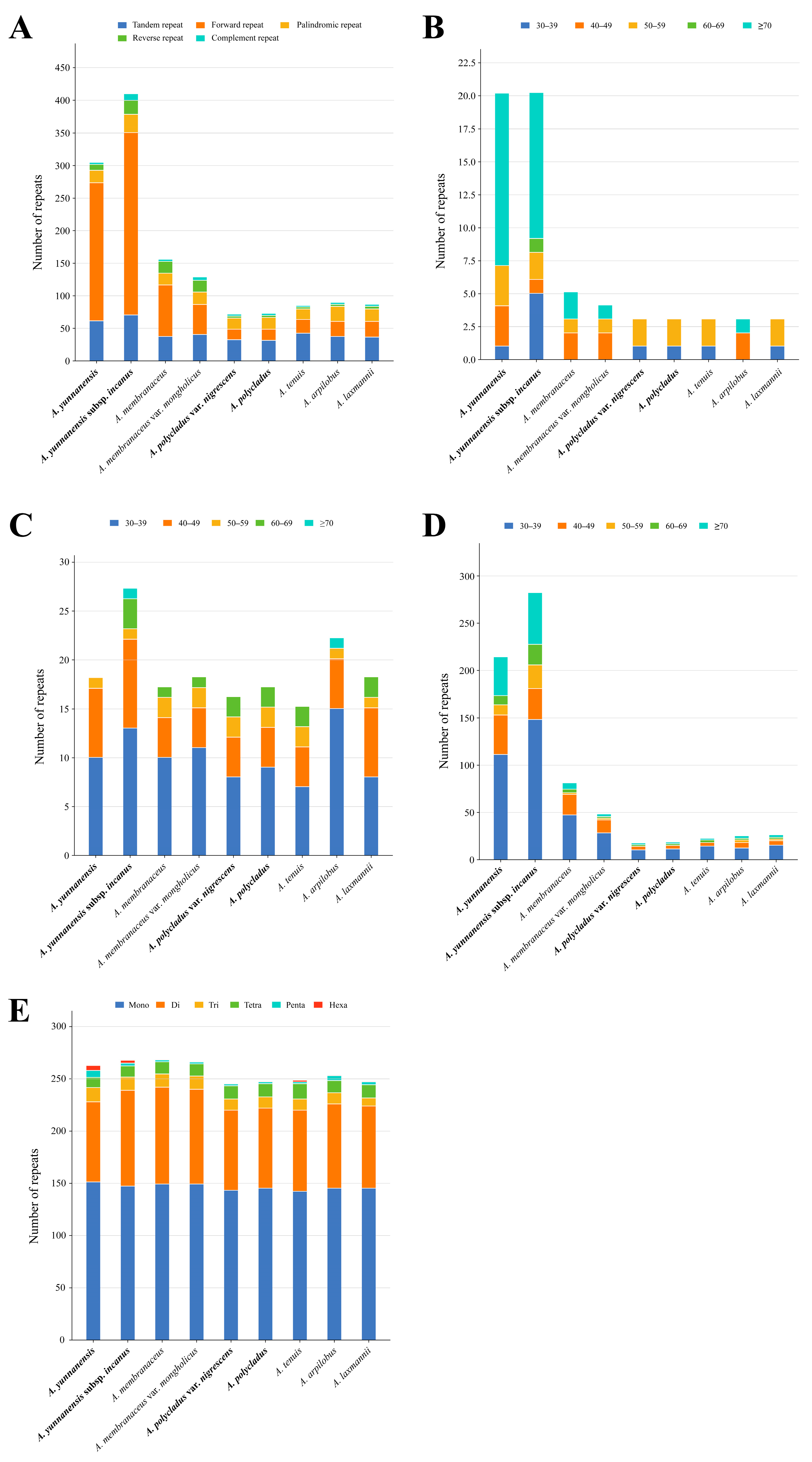
3.3. Repeat Sequence and SSR Analyses
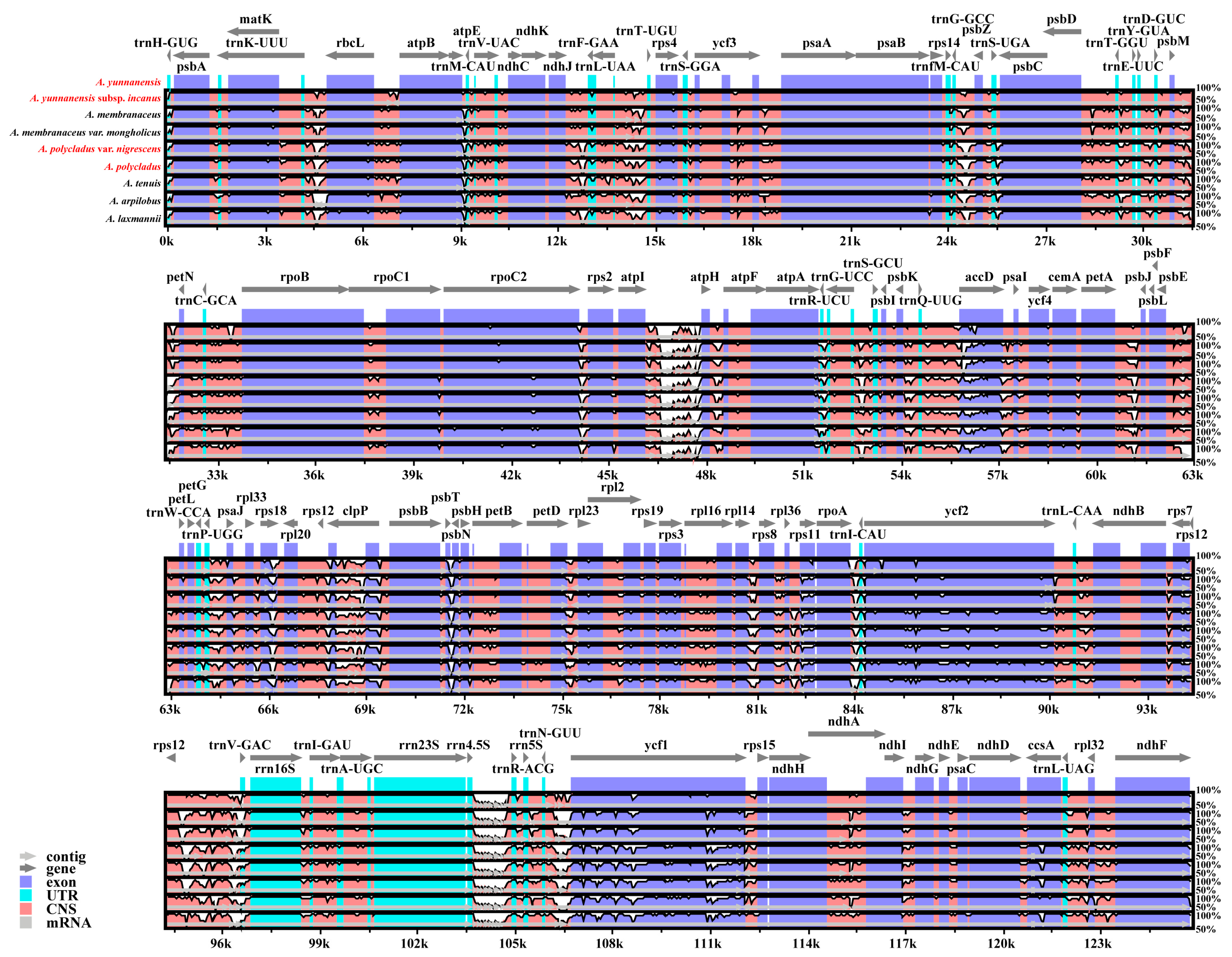
3.4. Sequence Divergence Analysis
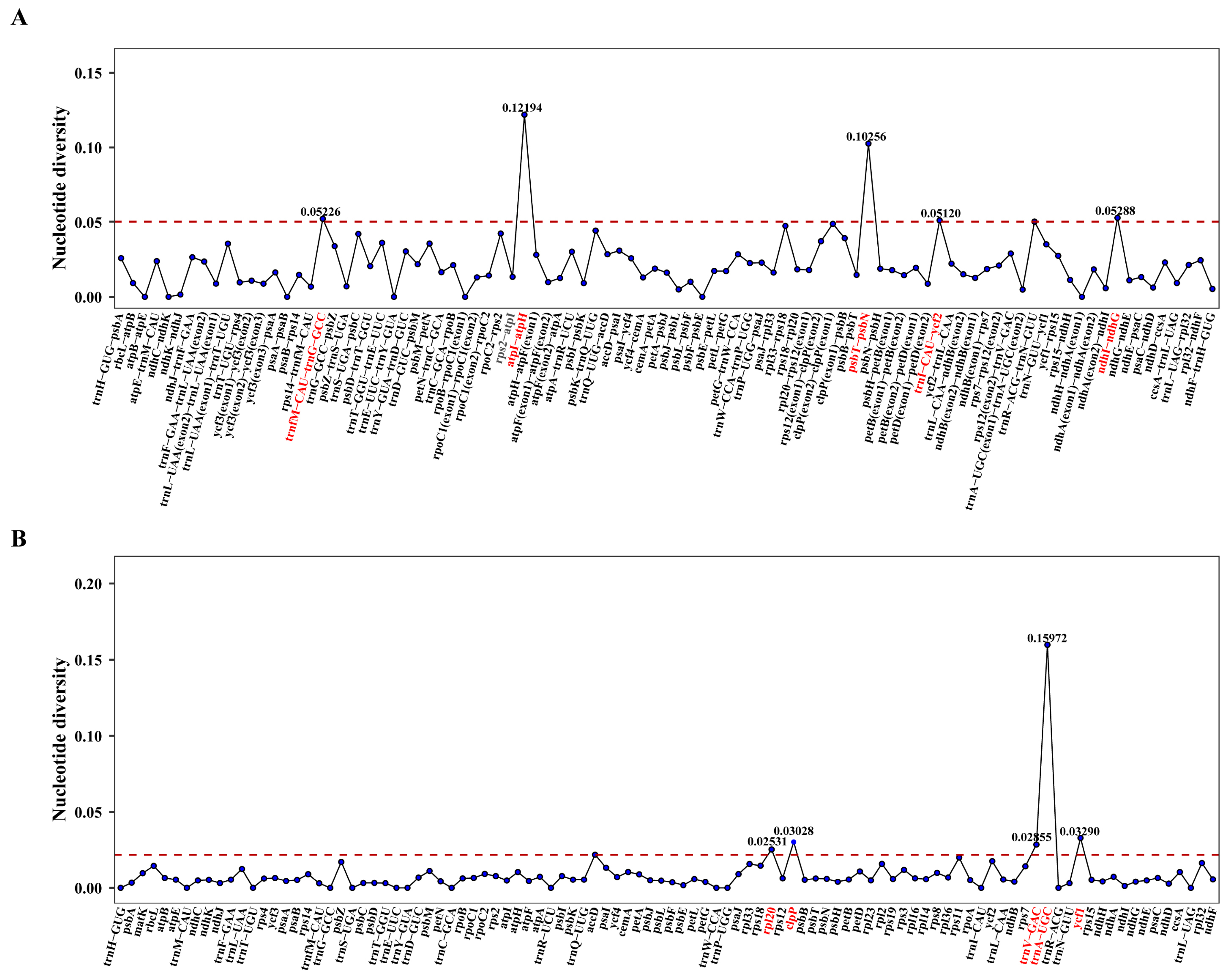
3.5. Nucleic Acid Polymorphism Analysis
3.6. Selective Pressure Analysis
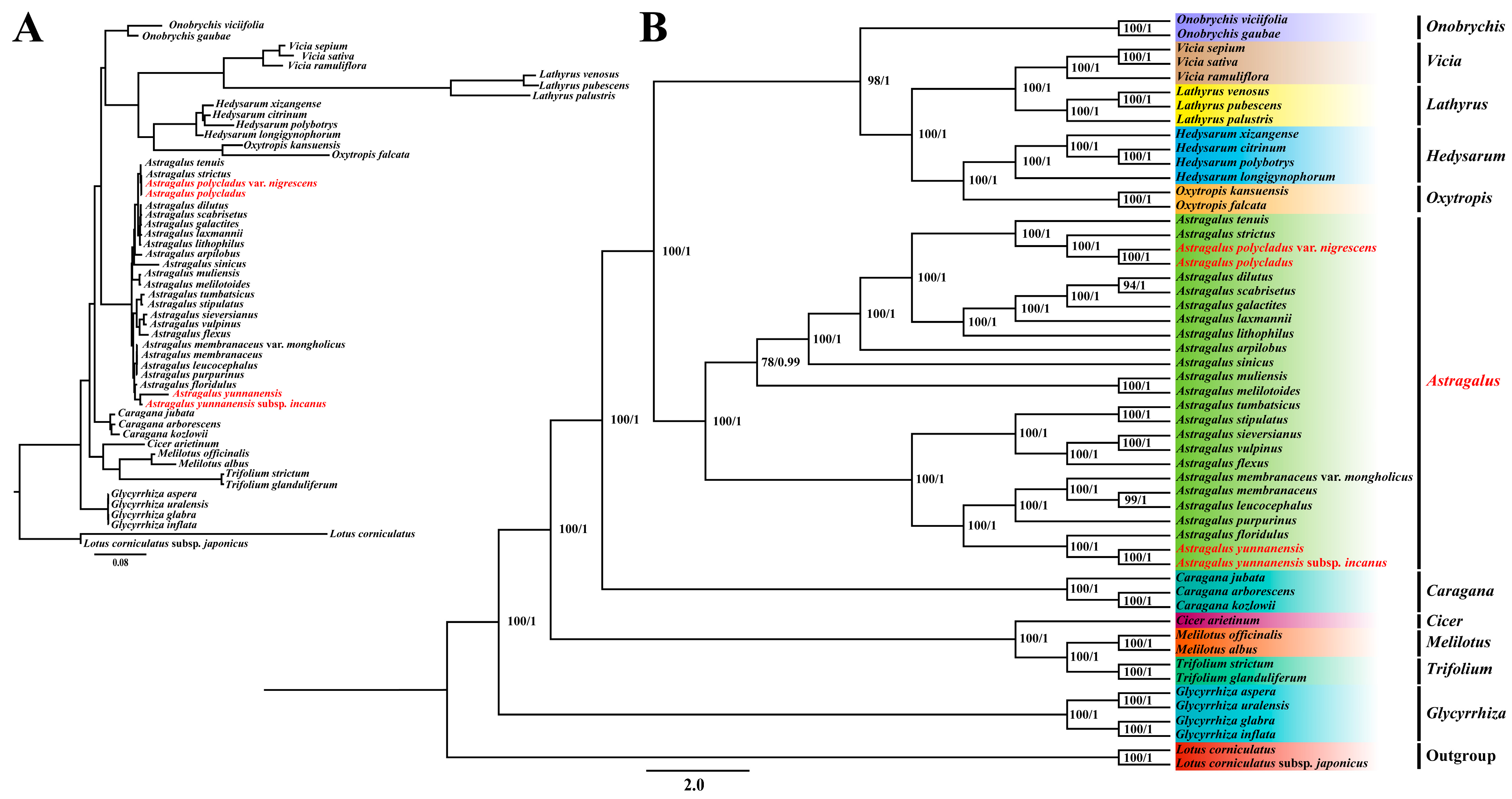
3.7. Phylogenetic Relationship Analysis
4. Discussion
4.1. Chloroplast Genome Structure
4.2. Characteristics of Codon Usage and Repetitive Sequences
4.3. Comparative Genomic Analysis and Nucleotide Diversity
4.4. Analysis of Selection Pressure
4.5. Phylogenetic Relationships of IR-Lacking Clades
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IRs | Inverted repeat regions |
| LSC | Large single-copy region |
| SSC | Small single-copy region |
| SSRs | Simple sequence repeats |
| IRLC | Inverted repeat lacking clade |
| PCGs | Protein-coding genes |
| IGS | Intergenic spacer region |
| Ka | Non-synonymous |
| Ks | Synonymous |
| ML | Maximum Likelihood |
| BI | Bayesian Inference |
References
- Li, X.X.; Qu, L.; Dong, Y.Z.; Han, L.F.; Liu, E.W.; Fang, S.M.; Zhang, Y.; Wang, T. A Review of Recent Research Progress on the Astragalus Genus. Molecules 2014, 19, 18850–18880. [Google Scholar] [CrossRef]
- Bagheri, A.; Maassoumi, A.A.; Rahiminejad, M.R.; Brassac, J.; Blattner, F.R. Molecular Phylogeny and Divergence Times of Astragalus Section Hymenostegis: An Analysis of a Rapidly Diversifying Species Group in Fabaceae. Sci. Rep. 2017, 7, 14033. [Google Scholar] [CrossRef]
- Xu, L.R.; Podlech, D. Astragalus L. In Flora of China: Fabaceae; Miss. Bot. Gard.: St. Louis, MO, USA; Science Press: Beijing, China, 2010; Volume 10, pp. 328–453. [Google Scholar]
- Wojciechowski, M.F.; Sanderson, M.J.; Hu, J.M. Evidence on the Monophyly of Astragalus (Fabaceae) and Its Major Subgroups Based on Nuclear Ribosomal DNA ITS and Chloroplast DNA trnL Intron Dat. Syst. Bot. 1999, 24, 409–437. [Google Scholar] [CrossRef]
- Maassoumi, A.A. A Checklist of Astragalus in the World: New Grouping, New Changes, and Additional Species with Augmented Data; Research Institute of Forests and Rangelands: Tehran, Iran, 2022; pp. 1–563. [Google Scholar]
- Lin, L.Z.; He, X.G.; Lindenmaier, M.; Nolan, G.; Yang, J.; Cleary, M.; Qiu, S.X.; Cordell, G.A. Liquid Chromatography-Electrospray Ionization Mass Spectrometry Study of the Flavonoids of the Roots of Astragalus mongholicus and A. membranaceus. J. Chromatogr. A 2000, 876, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Wang, Z.H.; Huang, L.F.; Zheng, S.H.; Wang, D.M.; Chen, S.L.; Zhang, H.T.; Yang, S.H. Review of the Botanical Characteristics, Phytochemistry, and Pharmacology of Astragalus membranaceus (Huangqi). Phytother. Res. 2014, 28, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.Z.; Lou, Y.M.; Kong, M.Y.; Luo, Q.; Liu, Z.Q.; Wu, J.J. A Systematic Review of Phytochemistry, Pharmacology and Pharmacokinetics on Astragali Radix: Implications for Astragali Radix as a Personalized Medicine. Int. J. Mol. Sci. 2019, 20, 1463. [Google Scholar] [CrossRef]
- Auyeung, K.K.; Han, Q.B.; Ko, J.K. Astragalus membranaceus: A Review of Its Protection Against Inflammation and Gastrointestinal Cancers. Am. J. Chin. Med. 2016, 44, 1–22. [Google Scholar] [CrossRef]
- Durazzo, A.; Nazhand, A.; Lucarini, M.; Amelia, M.S.; Selma, B.S.; Fabrizia, G.; Patricia, S.; Massimo, Z.; Eliana, B.S.; Antonello, S. Astragalus (Astragalus membranaceus Bunge): Botanical, Geographical, and Historical Aspects to Pharmaceutical Components and Beneficial Role. Rend. Fis. Acc. Lincei 2021, 32, 625–642. [Google Scholar] [CrossRef]
- Sheik, A.; Kim, K.; Varaprasad, G.L.; Lee, H.; Kim, S.; Kim, E.; Shin, J.Y.; Oh, S.Y.; Huh, Y.S. The Anti-Cancerous Activity of Adaptogenic Herb Astragalus membranaceus. Phytomedicine 2021, 91, 153698. [Google Scholar] [CrossRef]
- Zarre, S.; Azani, N. Perspectives in Taxonomy and Phylogeny of the Genus Astragalus (Fabaceae): A Review. Prog. Biol. Sci. 2013, 3. [Google Scholar]
- Osaloo, S.; Maassoumi, A.; Murakami, N. Molecular Systematics of the Genus Astragalus L. (Fabaceae): Phylogenetic Analyses of Nuclear Ribosomal DNA Internal Transcribed Spacers and Chloroplast Gene ndhF Sequences. Plant Syst. Evol. 2003, 242, 1–32. [Google Scholar] [CrossRef]
- Lin, C.P.; Huang, J.P.; Wu, C.S.; Hsu, C.Y.; Chaw, S.M. Comparative Chloroplast Genomics Reveals the Evolution of Pinaceae Genera and Subfamilies. Genome Biol. Evol. 2010, 2, 504–517. [Google Scholar] [CrossRef]
- Zha, X.; Wang, X.Y.; Li, J.R.; Gao, F.; Zhou, Y.J. Complete Chloroplast Genome of Sophora alopecuroides (Papilionoideae): Molecular Structures, Comparative Genome Analysis and Phylogenetic Analysis. J. Genet. 2020, 99, 13. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.W.; Cai, M.X.; Yan, G.X.; Wang, N.; Li, F.; Chen, B.Y.; Gao, G.Z.; Xu, K.; Li, J.; Wu, X.M. High-Throughput Multiplex cpDNA Resequencing Clarifies the Genetic Diversity and Genetic Relationships among Brassica napus, Brassica rapa and Brassica oleracea. Plant Biotechnol. J. 2016, 14, 409–418. [Google Scholar] [CrossRef]
- Compton, J.A.; Schrire, B.D.; Könyves, K.; Forest, F.; Malakasi, P.; Mattapha, S.; Sirichamorn, Y. The Callerya Group Redefined and Tribe Wisterieae (Fabaceae) Emended Based on Morphology and Data from Nuclear and Chloroplast DNA Sequences. PhytoKeys 2019, 125, 1–112. [Google Scholar] [CrossRef]
- Duan, L.; Li, S.J.; Su, C.; Sirichamorn, Y.; Han, L.N.; Ye, W.; Lôc, P.K.; Wen, J.; Compton, J.A.; Schrire, B.; et al. Phylogenomic Framework of the IRLC Legumes (Leguminosae Subfamily Papilionoideae) and Intercontinental Biogeography of Tribe Wisterieae. Mol. Phylogenet. Evol. 2021, 163, 107235. [Google Scholar] [CrossRef]
- Palmer, J.D.; Thompson, W.F. Chloroplast DNA Rearrangements Are More Frequent When a Large Inverted Repeat Sequence Is Lost. Cell 1982, 29, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Lavin, M.; Doyle, J.J.; Palmer, J.D. Evolutionary Significance of the Loss of the Chloroplast-DNA Inverted Repeat in the Leguminosae Subfamily Papilionoideae. Evolution 1990, 44, 390–402. [Google Scholar] [CrossRef]
- Jansen, R.K.; Wojciechowski, M.F.; Sanniyasi, E.; Lee, S.B.; Daniell, H. Complete Plastid Genome Sequence of the Chickpea (Cicer arietinum) and the Phylogenetic Distribution of rps12 and clpP Intron Losses among Legumes (Leguminosae). Mol. Phylogenet. Evol. 2008, 48, 1204–1217. [Google Scholar] [CrossRef]
- Wojciechowski, M.F.; Lavin, M.; Sanderson, M.J. A Phylogeny of Legumes (Leguminosae) Based on Analysis of the Plastid matK Gene Resolves Many Well-Supported Subclades within the Family. Am. J. Bot. 2004, 91, 1846–1862. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L.; Ballenger, J.A.; Palmer, J.D. The Distribution and Phylogenetic Significance of a 50-Kb Chloroplast DNA Inversion in the Flowering Plant Family Leguminosae. Mol. Phylogenet. Evol. 1996, 5, 429–438. [Google Scholar] [CrossRef]
- Millen, R.S.; Olmstead, R.G.; Adams, K.L.; Palmer, J.D.; Lao, N.T.; Heggie, L.; Kavanagh, T.A.; Hibberd, J.M.; Gray, J.C.; Morden, C.W.; et al. Many Parallel Losses of infA from Chloroplast DNA during Angiosperm Evolution with Multiple Independent Transfers to the Nucleus. Plant Cell 2001, 13, 645–658. [Google Scholar] [CrossRef]
- Doyle, J.J. DNA Data and Legume Phylogeny: A Progress Report. In Phylogeny; Crisp, M.D., Ed.; Royal Botanic Gardens: London, UK, 1995; pp. 11–30. [Google Scholar]
- Doyle, J.J.; Doyle, J.L.; Palmer, J.D. Multiple Independent Losses of Two Genes and One Intron from Legume Chloroplast Genomes. Syst. Bot. 1995, 20, 272–294. [Google Scholar] [CrossRef]
- Wojciechowski, M.F.; Sanderson, M.J.; Baldwin, B.G.; Donoghue, M.J. Monophyly of Aneuploid Astragalus (Fabaceae): Evidence from Nuclear Ribosomal DNA Internal Transcribed Spacer Sequences. Am. J. Bot. 1993, 80, 711–722, Erratum in Am. J. Bot. 1993, 80, 1099. [Google Scholar] [CrossRef]
- Yang, S.P.; Ma, J.H. Progress in Molecular Pharmacognosy Research on Astragalus. Tradit. Med. Asia-Pac. 2024, 20, 244–251. [Google Scholar]
- Pahlich, E.; Gerlitz, C. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue Semantic Scholar. Phytochemistry 1980, 19, 11–13. [Google Scholar] [CrossRef]
- Chen, S.F. Ultrafast One-Pass FASTQ Data Preprocessing, Quality Control, and Deduplication Using Fastp. Imeta 2023, 2, e107. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A Fast and Versatile Toolkit for Accurate de Novo Assembly of Organelle Genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Shi, L.C.; Chen, H.M.; Jiang, M.; Wang, L.Q.; Wu, X.; Huang, L.F.; Liu, C. CPGAVAS2, an Integrated Plastome Sequence Annotator and Analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Ralph, B. OrganellarGenomeDRAW (OGDRAW) Version 1.3.1: Expanded Toolkit for the Graphical Visualization of Organellar Genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High Speed BLASTN: An Accelerated MegaBLAST Search Tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef]
- Liu, S.Y.; Ni, Y.; Li, J.L.; Zhang, X.Y.; Yang, H.Y.; Chen, H.M.; Liu, C. CPGView: A Package for Visualizing Detailed Chloroplast Genome Structures. Mol. Ecol. Resour. 2023, 23, 694–704. [Google Scholar] [CrossRef]
- Lewis, S.E.; Searle, S.M.J.; Harris, N.; Gibson, M.; Lyer, V.; Richter, J.; Wiel, C.; Bayraktaroglu, L.; Birney, E.; Crosby, M.A.; et al. Apollo: A Sequence Annotation Editor. Genome Biol. 2002, 3, RESEARCH0082. [Google Scholar] [CrossRef]
- Sharp, P.M.; Li, W.H. An Evolutionary Perspective on Synonymous Codon Usage in Unicellular Organisms. J. Mol. Evol. 1986, 24, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The Manifold Applications of Repeat Analysis on a Genomic Scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Faircloth, B.C. Msatcommander: Detection of Microsatellite Repeat Arrays and Automated, Locus-Specific Primer Design. Mol. Ecol. Resour. 2008, 8, 92–94. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational Tools for Comparative Genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through Model Selection and Model Averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.L.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An Integrated and Scalable Desktop Platform for Streamlined Molecular Sequence Data Management and Evolutionary Phylogenetics Studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v 1.4.4; University of Edinburgh: Edinburgh, UK, 2018. [Google Scholar]
- Cook, D.; Gardner, D.R.; Pfister, J.A.; Lee, S.T.; Welch, K.D.; Welsh, S.L. A Screen for Swainsonine in Select North American Astragalus Species. Chem. Biodivers. 2017, 14, e1600364. [Google Scholar] [CrossRef]
- Tian, C.Y.; Li, X.S.; Wu, Z.N.; Li, Z.Y.; Hou, X.Y.; Li, F.Y.H. Characterization and Comparative Analysis of Complete Chloroplast Genomes of Three Species From the Genus Astragalus (Leguminosae). Front. Genet. 2021, 12, 705482. [Google Scholar] [CrossRef]
- Moghaddam, M.; Wojciechowski, M.F.; Kazempour-Osaloo, S. Characterization and Comparative Analysis of the Complete Plastid Genomes of Four Astragalus Species. PLoS ONE 2023, 18, e0286083. [Google Scholar] [CrossRef] [PubMed]
- Bonnefoy, E. The Ribosomal S16 Protein of Escherichia Coli Displaying a DNA-Nicking Activity Binds to Cruciform DNA. Eur. J. Biochem. 1997, 247, 852–859. [Google Scholar] [CrossRef]
- Gantt, J.S.; Baldauf, S.L.; Calie, P.J.; Weeden, N.F.; Palmer, J.D. Transfer of rpl22 to the Nucleus Greatly Preceded Its Loss from the Chloroplast and Involved the Gain of an Intron. EMBO J. 1991, 10, 3073–3078. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Fujimoto, M.; Arimura, S.I.; Tsutsumi, N.; Kadowaki, K.I. Presence of a Latent Mitochondrial Targeting Signal in Gene on Mitochondrial Genome. Mol. Biol. Evol. 2008, 25, 1791–1793. [Google Scholar] [CrossRef]
- Jia, J.; Xue, Q. Codon Usage Biases of Transposable Elements and Host Nuclear Genes in Arabidopsis thaliana and Oryza sativa. Genom. Proteom. Bioinform. 2009, 7, 175–184. [Google Scholar] [CrossRef]
- Leffler, E.M.; Bullaughey, K.; Matute, D.R.; Meyer, W.K.; Ségurel, L.; Venkat, A.; Andolfatto, P.; Przeworski, M. Revisiting an Old Riddle: What Determines Genetic Diversity Levels within Species? PLoS Biol. 2012, 10, e1001388. [Google Scholar] [CrossRef]
- Li, C.J.; Wang, R.N.; Li, D.Z. Comparative Analysis of Plastid Genomes within the Campanulaceae and Phylogenetic Implications. PLoS ONE 2020, 15, e0233167. [Google Scholar] [CrossRef]
- Xu, C.; Cai, X.N.; Chen, Q.Z.; Zhou, H.X.; Cai, Y.; Ben, A. Factors Affecting Synonymous Codon Usage Bias in Chloroplast Genome of Oncidium Gower Ramsey. Evol. Bioinform. 2011, 7, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Mawal, P.; Sharma, V.; Gupta, R.C. Analysis of Genetic Diversity and Population Structure in Asparagus Species Using SSR Markers. J. Genet. Eng. Biotechnol. 2020, 18, 50. [Google Scholar] [CrossRef]
- Souza, U.J.B.d.; Nunes, R.; Targueta, C.P.; Diniz-Filho, J.A.F.; Telles, M.P.d.C. The Complete Chloroplast Genome of Stryphnodendron adstringens (Leguminosae—Caesalpinioideae): Comparative Analysis with Related Mimosoid Species. Sci. Rep. 2019, 9, 14206. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.F.; Zang, M.Y.; Li, M.Z.; Fang, Y.M. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Quercus acutissima. Int. J. Mol. Sci. 2018, 19, 2443. [Google Scholar] [CrossRef]
- Xiong, Y.L.; Xiong, Y.; He, J.; Yu, Q.Q.; Zhao, J.M.; Lei, X.; Dong, Z.X.; Yang, J.; Peng, Y.; Zhang, X.Q.; et al. The Complete Chloroplast Genome of Two Important Annual Clover Species, Trifolium alexandrinum and T. resupinatum: Genome Structure, Comparative Analyses and Phylogenetic Relationships with Relatives in Leguminosae. Plants 2020, 9, 478. [Google Scholar] [CrossRef]
- Kimura, M. The Neutral Theory of Molecular Evolution. Sci. Am. 1979, 241, 98–100, 102, 108 passim. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, S.; He, F.; Zhu, J.; Hu, S.; Yu, J. How Do Variable Substitution Rates Influence Ka and Ks Calculations? Genom. Proteom. Bioinform. 2009, 7, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Zong, D.; Zhou, A.; Zhang, Y.; Zou, X.; Li, D.; Duan, A.; He, C. Characterization of the Complete Chloroplast Genomes of Five Populus Species from the Western Sichuan Plateau, Southwest China: Comparative and Phylogenetic Analyses. PeerJ 2019, 7, e6386. [Google Scholar] [CrossRef]
- Gu, X.L.; Li, L.L.; Li, S.C.; Shi, W.X.; Zhong, X.N.; Su, Y.J.; Wang, T. Adaptive Evolution and Co-Evolution of Chloroplast Genomes in Pteridaceae Species Occupying Different Habitats: Overlapping Residues Are Always Highly Mutated. BMC Plant Biol. 2023, 23, 511. [Google Scholar] [CrossRef]
- Wen, J.; Zhu, J.W.; Ma, X.D.; Li, H.M.; Wu, B.C.; Zhou, W.; Yang, J.X.; Song, C.F. Phylogenomics and Adaptive Evolution of Hydrophytic Umbellifers (Tribe Oenantheae, Apioideae) Revealed from Chloroplast Genomes. BMC Plant Biol. 2024, 24, 1140. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast Genomes: Diversity, Evolution, and Applications in Genetic Engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Liu, L.E.; Li, H.Y.; Li, J.X.; Li, X.J.; Hu, N.; Sun, J.; Zhou, W. Chloroplast Genomes of Caragana tibetica and Caragana turkestanica: Structures and Comparative Analysis. BMC Plant Biol. 2024, 24, 254. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An Empirical Test of Bootstrapping as a Method for Assessing Confidence in Phylogenetic Analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Accuracies and Statistical Tests of Phylogenetic Trees. In Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000; pp. 162–186. [Google Scholar]
- Clark, J.W.; Hetherington, A.J.; Morris, J.L.; Pressel, S.; Duckett, J.G.; Puttick, M.N.; Schneider, H.; Kenrick, P.; Wellman, C.H.; Donoghue, P.C.J. Evolution of Phenotypic Disparity in the Plant Kingdom. Nat. Plants 2023, 9, 1618–1626. [Google Scholar] [CrossRef]
- Han, G.L.; Li, Y.X.; Yang, Z.R.; Wang, C.F.; Zhang, Y.Y.; Wang, B.S. Molecular Mechanisms of Plant Trichome Development. Front. Plant Sci. 2022, 13, 910228. [Google Scholar] [CrossRef]
- Wu, R.; Cun, S.; Gao, Y.Q.; Ma, R.; Zhang, L.; Lev-Yadun, S.; Sun, H.; Song, B. Distribution Patterns of Glandular Trichomes in the Flora of the Hengduan Mountains, Southwestern China. Bot. J. Linn. Soc. 2025, 207, 83–94. [Google Scholar] [CrossRef]
- Wang, X.J.; Shen, C.; Meng, P.H.; Tan, G.F.; Lv, L.T. Analysis and Review of Trichomes in Plants. BMC Plant Biol. 2021, 21, 70. [Google Scholar] [CrossRef]
- Maes, L.; Goossens, A. Hormone-Mediated Promotion of Trichome Initiation in Plants Is Conserved but Utilizes Species and Trichome-Specific Regulatory Mechanisms. Plant Signal. Behav. 2010, 5, 205–207. [Google Scholar] [CrossRef] [PubMed]
- Matías-Hernández, L.; Aguilar-Jaramillo, A.E.; Cigliano, R.A.; Sanseverino, W.; Pelaz, S. Flowering and Trichome Development Share Hormonal and Transcription Factor Regulation. J. Exp. Bot. 2016, 67, 1209–1219. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, P.; Bai, W.H.; Deng, Y.; Cheng, Z.K.; Su, L.W.; Nong, L.F.; Liu, T.; Yang, W.R.; Yang, X.P.; et al. Quantitative Trait Loci Sequencing and Genetic Mapping Reveal Two Main Regulatory Genes for Stem Color in Wax Gourds. Plants 2024, 13, 1804. [Google Scholar] [CrossRef]
- Nong, L.F.; Wang, P.; Yang, W.R.; Liu, T.; Su, L.W.; Cheng, Z.K.; Bai, W.H.; Deng, Y.; Chen, Z.H.; Liu, Z.G. Analysis of BhAPRR2 Allele Variation, Chlorophyll Content, and Chloroplast Structure of Different Peel Colour Varieties of Wax Gourd (Benincasa hispida) and Development of Molecular Markers. Euphytica 2023, 219, 107. [Google Scholar] [CrossRef]
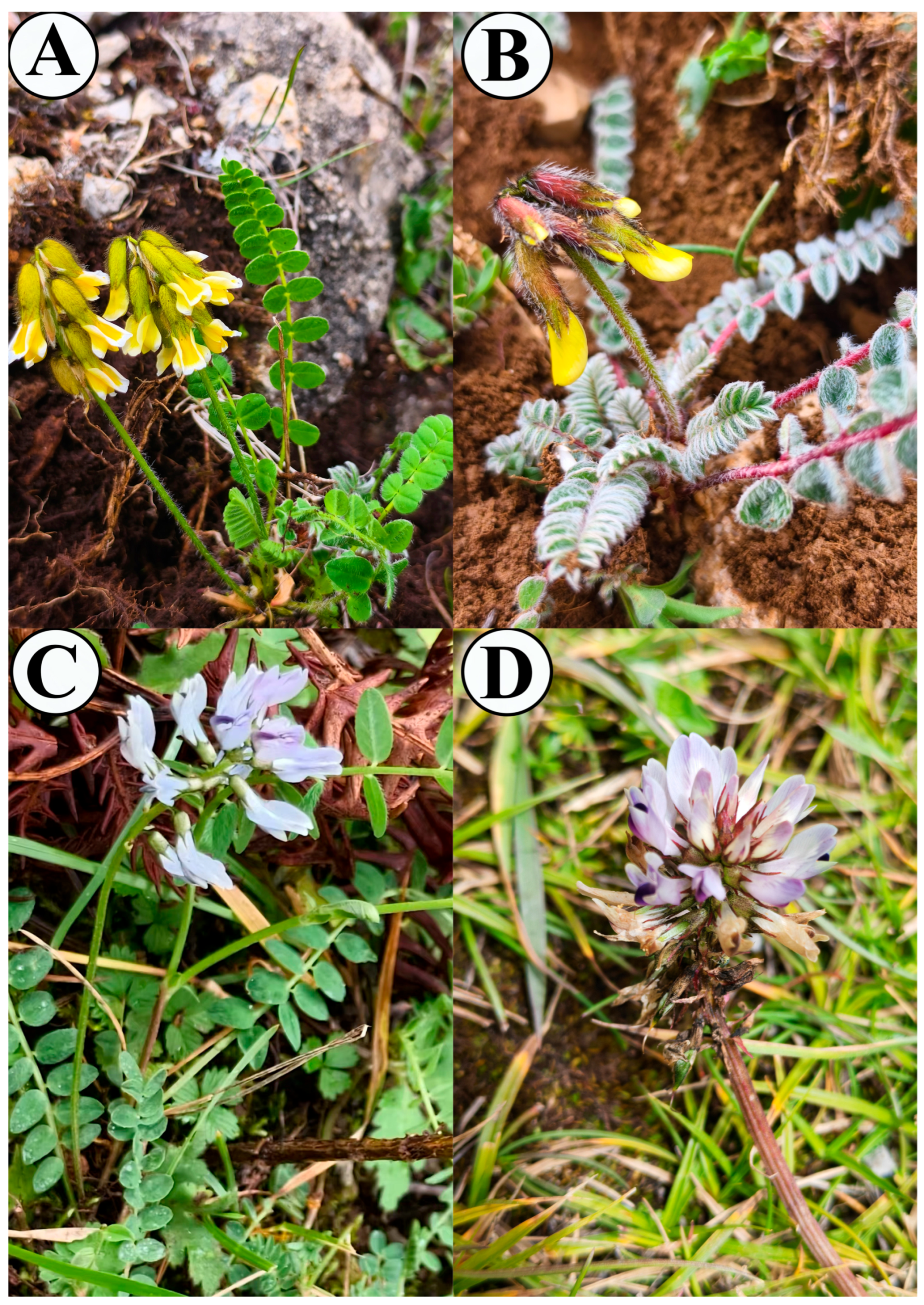
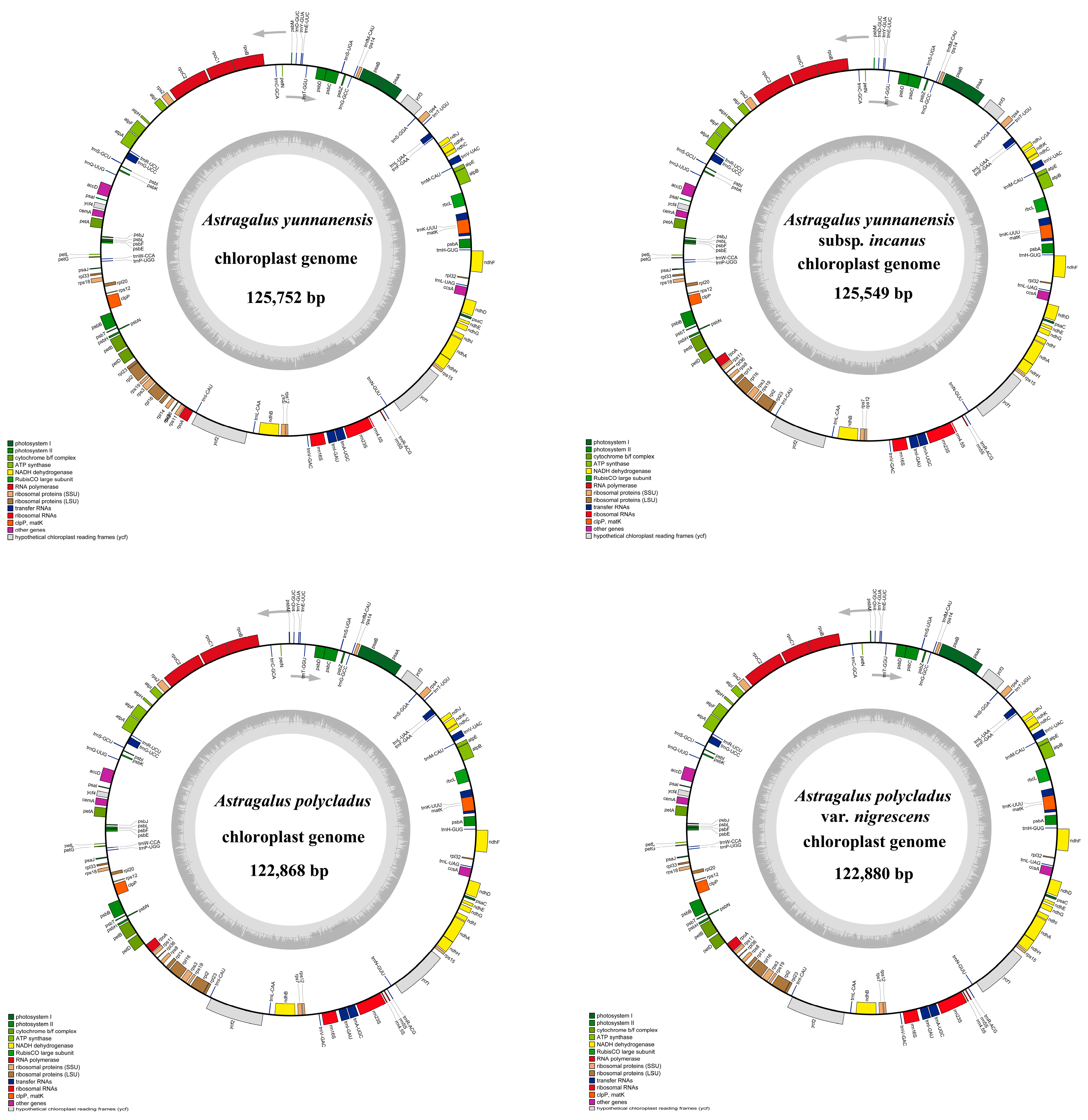

| Plastome Characteristics | A. yunnanensis | A. yunnanensis Subsp. incanus | A. polycladus | A. polycladus var. nigrescens | A. membranaceus | Astragalus membranaceus var. mongholicus | A. tenuis | A. laxmannii | A. arpilobus | |
|---|---|---|---|---|---|---|---|---|---|---|
| GenBank accession | PV156652.1 | PV156653.1 | PV910878 | PV910879 | OR528897.1 | OR712437.1 | OP723862.1 | NC_085710.1 | NC_077549.1 | |
| Protein coding genes (PCG) | Length (bp) | 66,111 | 65,994 | 65,916 | 65,916 | 65,745 | 65,742 | 65,922 | 65,922 | 65,892 |
| GC (%) | 36.53 | 36.54 | 36.43 | 36.44 | 36.53 | 36.52 | 36.42 | 36.43 | 36.36 | |
| Length (%) | 52.57 | 52.56 | 53.65 | 53.64 | 53.21 | 53.3 | 53.59 | 53.68 | 53.55 | |
| Number | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | 76 | |
| tRNA | Length (bp) | 2268 | 2268 | 2296 | 2296 | 2066 | 2194 | 2309 | 1999 | 2269 |
| GC (%) | 52.51 | 52.82 | 52.7 | 52.7 | 51.84 | 52.64 | 51.41 | 51.98 | 52.67 | |
| Length (%) | 1.8 | 1.81 | 1.87 | 1.87 | 1.67 | 1.78 | 1.88 | 1.63 | 1.84 | |
| Number | 30 | 30 | 30 | 30 | 27 | 29 | 30 | 26 | 30 | |
| rRNA | Length (bp) | 4507 | 4509 | 4509 | 4509 | 4509 | 4512 | 4509 | 4509 | 4512 |
| GC (%) | 54.14 | 54.18 | 54.25 | 54.25 | 54.22 | 54.26 | 54.22 | 54.2 | 54.12 | |
| Length (%) | 3.58 | 3.59 | 3.67 | 3.67 | 3.65 | 3.66 | 3.67 | 3.67 | 3.67 | |
| Number | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | |
| Total | Length (bp) | 125,752 | 125,549 | 122,868 | 122,880 | 123,548 | 123,349 | 123,012 | 122,796 | 123,054 |
| Number of genes | 110 | 110 | 110 | 110 | 107 | 109 | 110 | 106 | 110 | |
| GC (%) | 34.22 | 34.18 | 34.14 | 34.15 | 34.07 | 34.1 | 34.1 | 34.11 | 33.97 |
| Category for Gene | Group of Genes | Name of Genes |
|---|---|---|
| Gene for photosynthesis | Subunits of NADH-dehydrogenase | ndhA *, ndhB *, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of cytochrome b/f complex | petA, petB *, petD *, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | |
| Large subunit of rubisco | rbcL | |
| Self-replication | Small subunit of ribosome | rps2, rps3, rps4, rps7, rps8, rps11, rps12 *, rps14, rps15, rps18, rps19 |
| Large subunit of ribosome | rpl2 *, rpl14, rpl16 *, rpl20, rpl23, rpl32, rpl33, rpl36 | |
| DNA dependent RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | |
| tRNA genes | trnA-UGC *, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC *, trnH-GUG, trnI-CAU, trnI-GAU *, trnK-UUU *, trnL-CAA, trnL-UAA *, trnL-UAG, trnM-CAU, trnN-GUU, trnP-UGG, trnQ-UUG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC, trnV-UAC *, trnW-CCA, trnY-GUA, trnfM-CAU | |
| rRNA genes | rrn4.5S, rrn5S, rrn16S, rrn23S | |
| Other genes | Maturase | matK |
| c-type cytochrom synthesis gene | ccsA | |
| Envelope membrane protein | cemA | |
| Protease | clpP * | |
| Subunit of Acetyl-CoA-carboxylase | accD | |
| Genes of unknown function | Conserved hypothetical chloroplast ORF | ycf1, ycf2, ycf3 **, ycf4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, H.-T.; Chen, Q.-Y.; Rao, J.-H.; Wang, K.-L.; Jiang, B.; Zhang, Y.-Z. A Comprehensive Comparative Analysis and Phylogenetic Investigation of the Chloroplast Genome Sequences in Four Astragalus Species. Curr. Issues Mol. Biol. 2025, 47, 978. https://doi.org/10.3390/cimb47120978
Ma H-T, Chen Q-Y, Rao J-H, Wang K-L, Jiang B, Zhang Y-Z. A Comprehensive Comparative Analysis and Phylogenetic Investigation of the Chloroplast Genome Sequences in Four Astragalus Species. Current Issues in Molecular Biology. 2025; 47(12):978. https://doi.org/10.3390/cimb47120978
Chicago/Turabian StyleMa, Hai-Tao, Qi-Yin Chen, Jie-Hua Rao, Kai-Ling Wang, Bei Jiang, and Yong-Zeng Zhang. 2025. "A Comprehensive Comparative Analysis and Phylogenetic Investigation of the Chloroplast Genome Sequences in Four Astragalus Species" Current Issues in Molecular Biology 47, no. 12: 978. https://doi.org/10.3390/cimb47120978
APA StyleMa, H.-T., Chen, Q.-Y., Rao, J.-H., Wang, K.-L., Jiang, B., & Zhang, Y.-Z. (2025). A Comprehensive Comparative Analysis and Phylogenetic Investigation of the Chloroplast Genome Sequences in Four Astragalus Species. Current Issues in Molecular Biology, 47(12), 978. https://doi.org/10.3390/cimb47120978





