Comparative Multi-Omics Analysis Identifies Shared Transcriptomic Signatures and Therapeutic Targets in Alzheimer’s, Parkinson’s, and Huntington’s Diseases
Abstract
1. Introduction
1.1. Background and Motivation
- Functional enrichment and pathway analysis to identify shared biological processes.
- Construction of protein–protein interaction (PPI) networks and hub gene identification.
- Gene–disease association analysis to contextualize shared genes within known disease frameworks.
- Protein–drug interaction analysis to identify potential therapeutic targets.
1.2. State of the Art
1.3. Aim of This Research
- Identify DEGs for AD, PD, and HD using RNA-seq datasets (GSE53697, GSE68719, and GSE64810).
- Determine overlapping DEGs shared among the three diseases.
- Perform functional enrichment to identify shared pathways and ontologies.
- Construct PPI networks, prioritize hub genes, and explore their biological significance using Cytoscape (version 3.10.3) and CytoHubba (https://apps.cytoscape.org/apps/cytohubba accessed on 15 November 2024).
- Analyze gene–disease and protein–drug interactions to link transcriptomic patterns to functional and therapeutic contexts.
- Present a comprehensive workflow integrating these analyses to provide a unified view of molecular convergence among AD, PD, and HD.
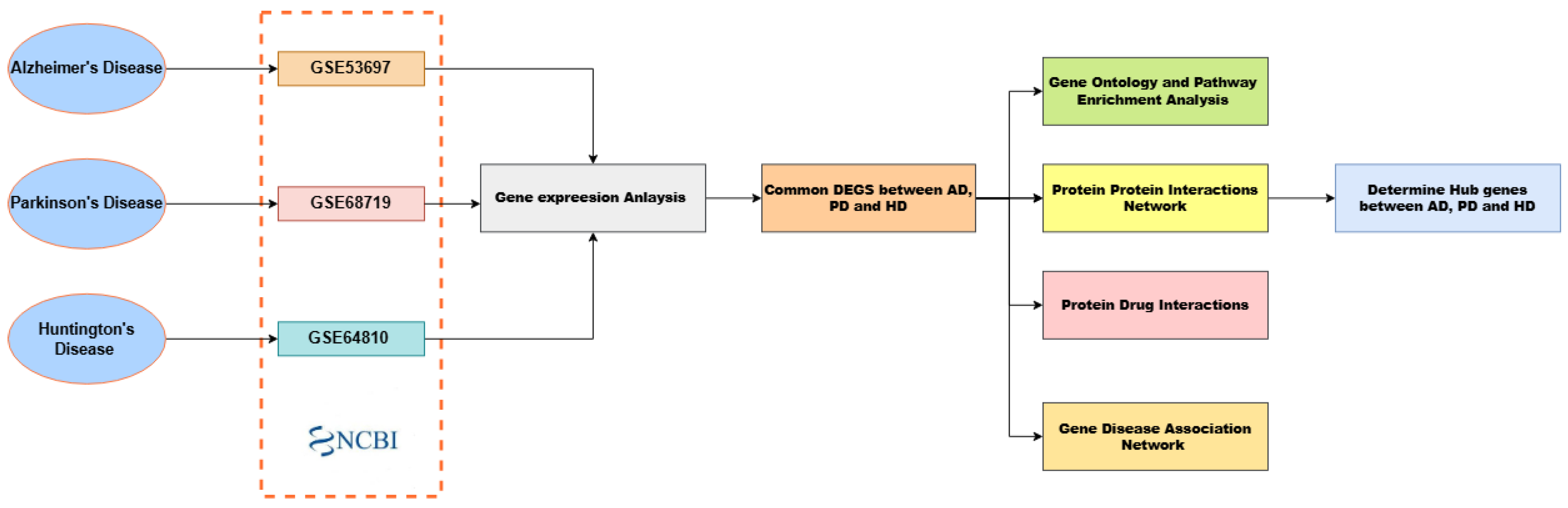
2. Materials and Methods
2.1. Datasets
- AD (GSE53697): This dataset includes 9 control and 8 AD postmortem human brain samples from the Brodmann area 9 (BA9) region. Sequencing was performed using the Illumina HiSeq platform for Homo sapiens [29].
- PD (GSE68719): Contains 44 neurologically healthy and 29 PD postmortem BA9 samples. Differential expression for this dataset was initially assessed using the GEO2R tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/; accessed on 15 August 2024) [30].
- HD (GSE64810): Contributed by Labadorf et al. [31], this dataset includes 49 control and 20 HD postmortem BA9 brain samples.
2.2. Identification of Differentially Expressed Genes (DEGs)
2.3. Functional and Pathway Enrichment Analysis
2.4. Protein–Protein Interaction (PPI) Network Construction
2.5. Protein Drug Interactions Assessment
2.6. Gene Disease Association Analysis
3. Results
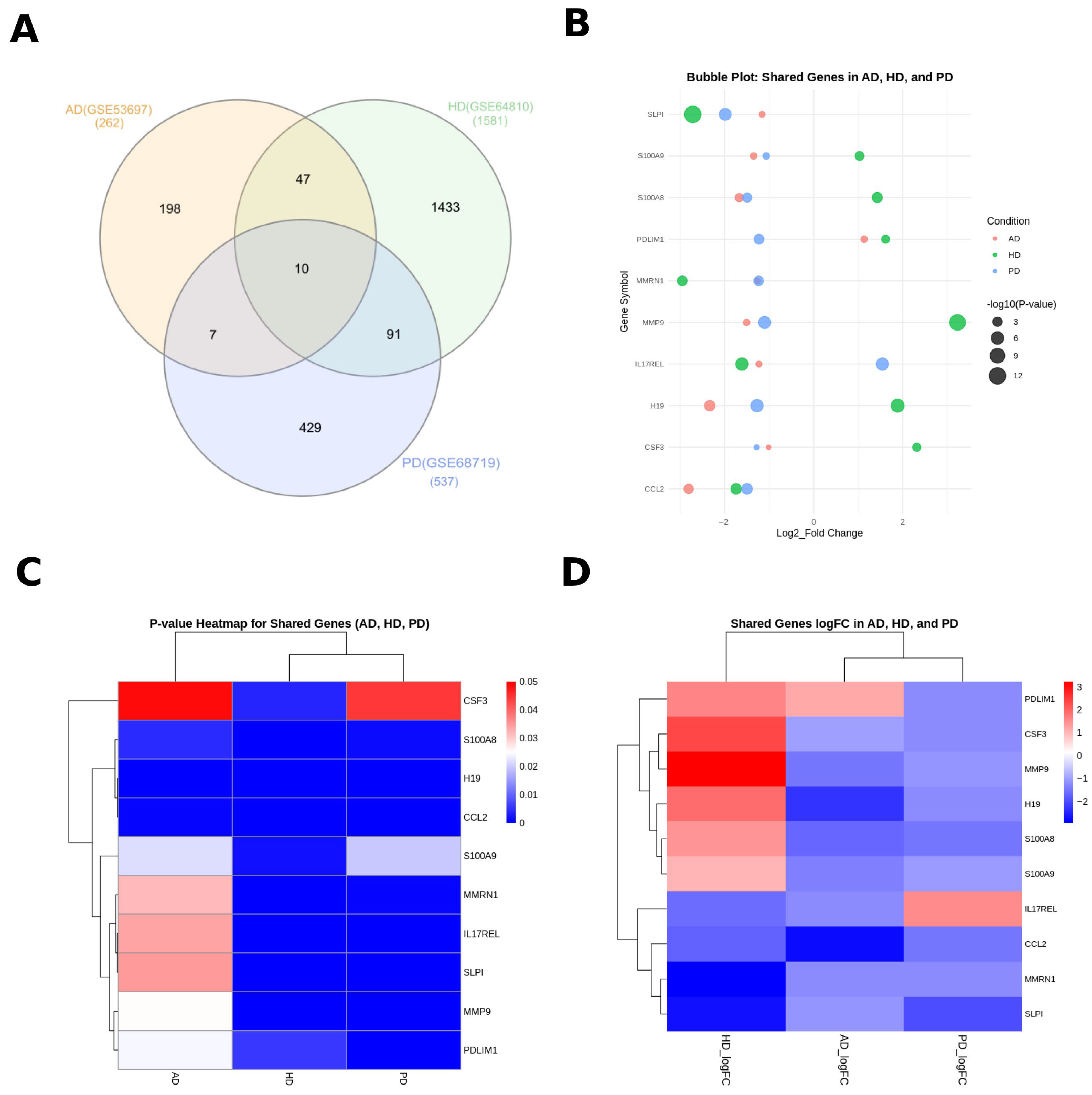
3.1. Identification of Differentially Expressed Genes (DEGs) and Common Signatures Among PD, HD, and AD
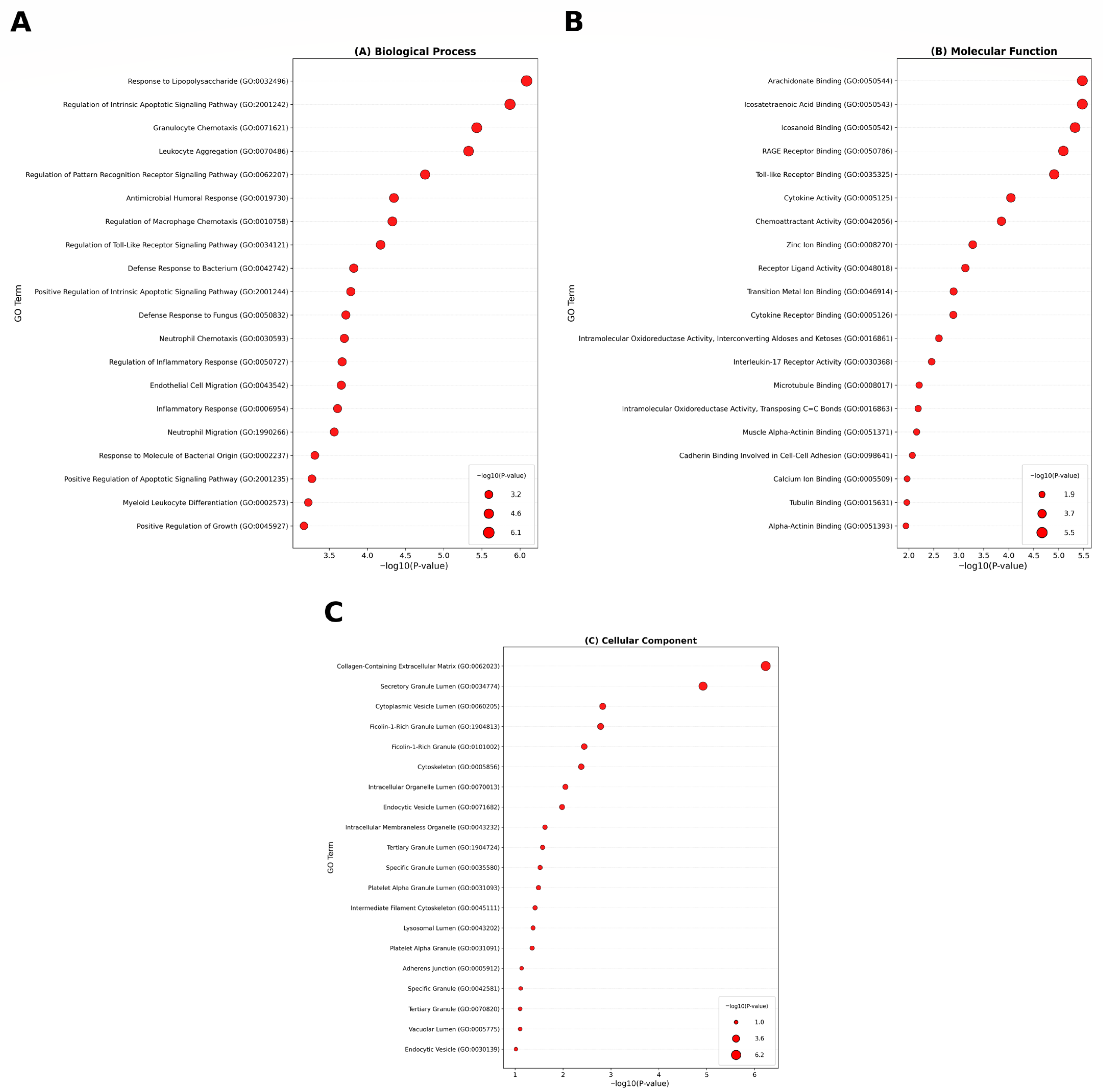
3.2. Functional and Pathway Enrichment Analysis
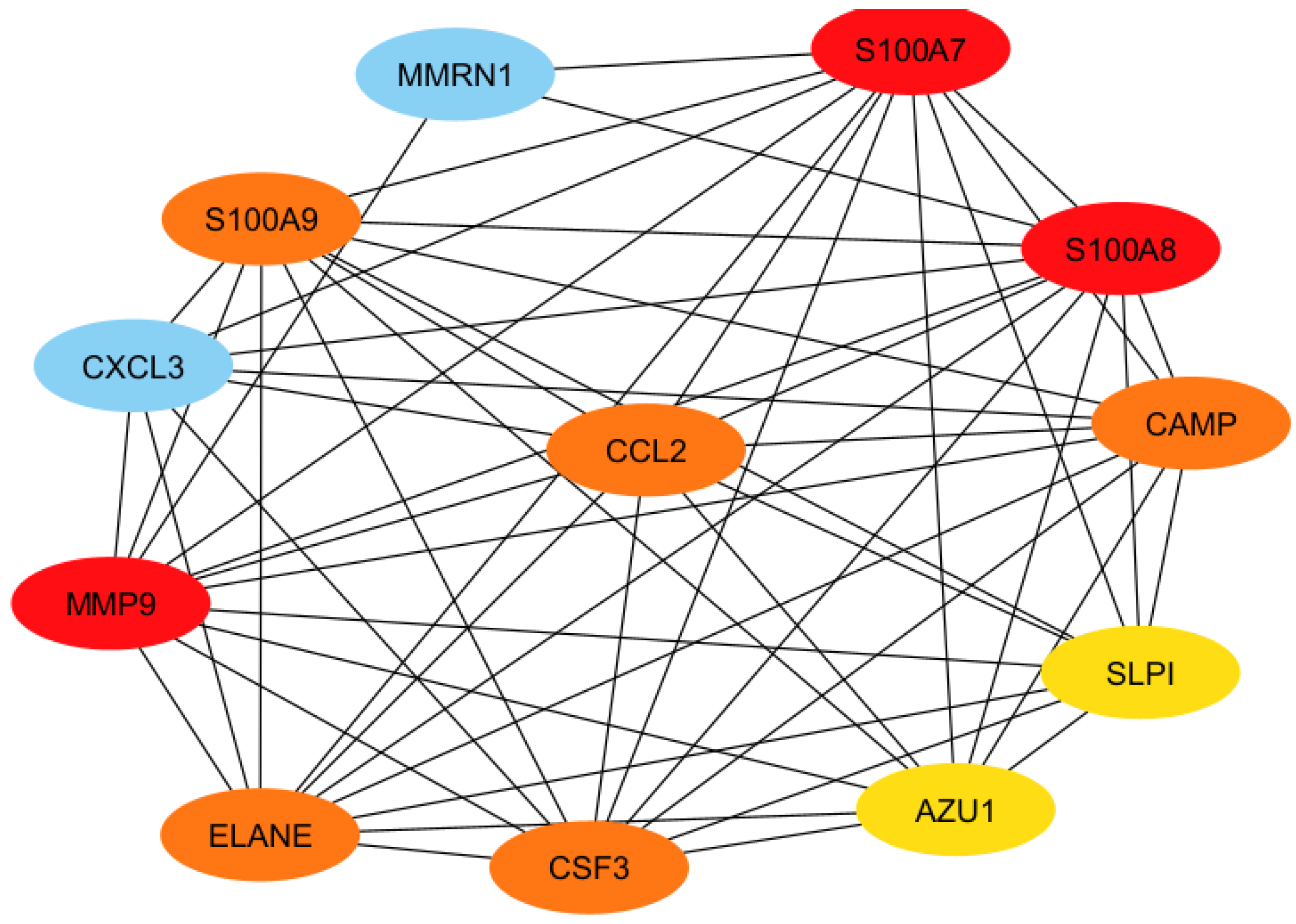
3.3. Protein–Protein Interactions Network and Identification of Hub Proteins
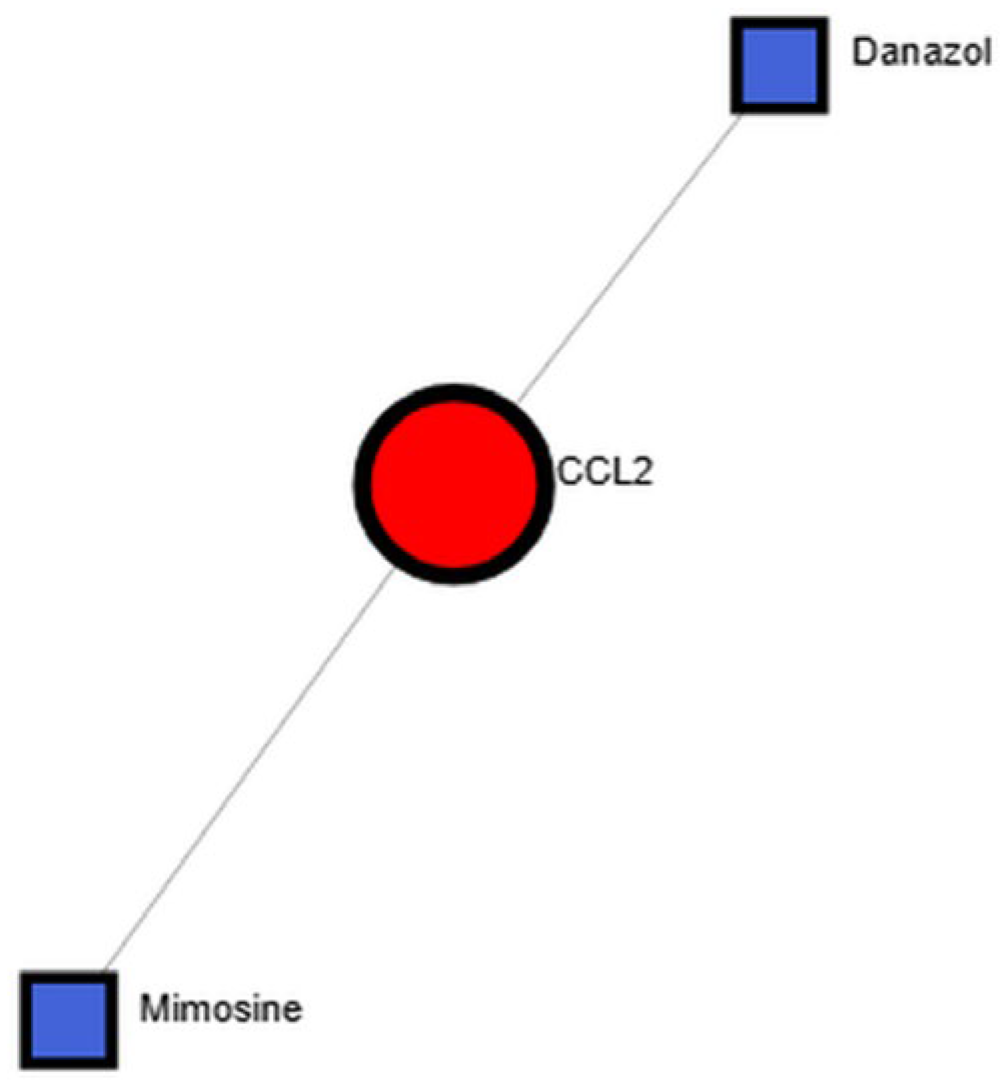
3.4. Identification of Candidate Therapeutic Compounds
3.5. Validation and Functional Interpretation of Hub Genes
3.6. Gene–Disease Association Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alzheimer’s Association. 2019 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2019, 15, 321–387. [Google Scholar] [CrossRef]
- Scheltens, P.; De Strooper, B.; Kivipelto, M.; Holstege, H.; Chételat, G.; Teunissen, C.E.; Cummings, J.; van der Flier, W.M. Alzheimer’s disease. Lancet 2021, 397, 1577–1590. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.H.; Rana, H.K.; Peng, S.; Kibria, M.G.; Islam, M.Z.; Mahmud, S.M.H.; Moni, M.A. Bioinformatics and system biology approaches to identify pathophysiological impact of COVID-19 to the progression and severity of neurological diseases. Comput. Biol. Med. 2021, 138, 104859. [Google Scholar] [CrossRef]
- Ferri, C.P.; Prince, M.; Brayne, C.; Brodaty, H.; Fratiglioni, L.; Ganguli, M.; Hall, K.; Hasegawa, K.; Hendrie, H.; Huang, Y.; et al. Global prevalence of dementia: A Delphi consensus study. Lancet 2005, 366, 2112–2117. [Google Scholar] [CrossRef]
- Ayton, S.; Lei, P.; Bush, A.I. Metallostasis in Alzheimer’s disease. Free Radic. Biol. Med. 2013, 62, 76–89. [Google Scholar] [CrossRef]
- Tagarelli, A.; Tagarelli, G.; Laganà, G.; Condorelli, D.F.; Ferri, C. Alois Alzheimer: A Hundred Years after the Discovery of the Eponymous Disorder. Int. J. Biomed. Sci. 2006, 2, 196–204. [Google Scholar] [CrossRef]
- Zhao, Y.; Xie, Y.Z.; Liu, Y.S. Accelerated aging. Front. Aging Neurosci. 2022, 14, 949074. [Google Scholar]
- Bates, G.P.; Dorsey, R.; Gusella, J.F.; Hayden, M.R.; Kay, C.; Leavitt, B.R.; Nance, M.; Ross, C.A.; Scahill, R.; Wetzel, R.; et al. Huntington disease. Nat. Rev. Dis. Primers 2015, 1, 15005. [Google Scholar] [CrossRef]
- McColgan, P.; Tabrizi, S.J. Huntington’s disease: A clinical review. Eur. J. Neurol. 2018, 25, 24–34. [Google Scholar] [CrossRef]
- Walker, F.O. Huntington’s disease. Lancet 2007, 369, 218–228. [Google Scholar] [CrossRef]
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef] [PubMed]
- Tanner, C.M. Epidemiology of Parkinson’s disease. Neurol. Clin. 1992, 10, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Rl, N. Alzheimer’s disease and Parkinson’s disease. N. Engl. J. Med. 2003, 348, 1356–1364. [Google Scholar]
- Chen, R.C.; Chang, S.F.; Su, C.L.; Chen, T.H.H.; Yen, M.F.; Wu, H.M.; Chen, Z.Y.; Liou, H.H. Prevalence, incidence, and mortality of PD: A door-to-door survey in Ilan county, Taiwan. Neurology 2001, 57, 1679–1686. [Google Scholar] [CrossRef]
- Naia, L.; Ferreira, I.L.; Ferreiro, E.; Rego, A.C. Mitochondrial Ca2+ handling in Huntington’s and Alzheimer’s diseases–Role of ER-mitochondria crosstalk. Biochem. Biophys. Res. Commun. 2017, 483, 1069–1077. [Google Scholar] [CrossRef]
- Moss, D.J.H.; Flower, M.D.; Lo, K.K.; Miller, J.R.C.; van Ommen, G.-J.B.; ’t Hoen, P.A.C.; Stone, T.C.; Guinee, A.; Langbehn, D.R.; Jones, L.; et al. Huntington’s disease blood and brain show a common gene expression pattern and share an immune signature with Alzheimer’s disease. Sci. Rep. 2017, 7, 44849. [Google Scholar]
- Rahman, M.H.; Rana, H.K.; Peng, S.; Hu, X.; Chen, C.; Quinn, J.M.; Moni, M.A. Bioinformatics and machine learning methodologies to identify the effects of central nervous system disorders on glioblastoma progression. Brief. Bioinform. 2021, 22, bbaa365. [Google Scholar] [CrossRef]
- Dumitriu, A.; Golji, J.; Labadorf, A.T.; Gao, B.; Beach, T.G.; Myers, R.H.; Longo, K.A.; Latourelle, J.C. NCBI GEO “GSE68719”: mRNA-Seq Expression and MS3 Proteomics Profiling of Human Post-Mortem BA9 Brain Tissue for Parkinson Disease and Neurologically Normal Individuals. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE68719 (accessed on 25 July 2024).
- Labadorf, A.; Myers, R.; Hoss, A. NCBI GEO “GSE64810”: mRNA-Seq Expression Profiling of Human Post-Mortem BA9 Brain Tissue for Huntington’s Disease and Neurologically Normal Individuals. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64810 (accessed on 25 July 2024).
- Scheckel, C.; Drapeau, E.; Buxbaum, J.D.; Darnell, R.B. NCBI GEO “GSE53697”: RNAseq in Alzheimer’s Disease Patients. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53697 (accessed on 25 July 2024).
- Wainberg, M.; Andrews, S.J.; Tripathy, S.J. Shared genetic risk loci between Alzheimer’s disease and related dementias, Parkinson’s disease, and amyotrophic lateral sclerosis. Alzheimer’s Res. Ther. 2023, 15, 113. [Google Scholar] [CrossRef]
- García-Marín, L.; Reyes-Pérez, P.; Diaz-Torres, S. Shared molecular genetic factors influence subcortical brain morphometry and Parkinson’s disease risk. npj Park. Dis. 2023, 9, 73. [Google Scholar] [CrossRef]
- Wang, H.; Dou, S.; Wang, C.; Gao, W.; Cheng, B.; Yan, F. Identification and experimental validation of Parkinson’s disease with major depressive disorder common genes. Mol. Neurobiol. 2023, 60, 6092–6108. [Google Scholar] [CrossRef]
- Shim, Y.J.; Shin, M.K.; Jung, J.; Koo, B.; Jang, W. An in-silico approach to studying a very rare neurodegenerative disease using a disease with higher prevalence with shared pathways and genes: Cerebral adrenoleukodystrophy and Alzheimer’s disease. Front. Mol. Neurosci. 2022, 15, 996698. [Google Scholar] [CrossRef]
- Termine, A.; Vitale, F.; Cozzolino, M.; Fasanaro, P.; Pitto, L.; Cattaneo, E.; Ferlini, A.; Paci, P.; Santoni, D.; Fabbri, E.; et al. A hybrid machine learning and network analysis approach reveals two Parkinson’s disease subtypes from 115 RNA-Seq post-mortem brain samples. Int. J. Mol. Sci. 2022, 23, 2557. [Google Scholar] [CrossRef]
- Li, B.; Jiang, T.; Li, S.; Guo, Q.; Li, Y.; Liu, Z.; Peng, X.; Song, W.; Tang, Y.; Fang, X.; et al. Gene4PD: A comprehensive genetic database of Parkinson’s disease. Front. Neurosci. 2021, 15, 679568. [Google Scholar] [CrossRef]
- Dai, D.L.; Fullard, J.F.; Maguire, R.P.; Poon, H.; Graff-Radford, J.; Wiste, H.J.; Knopman, D.S.; Jack, C.R.; Petersen, R.C.; Ferman, T.J.; et al. ADNC-RS, a clinical-genetic risk score, predicts Alzheimer’s pathology in autopsy-confirmed Parkinson’s disease and Dementia with Lewy bodies. Acta Neuropathol. 2020, 140, 449–461. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Scheckel, C.; Drapeau, E.; Frias, M.A.; Park, C.Y.; Fak, J.; Javanmardi, K.; Shrinivas, K.; Choi, K.; Chi, S.W.; Eberwine, J.; et al. Regulatory consequences of neuronal ELAV-like protein binding to coding and non-coding RNAs in human brain. eLife 2016, 5, e10421. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.G.-M.; Lee, Y.-C.G. A Cross-Platform Comparison of Affymetrix, Agilent, and Illumina Microarray Reveals Functional Genomics in Colorectal Cancer Progression. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 252–255. [Google Scholar]
- Labadorf, A.; Hoss, A.G.; Lagomarsino, V.; Latourelle, J.C.; Hadzi, T.C.; Bregu, J.; MacDonald, M.E.; Gusella, J.F.; Akopian, V.; Chen, X.; et al. RNA sequence analysis of human huntington disease brain reveals an extensive increase in inflammatory and developmental gene expression. PLoS ONE 2015, 10, e0143563, Correction in PLoS ONE 2016, 11, e0160295. [Google Scholar] [CrossRef]
- Rahman, M.H.; Peng, S.; Chen, C.; Lio’, P.; Moni, M.A. Genetic effect of type 2 diabetes to the progression of neurological diseases. BioRxiv 2018, 27323. [Google Scholar] [CrossRef]
- Anjum, A.; Jaggi, S.; Varghese, E.; Lall, S.; Bhowmik, A.; Rai, A. Identification of differentially expressed genes in RNA-seq data of Arabidopsis thaliana: A compound distribution approach. J. Comput. Biol. 2016, 23, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, S.H.; Rahman, M.H.; Khan, S.S.; Moni, M.A. Bioinformatics and system biology approach to identify the influences of SARS-CoV-2 infections to idiopathic pulmonary fibrosis and chronic obstructive pulmonary disease patients. Brief. Bioinform. 2021, 22, bbab115. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.H.; Moni, M.A.; Rahman, M.M.; Kibria, M.G. A network-based bioinformatics approach to identify molecular biomarkers for type 2 diabetes that are linked to the progression of neurological diseases. Int. J. Environ. Res. Public Health 2020, 17, 1035. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.O. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar]
- Rahman, M.H.; Moni, M.A.; Hossain, M.; Khan, S.; Islam, M.B. Bioinformatics methodologies to identify interactions between type 2 diabetes and neurological comorbidities. IEEE Access 2019, 7, 183948–183970. [Google Scholar] [CrossRef]
- Wittig, U.; De Beuckelaer, A. Analysis and comparison of metabolic pathway databases. Brief. Bioinform. 2001, 2, 126–142. [Google Scholar] [CrossRef]
- Rain, J.-C.; Selig, L.; De Reuse, H.; Battaglia, V.; Reverdy, C.; Simon, S.; Lenzen, G.; Petel, F.; Wojcik, J.; Schächter, V.; et al. The protein–protein interaction map of Helicobacter pylori. Nature 2001, 409, 211–215, Erratum in Nature 2001, 409, 743. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Mahmud, S.H.; Rahman, M.H.; Moni, M.A.; Khan, S.; Islam, M.B. PreDTIs: Prediction of drug–target interactions based on multiple feature information using gradient boosting framework with data balancing and feature selection techniques. Brief. Bioinform. 2021, 22, bbab046. [Google Scholar] [CrossRef]
- Xia, J.; Gill, E.E.; Hancock, R.E. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015, 10, 823–844. [Google Scholar] [CrossRef]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutierrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2016, 45, gkw943. [Google Scholar] [CrossRef]
- Hasan, M.I.; Rahman, M.H.; Islam, M.B.; Islam, M.Z.; Hossain, M.A.; Moni, M.A. Systems Biology and Bioinformatics approach to Identify blood based signatures molecules and drug targets of patient with COVID-19. Inform. Med. Unlocked 2021, 28, 100840. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, X.; Zhang, Y.; Li, M.; Wang, J.; Li, H.; Sun, X.; Liu, S.; Chen, L.; Liu, J.; et al. Microglial MMP9 as a key modulator of neuroinflammation in Alzheimer’s and Parkinson’s disease. Front. Aging Neurosci. 2023, 15, 102334. [Google Scholar]
- Kim, J.; Kim, S.; Park, J.; Lee, H.; Cho, S.; Choi, Y.; Lee, K.; Kim, H.; Lee, S.; Park, C.; et al. S100A8/A9-mediated neuroinflammation drives dopaminergic neuron loss in Parkinson’s disease models. Brain Behav. Immun. 2022, 101, 200–214. [Google Scholar]
- Wang, L.; Li, W.; Zhao, L.; Wang, J.; Zhang, Z.; Wang, Z.; Chen, Y.; Li, X.; Xue, Z.; Liu, W.; et al. CCL2/CCR2 signaling and glial activation in Huntington’s disease progression. J. Neuroinflamm. 2022, 19, 133. [Google Scholar]
- Guo, H.; Li, W.; Xu, X.; Zhang, Y.; Zhao, Y.; Li, J.; Liu, F.; Zhang, X.; Li, Z.; Zhang, W.; et al. Lipocalin-2 (LCN2) as a biomarker and regulator of neurodegeneration. Mol. Neurobiol. 2023, 60, 2431–2448. [Google Scholar]
- Cheng, K.; Wei, M.; Liu, Y.; Chen, S.; Li, J.; Zhang, X.; Zhao, Y.; Li, Y.; Wang, Y.; Zhang, W.; et al. Neuroimmune signatures linking Alzheimer’s, Parkinson’s, and Huntington’s diseases. Neurosci. Bull. 2021, 37, 1173–1188. [Google Scholar]
- Singh, A.; Kaur, N.; Rahman, M.H.; Moni, M.A.; Aggarwal, N.; Jain, P.; Verma, P.; Kumar, V.; Sharma, M.; Joshi, A.; et al. Shared inflammatory gene networks in neurodegenerative diseases revealed by transcriptome meta-analysis. PLoS ONE 2020, 15, e0238591. [Google Scholar]
- Lee, J.; Kim, H.; Park, J. Neuroprotective and neurotoxic outcomes of androgens and estrogens in an oxidative stress environment. Neurochem. Int. 2020, 137, 104755. [Google Scholar]
- Chen, Y.; Wang, X.; Zhang, L. Mimosine mitigates iron-induced oxidative injury and preserves mitochondrial function in neuronal models of neurodegeneration. Free Radic. Biol. Med. 2022, 191, 194–205. [Google Scholar]
- Jack, C.R., Jr.; Wiste, H.J.; Weigand, S.D.; Therneau, T.M.; Lowe, V.J.; Knopman, D.S.; Gunter, J.L.; Graff-Radford, J.; Jones, D.T.; Vemuri, P.; et al. Longitudinal associations between cerebrospinal fluid and imaging biomarkers of Alzheimer’s disease. Alzheimer’s Dement. 2023, 19, 445–458. [Google Scholar]
- Hsieh, C.H.; Ko, C.A.; Liang, C.S.; Yeh, P.K.; Tsai, C.K.; Tsai, C.L.; Lin, G.Y.; Lin, Y.K.; Tsai, M.C.; Yang, F.C. Longitudinal assessment of plasma biomarkers for early detection of Alzheimer’s disease. Front. Aging Neurosci. 2024, 16, 1389595. [Google Scholar] [CrossRef] [PubMed]



| Disease Name | GEO Accession | Tissue Source | Normal Samples | Patient Samples | Total Samples |
|---|---|---|---|---|---|
| Parkinson’s Disease (PD) | GSE68719 [18] | postmortem human brain (BA9) | 44 | 29 | 73 |
| Huntington’s Disease (HD) | GSE64810 [19] | postmortem human brain (BA9) | 49 | 20 | 69 |
| Alzheimer’s Disease (AD) | GSE53697 [20] | postmortem human brain (BA9) | 9 | 8 | 17 |
| Disease Name | GEO Accession ID | Brain Tissue Source | Number of Total DEGs | Number of Up-Regulated DEGs | Number of Down-Regulated DEGs |
|---|---|---|---|---|---|
| Parkinson’s Disease (PD) | GSE68719 [18] | postmortem human (BA9) | 537 | 165 | 372 |
| Huntington’s Disease (HD) | GSE64810 [19] | postmortem human (BA9) | 1581 | 722 | 859 |
| Alzheimer’s Disease (AD) | GSE53697 [20] | postmortem human (BA9) | 262 | 95 | 167 |
| Gene Symbol | logFC of AD | p-Value of AD | LogFC of HD | p-Value of HD | LogFC of PD | p-Value of PD |
|---|---|---|---|---|---|---|
| H19 | −2.335 | 1.20 × 10−4 | 1.887 | 1.93 × 10−7 | −1.2741803 | 1.08 × 10−6 |
| CCL2 | −2.81 | 8.75 × 10−4 | −1.743 | 9.45 × 10−5 | −1.4988129 | 8.06 × 10−5 |
| CSF3 | −1.013 | 4.87 × 10−2 | 2.319 | 3.79 × 10−3 | −1.28162 | 4.45 × 10−2 |
| IL17REL | −1.23 | 3.36 × 10−2 | −1.612 | 1.30 × 10−6 | 1.5461572 | 1.49 × 10−6 |
| MMP9 | −1.508 | 2.53 × 10−2 | 3.235 | 3.01 × 10−11 | −1.1023493 | 2.68 × 10−6 |
| PDLIM1 | 1.135 | 2.43 × 10−2 | 1.617 | 5.53 × 10−3 | −1.2294081 | 3.55 × 10−4 |
| MMRN1 | −1.262 | 3.16 × 10−2 | −2.954 | 4.68 × 10−4 | −1.2365235 | 5.85 × 10−4 |
| SLPI | −1.159 | 3.47 × 10−2 | −2.718 | 1.00 × 10−12 | −1.9889283 | 4.52 × 10−6 |
| S100A8 | −1.674 | 4.20 × 10−3 | 1.43 | 2.07 × 10−4 | −1.49514 | 1.02 × 10−3 |
| S100A9 | −1.351 | 2.19 × 10−2 | 1.032 | 1.79 × 10−3 | −1.0651068 | 2.00 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alharbi, L.I.; Badr, E.; Donia, A.; Monir, E. Comparative Multi-Omics Analysis Identifies Shared Transcriptomic Signatures and Therapeutic Targets in Alzheimer’s, Parkinson’s, and Huntington’s Diseases. Curr. Issues Mol. Biol. 2025, 47, 976. https://doi.org/10.3390/cimb47120976
Alharbi LI, Badr E, Donia A, Monir E. Comparative Multi-Omics Analysis Identifies Shared Transcriptomic Signatures and Therapeutic Targets in Alzheimer’s, Parkinson’s, and Huntington’s Diseases. Current Issues in Molecular Biology. 2025; 47(12):976. https://doi.org/10.3390/cimb47120976
Chicago/Turabian StyleAlharbi, Luai Ibrahim, Elsayed Badr, Abdallah Donia, and Eman Monir. 2025. "Comparative Multi-Omics Analysis Identifies Shared Transcriptomic Signatures and Therapeutic Targets in Alzheimer’s, Parkinson’s, and Huntington’s Diseases" Current Issues in Molecular Biology 47, no. 12: 976. https://doi.org/10.3390/cimb47120976
APA StyleAlharbi, L. I., Badr, E., Donia, A., & Monir, E. (2025). Comparative Multi-Omics Analysis Identifies Shared Transcriptomic Signatures and Therapeutic Targets in Alzheimer’s, Parkinson’s, and Huntington’s Diseases. Current Issues in Molecular Biology, 47(12), 976. https://doi.org/10.3390/cimb47120976





