Analysis of Cytosine Base Editors in Bovine Zygotes: Efficiency and Editing Window Characterization Through Targeting the MYO7A Gene
Abstract
1. Introduction
2. Materials and Methods
2.1. Injection Material Preparation
2.2. Production of Bovine Zygotes and Cytoplasmic Injection
2.3. Editing Efficiency Confirmation
3. Results
3.1. Editing Efficiency of sgRNA with Cas9 Microinjection
3.2. C-to-T Conversion Efficiency and Window Comparison
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Simerly, C.; Dominko, T.; Navara, C.; Payne, C.; Capuano, S.; Gosman, G.; Chong, K.Y.; Takahashi, D.; Chace, C.; Compton, D.; et al. Molecular correlates of primate nuclear transfer failures. Science 2003, 300, 297. [Google Scholar] [CrossRef]
- Liu, Z.; Cai, Y.; Wang, Y.; Nie, Y.; Zhang, C.; Xu, Y.; Zhang, X.; Lu, Y.; Wang, Z.; Poo, M.; et al. Cloning of Macaque Monkeys by Somatic Cell Nuclear Transfer. Cell 2018, 174, 245. [Google Scholar] [CrossRef]
- Ryu, J.; Statz, J.P.; Chan, W.; Burch, F.C.; Brigande, J.V.; Kempton, B.; Porsov, E.V.; Renner, L.; McGill, T.; Burwitz, B.J.; et al. CRISPR/Cas9 editing of the MYO7A gene in rhesus macaque embryos to generate a primate model of Usher syndrome type 1B. Sci. Rep. 2022, 12, 10036. [Google Scholar] [CrossRef] [PubMed]
- Carey, K.; Ryu, J.; Uh, K.; Lengi, A.J.; Clark-Deener, S.; Corl, B.A.; Lee, K. Frequency of off-targeting in genome edited pigs produced via direct injection of the CRISPR/Cas9 system into developing embryos. BMC Biotechnol. 2019, 19, 25. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Ryu, J.; Wen, K.; Twitchell, E.; Bui, T.; Ramesh, A.; Weiss, M.; Li, G.; Samuel, H.; Clark-Deener, S.; et al. Increased and prolonged human norovirus infection in RAG2/IL2RG deficient gnotobiotic pigs with severe combined immunodeficiency. Sci. Rep. 2016, 6, 25222. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Krisher, R.L.; Herrick, J.R. Bovine embryo production in vitro: Evolution of culture media and commercial perspectives. Anim. Reprod. 2024, 21, e20240051. [Google Scholar] [CrossRef]
- Gehrke, J.M.; Cervantes, O.; Clement, M.K.; Wu, Y.; Zeng, J.; Bauer, D.E.; Pinello, L.; Joung, J.K. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 2018, 36, 977–982. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, Y.; Li, M.; Gu, Z. Precise genome editing with base editors. Med. Rev. 2023, 3, 75–84. [Google Scholar] [CrossRef]
- Concordet, J.P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef]
- Wang, X.; Li, J.; Wang, Y.; Yang, B.; Wei, J.; Wu, J.; Wang, R.; Huang, X.; Chen, J.; Yang, L. Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat. Biotechnol. 2018, 36, 946–949. [Google Scholar] [CrossRef] [PubMed]
- Kuscu, C.; Parlak, M.; Tufan, T.; Yang, J.; Szlachta, K.; Wei, X.; Mammadov, R.; Adli, M. CRISPR-STOP: Gene silencing through base-editing-induced nonsense mutations. Nat. Methods 2017, 14, 710–712. [Google Scholar] [CrossRef] [PubMed]
- Zuo, E.; Sun, Y.; Yuan, T.; He, B.; Zhou, C.; Ying, W.; Liu, J.; Wei, W.; Zeng, R.; Li, Y.; et al. A rationally engineered cytosine base editor retains high on-target activity while reducing both DNA and RNA off-target effects. Nat. Methods 2020, 17, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xue, W.; Yan, L.; Li, X.; Wei, J.; Chen, M.; Wu, J.; Yang, B.; Yang, L.; Chen, J. Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor. Cell Res. 2017, 27, 1289–1292. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, M.; Sun, L.; Peng, Y.; Sun, Y.; Fan, Y.; Li, H.; Liu, D.; Lu, H. Synergistic optimization enhancing the precision and efficiency of cytosine base editors in poplar. Commun. Biol. 2025, 8, 904. [Google Scholar] [CrossRef]
- Varshney, G.K.; Burgess, S.M. CRISPR-based functional genomics tools in vertebrate models. Exp. Mol. Med. 2025, 57, 1355–1372. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Luo, L.; Shi, Y.; Wang, H.; Wang, Z.; Dang, Y.; Li, S.; Wang, S.; Zhang, K. Base editing in bovine embryos reveals a species-specific role of SOX2 in regulation of pluripotency. PLoS Genet. 2022, 18, e1010307. [Google Scholar] [CrossRef]
- Shi, Y.; Hu, B.; Wang, Z.; Wu, X.; Luo, L.; Li, S.; Wang, S.; Zhang, K.; Wang, H. Functional role of GATA3 and CDX2 in lineage specification during bovine early embryonic development. Reproduction 2023, 165, 325–333. [Google Scholar] [CrossRef]
- Hennig, S.L.; Owen, J.R.; Lin, J.C.; Young, A.E.; Ross, P.J.; Van Eenennaam, A.L.; Murray, J.D. Evaluation of mutation rates, mosaicism and off target mutations when injecting Cas9 mRNA or protein for genome editing of bovine embryos. Sci. Rep. 2020, 10, 22309. [Google Scholar] [CrossRef]
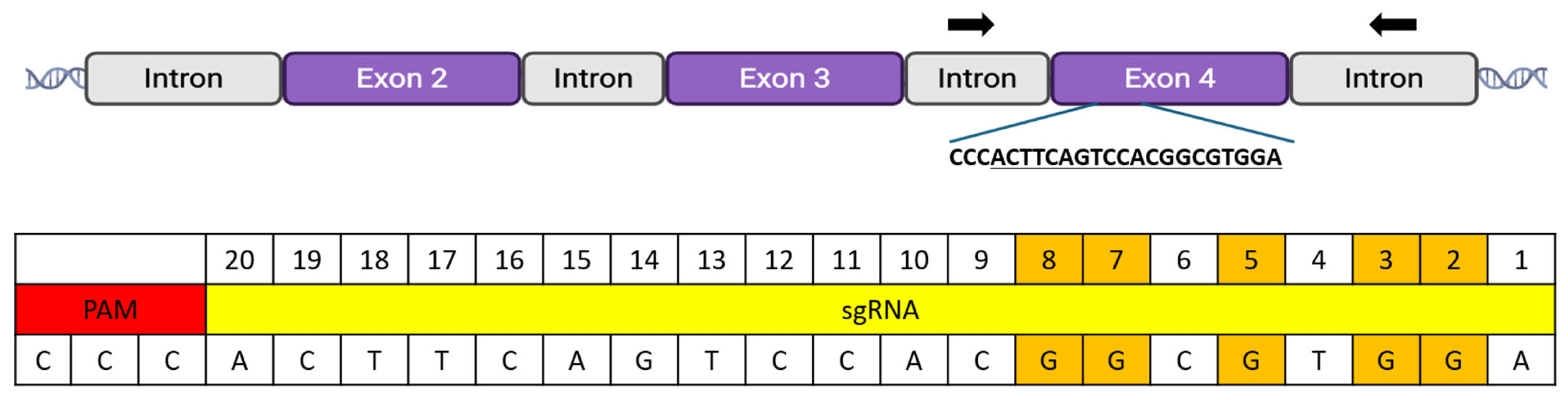


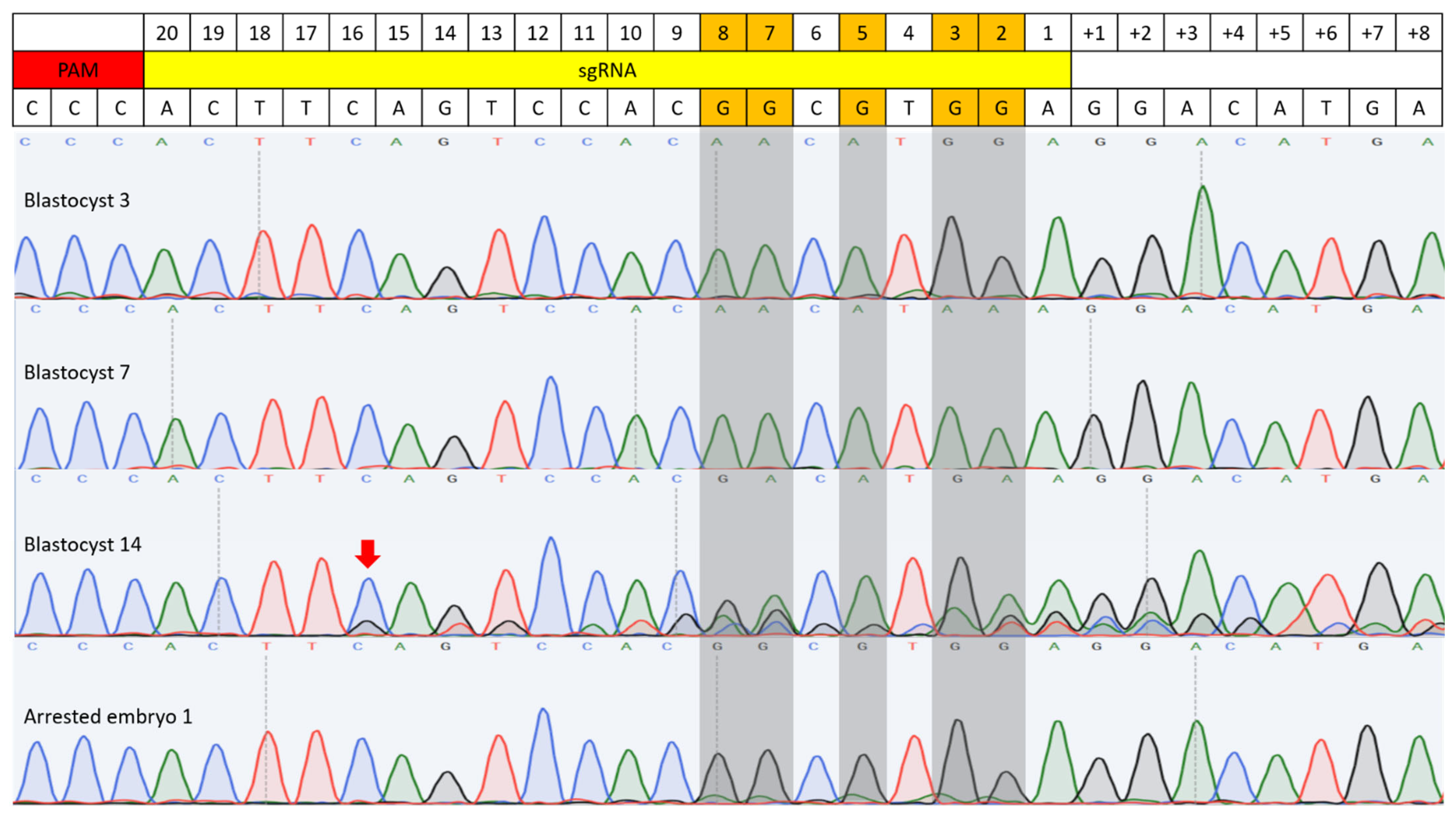
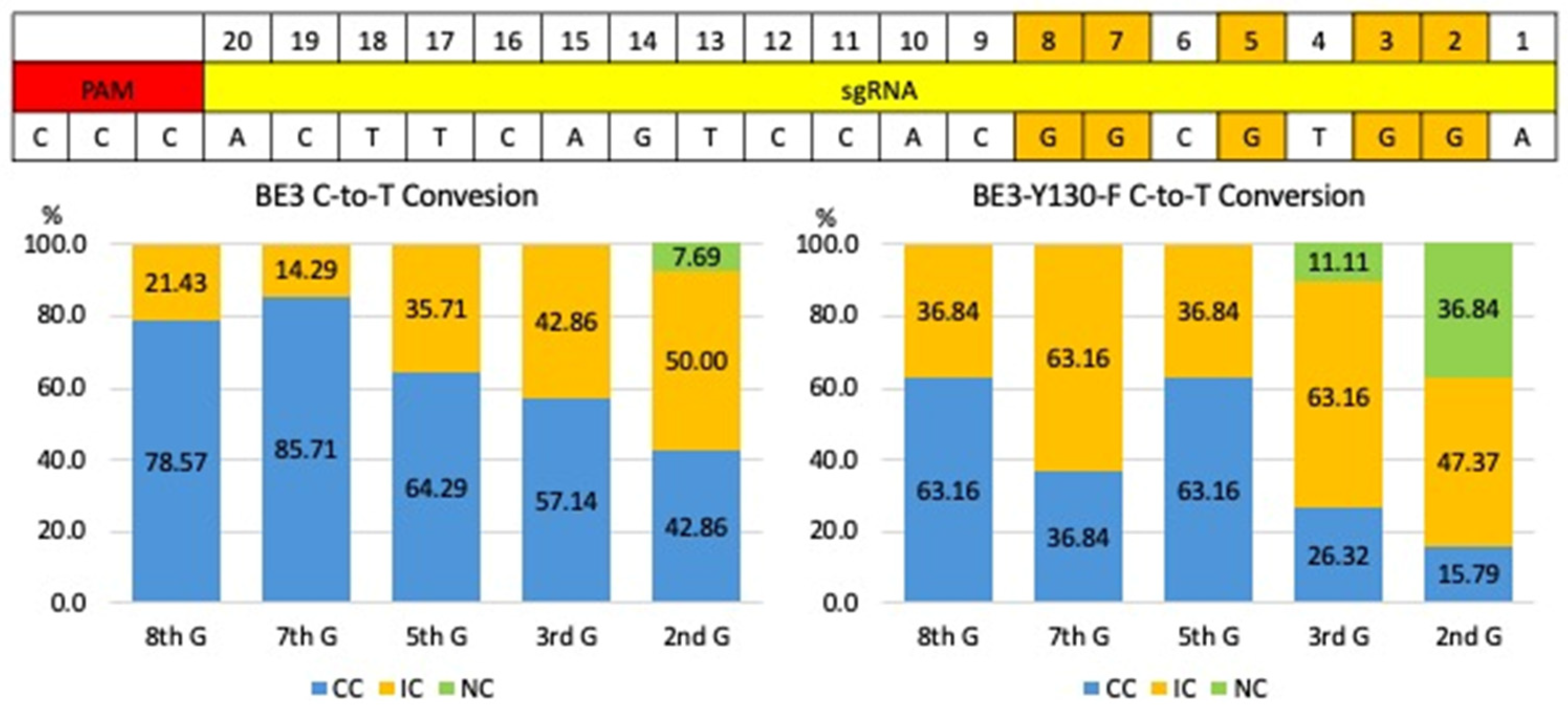
| Injection Material | # of Injected Zygotes | # of Blastocysts | # of Arrested Embryos Analyzed | Results | |
|---|---|---|---|---|---|
| # of Knockout Embryos | # of Embryos: Wild Type | ||||
| Cas9 mRNA + sgRNA | 95 | 6 | 2 | 7 | 0 |
| Injection Material | # of Injected Zygotes | # of Blastocysts | # of Arrested Embryos Analyzed | Results | |
|---|---|---|---|---|---|
| # with a C-to-T Conversion | # with no Conversion | ||||
| BE3 mRNA + sgRNA | 143 | 11 | 3 | 14 | 0 |
| BE3-Y130F + sgRNA | 171 | 16 | 3 | 19 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryu, J.; Tippner-Hedges, R.; Neuringer, M.; Hennebold, J.D. Analysis of Cytosine Base Editors in Bovine Zygotes: Efficiency and Editing Window Characterization Through Targeting the MYO7A Gene. Curr. Issues Mol. Biol. 2025, 47, 1033. https://doi.org/10.3390/cimb47121033
Ryu J, Tippner-Hedges R, Neuringer M, Hennebold JD. Analysis of Cytosine Base Editors in Bovine Zygotes: Efficiency and Editing Window Characterization Through Targeting the MYO7A Gene. Current Issues in Molecular Biology. 2025; 47(12):1033. https://doi.org/10.3390/cimb47121033
Chicago/Turabian StyleRyu, Junghyun, Rebecca Tippner-Hedges, Martha Neuringer, and Jon D. Hennebold. 2025. "Analysis of Cytosine Base Editors in Bovine Zygotes: Efficiency and Editing Window Characterization Through Targeting the MYO7A Gene" Current Issues in Molecular Biology 47, no. 12: 1033. https://doi.org/10.3390/cimb47121033
APA StyleRyu, J., Tippner-Hedges, R., Neuringer, M., & Hennebold, J. D. (2025). Analysis of Cytosine Base Editors in Bovine Zygotes: Efficiency and Editing Window Characterization Through Targeting the MYO7A Gene. Current Issues in Molecular Biology, 47(12), 1033. https://doi.org/10.3390/cimb47121033







