Genome-Edited Fish in the Field
Abstract
1. Introduction
2. Current Status of GED Fish Development and Field Trials
2.1. Expansion of Genome Editing Applications in Aquaculture
2.2. Trait Categories and Target Genes
2.3. Transition from Laboratory to Farm-Scale Evaluation
2.4. Emerging Field Trials and Research Gaps
- (1)
- (2)
- Multi-generational assessment: Most published studies report results only up to the F1 or F2 generations, with few tracking lineages over multiple generations to evaluate heritability, edit stability, and potential long-term fitness trade-offs [96].
- (3)
- Ecological interaction studies: Quantitative data on behavior, competition, reproductive success in mixed populations, and ecological fitness in open systems remain scarce. The ecological implications of potential escapes or gene flow under real-world conditions are largely unexplored in the context of GED fish [29,97].
- (4)
- Public transparency and data accessibility: Many genome editing projects remain documented only in internal reports, regulatory dossiers, or industry announcements rather than peer-reviewed publications. This restricts opportunities for independent validation, replication, and accumulation of scientific knowledge [98].
3. GED Fishes in the Field
3.1. Genome Edited Fish in the Field in Japan
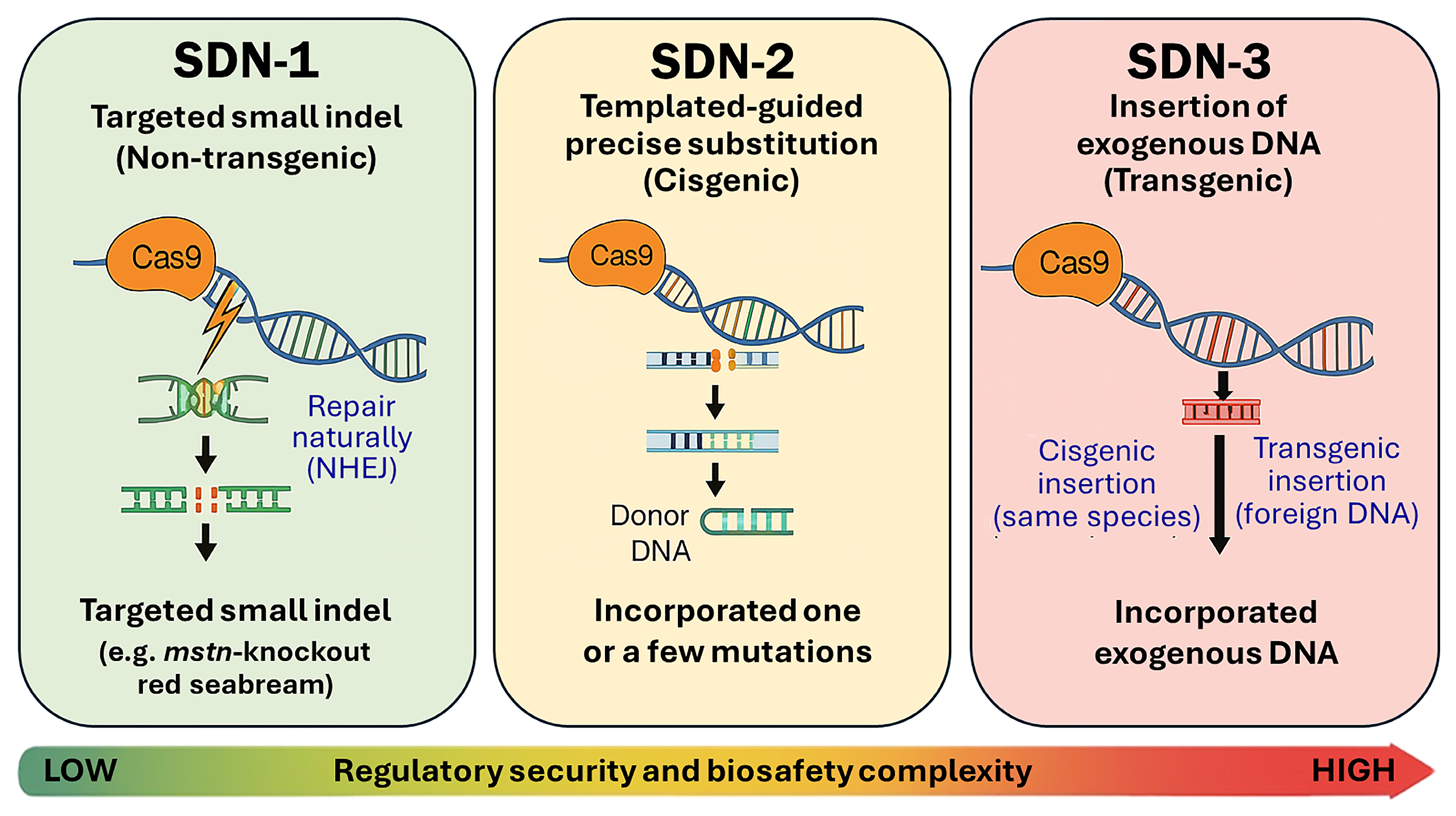
3.1.1. Unified Regulatory Pathway of SDN-1 GED Fish in Japan
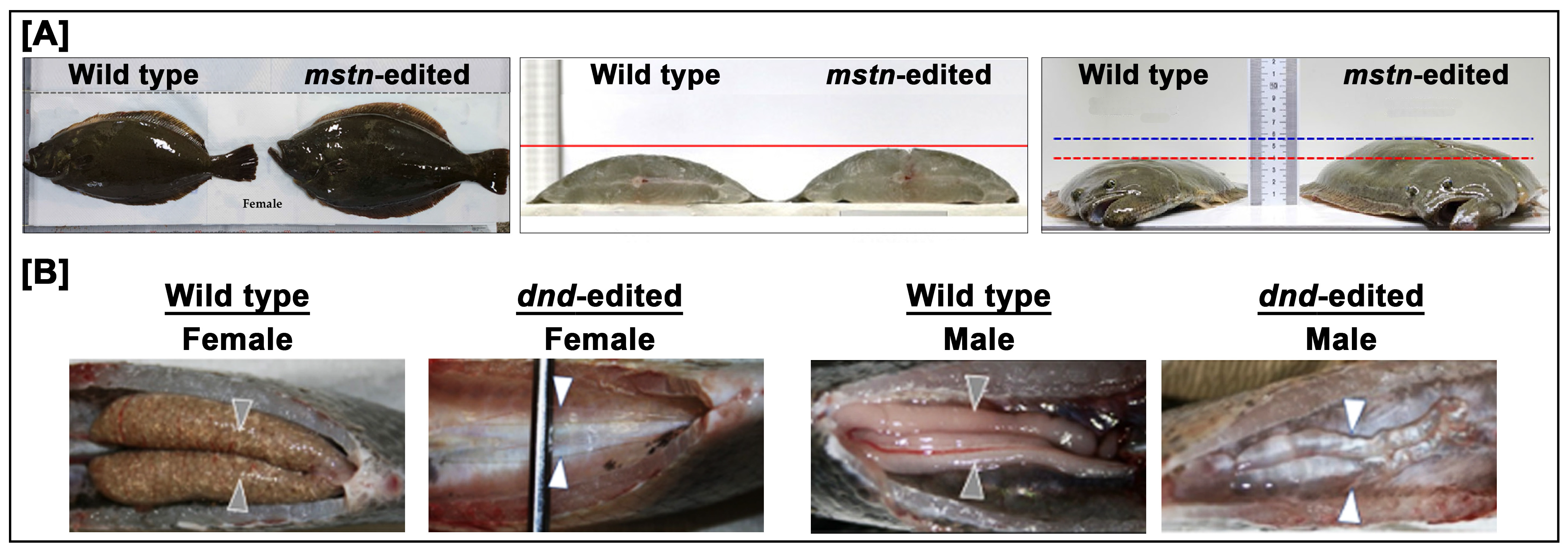
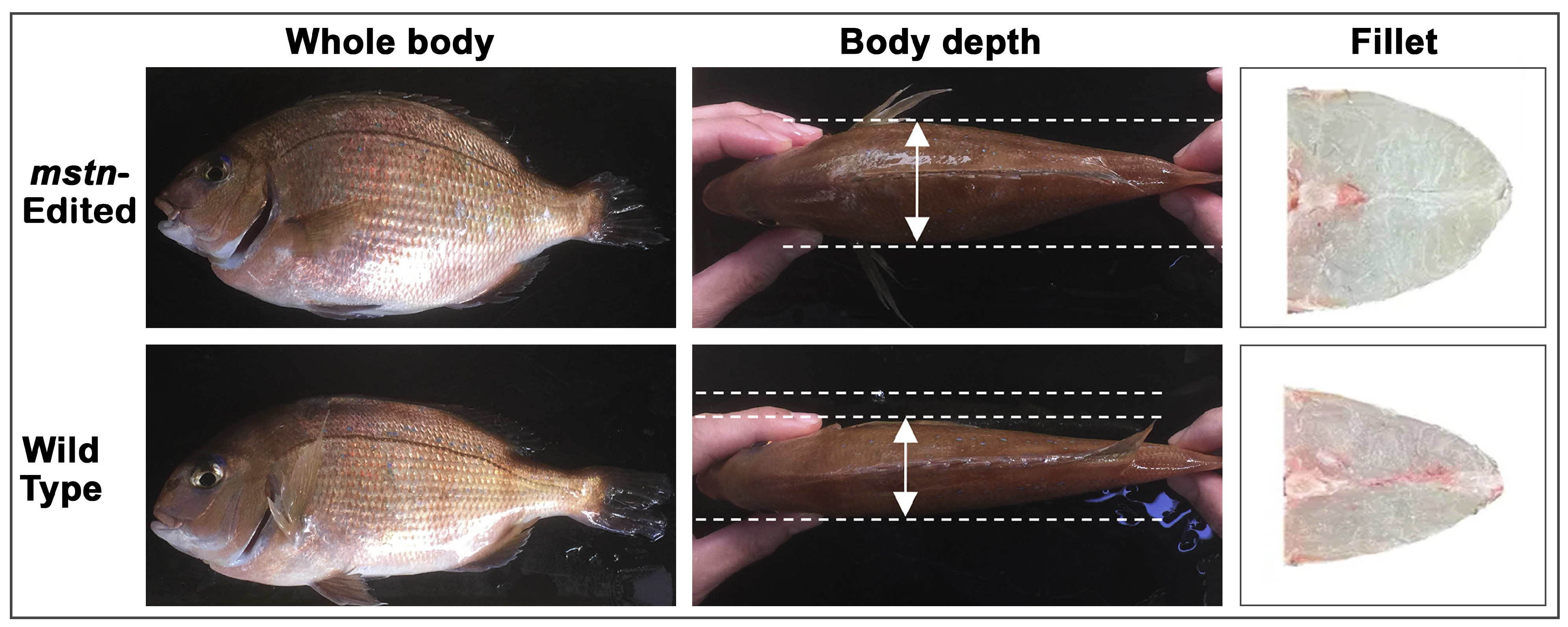
3.1.2. Red Seabream
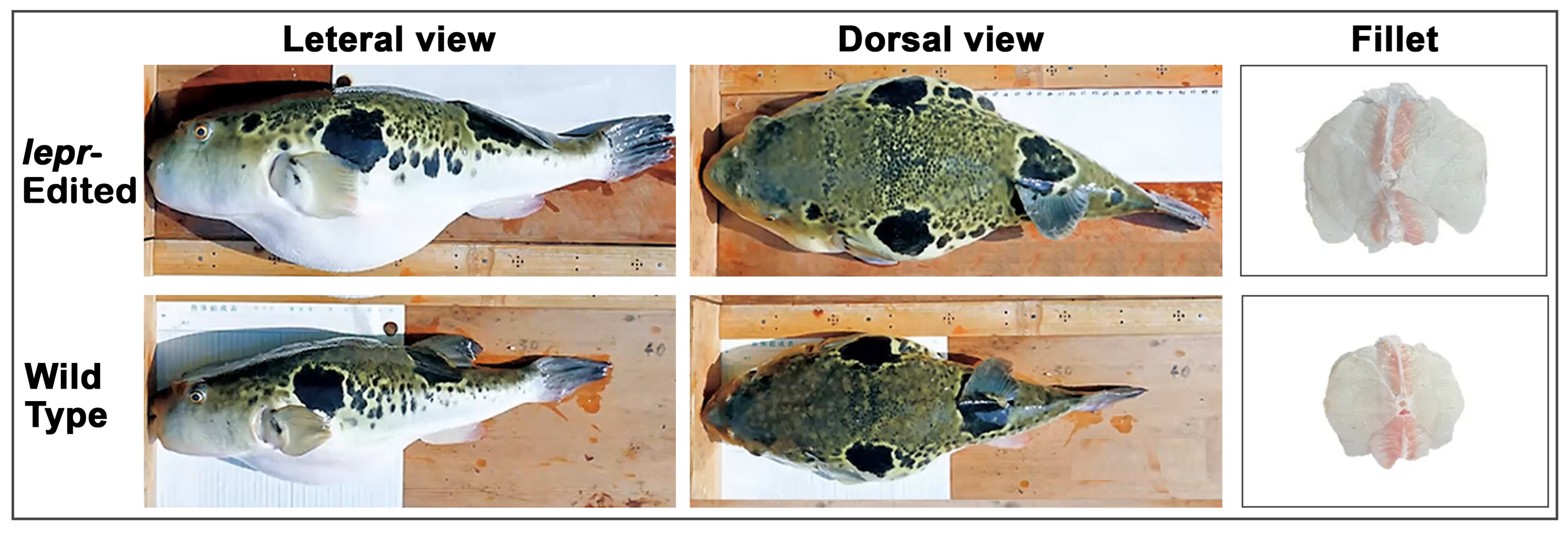
3.1.3. Tiger Puffer
3.1.4. Olive Flounder
3.2. Nile Tilapia in South America
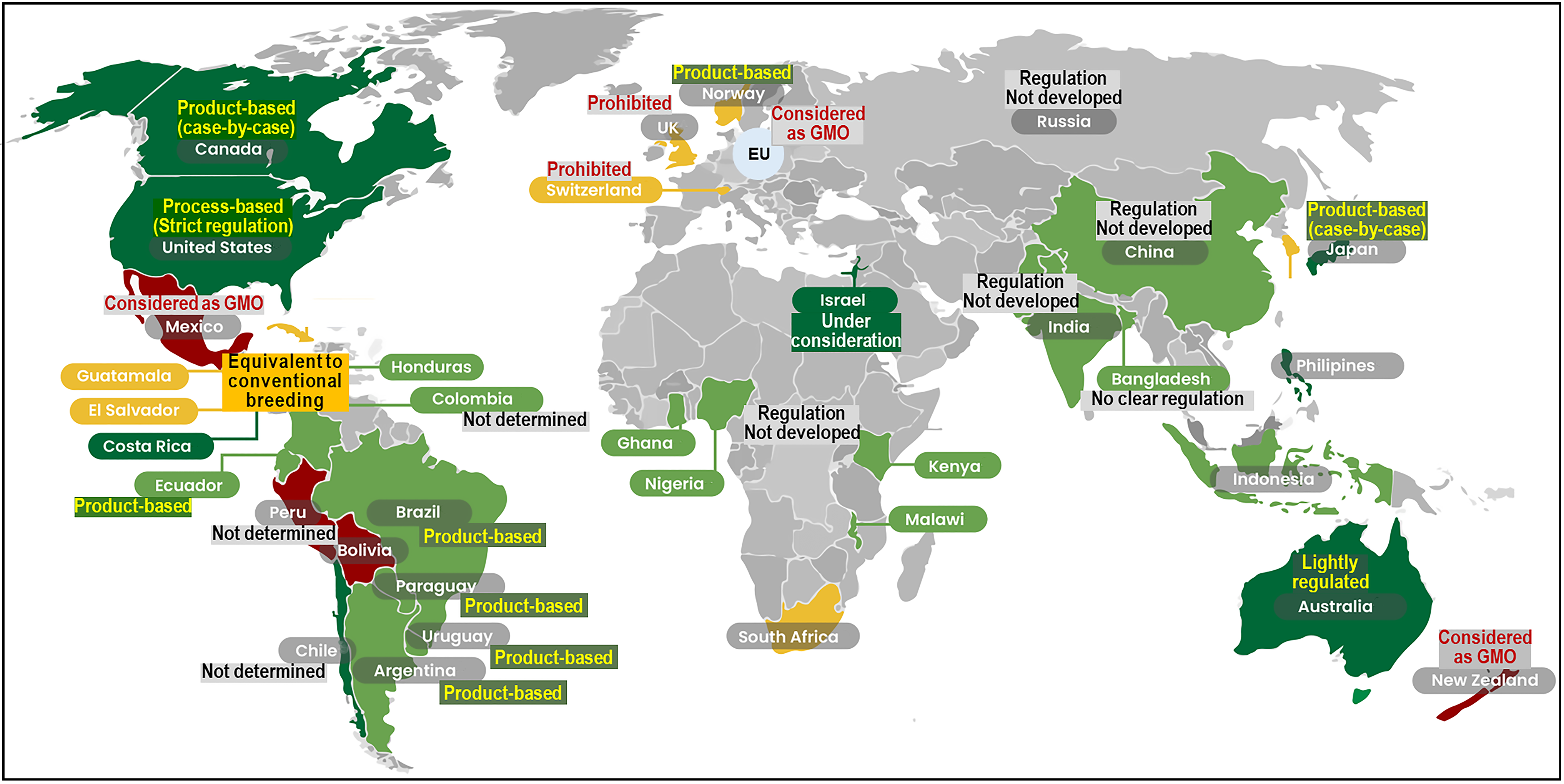
4. Regulatory and Policy Landscape of GED Fish
4.1. Diverging Global Approaches
4.2. Asia-Pacific Leadership: Japan and China
4.3. The Americas: Product-Based Oversight
4.4. Labeling and Traceability Practices
4.5. Ethical, Societal, and Trade Implications
5. Consumer Perception and Communication
5.1. Public Awareness and Knowledge Gaps
5.2. Factors Influencing Acceptance
5.2.1. Perceived Naturalness
5.2.2. Transparency
5.2.3. Perceived Benefits
5.3. Communication Strategies
5.3.1. Multi-Channel Transparency
5.3.2. Purpose Framing
5.3.3. Intermediary Empowerment
5.3.4. Responsive Labeling
5.3.5. Consistent and Carefully Chosen Language
5.3.6. Balanced Risk-Benefit Communication
5.3.7. Dialogue and Co-Creation
6. Framework and Reporting Checklist for Field Trials
6.1. Rationale for a Structured Framework
6.2. Core Components of the Framework
6.2.1. Molecular Characterization
6.2.2. Performance Evaluation
6.2.3. Welfare and Health Monitoring
6.2.4. Biosafety and Ecological Containment
6.2.5. Data Transparency and Reporting
6.3. Proposed Reporting Checklist
7. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| SDN | Site-Directed Nuclease |
| ARRIVE | Animal Research: Reporting of In Vivo Experiments |
| bmp6 | Bone morphogenetic protein 6 |
| Cas9 | CRISPR-associated protein 9 |
| CAT | Center for Aquaculture Technologies |
| cath | Cathelicidin |
| chi4 | Chitinase 4 |
| CONABIA | National Advisory Commission on Agricultural Biotechnology, Argentina |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| csf1ra | Colony stimulating factor 1 receptor a |
| CTNBio | National Technical Biosafety Commission, Brazil |
| dmrt1 | Doublesex and mab-3 related transcription factor 1 |
| DNA | Deoxyribonucleic Acid |
| dnd | Dead end homolog |
| eDNA | Environmental DNA |
| EU | European Union |
| FAIR | Findable, Accessible, Interoperable and Reusable |
| FAO | Food and Agriculture Organization of the United Nations |
| fgf | Fibroblast growth factor |
| foxl2 | Forkhead box L2 |
| fshb | Follicle-stimulating hormone beta subunit |
| GCRV | Grass Carp Reovirus |
| GED | Genome-edited |
| GMO | Genetically Modified Organism |
| gol | Golden (also known as slc24a5-like) |
| HDR | Homology Directed Repair |
| HSPs | Heat Shock Proteins |
| IBs | Intermuscular Bones |
| jam-a | Junctional adhesion molecule A |
| kctd10 | Potassium channel tetramerization domain containing 10 |
| KO | Knockout |
| lepr | Leptin receptor |
| lh | Luteinizing hormone |
| ltk | Leukocyte tyrosine kinase |
| LMO | Living Modified Organism |
| MAFF | Ministry of Agriculture, Forestry and Fisheries, Japan |
| MARA | Ministry of Agriculture and Rural Affairs, China |
| MHLW | Ministry of Health, Labour and Welfare, Japan |
| MOE | Ministry of Environment, Japan |
| mitfa | Microphthalmia-associated transcription factor a |
| mstn | Myostatin |
| nanos | Nanos homolog |
| NCBI | National Center for Biotechnology Information |
| NHEJ | Non-Homologous End Joining |
| oca2 | Oculocutaneous albinism II |
| OECD | Organisation for Economic Co-operation and Development |
| pax6 | Paired box gene 6 |
| PCR | Polymerase chain reaction |
| pi4kb | Phosphatidylinositol 4-kinase beta |
| RNA | Ribonucleic Acid |
| runx2b | Runt-related transcription factor 2b |
| RyR1b | Ryanodine receptor 1b |
| sdY | Sexually dimorphic on the Y-chromosome |
| sgRNA | Single-guide RNA |
| slc24a5 | Solute carrier family 24 member 5 |
| slc45a2 | Solute carrier family 45 member 2 |
| SNP | Single Nucleotide Polymorphism |
| sp7 | Sp7 transcription factor |
| SRA | Sequence Read Archive |
| tlr-22 | Toll-like receptor 22 |
| tyr | Tyrosinase |
| USDA | United States Department of Agriculture |
| v1a2 | Arginine vasotocin receptor |
| WGS | Whole Genome Sequencing |
| β-tubulin | Beta tubulin |
References
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.R.; Joung, J.K. Efficient Genome Editing in Zebrafish Using a CRISPR–Cas System. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef]
- Li, M.; Yang, H.; Zhao, J.; Fang, L.; Shi, H.; Li, M.; Sun, Y.; Zhang, X.; Jiang, D.; Zhou, L.; et al. Efficient and Heritable Gene Targeting in Tilapia by CRISPR/Cas9. Genetics 2014, 197, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yan, Y.; Tao, Y.; Lu, S.; Xu, P.; Qiang, J. Transcriptional Knock-Down of mstn Encoding Myostatin Improves Muscle Quality of Nile Tilapia (Oreochromis niloticus). Mar. Biotechnol. 2023, 25, 951–965. [Google Scholar] [CrossRef]
- Sukhan, Z.P.; Cho, Y.; Hossen, S.; Cho, D.H.; Kho, K.H. Molecular Characterization, Expression Analysis, and CRISPR/Cas9-Mediated Gene Disruption of Myogenic Regulatory Factor 4 (MRF4) in Nile Tilapia. Curr. Issues Mol. Biol. 2024, 46, 13725–13745. [Google Scholar] [CrossRef]
- Khalil, K.; Elayat, M.; Khalifa, E.; Daghash, S.; Elaswad, A.; Miller, M.; Abdelrahman, H.; Ye, Z.; Odin, R.; Drescher, D.; et al. Generation of Myostatin Gene-Edited Channel Catfish (Ictalurus punctatus) via Zygote Injection of CRISPR/Cas9 System. Sci. Rep. 2017, 7, 7301. [Google Scholar] [CrossRef]
- Zhang, X.; Li, M.; Ma, H.; Liu, X.; Shi, H.; Li, M.; Sun, Y.; Zhang, X.; Jiang, D.; Zhou, L.; et al. Targeted Genome Editing in Channel Catfish (Ictalurus punctatus) Using CRISPR/Cas9. Front. Genet. 2020, 11, 573. [Google Scholar] [CrossRef]
- Wargelius, A.; Leininger, S.; Skaftnesmo, K.O.; Kleppe, L.; Andersson, E.; Taranger, G.L.; Schulz, R.W.; Edvardsen, R.B. Dnd Knockout Ablates Germ Cells and Demonstrates Germ Cell Independent Sex Differentiation in Atlantic Salmon. Sci. Rep. 2016, 6, 21284. [Google Scholar] [CrossRef]
- Yano, A.; Nicol, B.; Jouanno, E.; Guiguen, Y. Heritable Targeted Inactivation of the Rainbow Trout (Oncorhynchus mykiss) Master Sex-Determining Gene Using Zinc-Finger Nucleases. Mar. Biotechnol. 2014, 16, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Niu, P.; Wang, M.; Huang, G.; Xu, S.; Sun, Y.; Xu, X.; Hou, Y.; Sun, X.; Yan, Y.; et al. Targeted Disruption of sp7 and Myostatin with CRISPR/Cas9 Results in Severe Bone Defects and More Muscular Cells in Common Carp. Sci. Rep. 2016, 6, 22953. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Shen, Y.; Cheng, J.; Zhang, L.; Qu, Z.; Li, W.; Liu, X.; Li, M.; Dai, S. Generation of Fast-Growth Grass Carp by Mutation of mstnb via CRISPR/Cas9 System. Reprod. Breed. 2025, 5, 163–170. [Google Scholar] [CrossRef]
- Kishimoto, K.; Washio, Y.; Yoshiura, Y.; Toyoda, A.; Ueno, T.; Fukuyama, H.; Kato, K.; Kinoshita, M. Production of a Breed of Red Sea Bream (Pagrus major) with an Increase of Skeletal Muscle Mass and Reduced Body Length by Genome Editing with CRISPR/Cas9. Aquaculture 2018, 495, 415–427. [Google Scholar] [CrossRef]
- Kim, J.; Cho, J.Y.; Kim, J.W.; Kim, H.C.; Noh, J.K.; Kim, Y.O.; Hwang, H.-K.; Kim, W.J.; Yeo, S.Y.; An, C.M.; et al. CRISPR/Cas9-Mediated Myostatin Disruption Enhances Muscle Mass in the Olive Flounder Paralichthys olivaceus. Aquaculture 2019, 512, 734336. [Google Scholar] [CrossRef]
- Kishimoto, K. Application of Genome Editing to Marine Aquaculture as a New Breeding Technology. No. 21827. Ph.D. Dissertation, Kyoto University, Kyoto, Japan, 2019. Available online: https://repository.kulib.kyoto-u.ac.jp/bitstream/2433/242704/2/gnogk02340.pdf (accessed on 29 November 2025).
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The New Frontier of Genome Engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Podevin, N.; Davies, H.V.; Hartung, F.; Nogué, F.; Casacuberta, J.M. Site-Directed Nucleases: A Paradigm Shift in Genetic Modification. Trends Biotechnol. 2013, 31, 375–383. [Google Scholar] [CrossRef]
- Ishii, T. Consumer Choices Regarding Genome-Edited Food Crops: Lessons from Japan. Front. Genome Ed. 2025, 7, 1672358. [Google Scholar] [CrossRef]
- Matsuo, M.; Tachikawa, M. Implications and Lessons from the Introduction of Genome-Edited Food Products in Japan. Front. Genome Ed. 2022, 4, 899154. [Google Scholar] [CrossRef]
- Whelan, A.I.; Gutti, P.; Lema, M.A. Gene Editing Regulation and Innovation Economics. Front. Bioeng. Biotechnol. 2020, 8, 303. [Google Scholar] [CrossRef] [PubMed]
- FAO. The State of World Fisheries and Aquaculture 2022. Towards Blue Transformation; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- Houston, R.D.; Bean, T.P.; Macqueen, D.J.; Gundappa, M.K.; Jin, Y.H.; Jenkins, T.L.; Selly, S.L.C.; Martin, S.A.M.; Stevens, J.R.; Santos, E.M.; et al. Harnessing Genomics to Fast-Track Genetic Improvement in Aquaculture. Nat. Rev. Genet. 2020, 21, 389–409. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Li, H.; Zhang, Y.; Wang, Y.; Wei, Y.; Fan, Y.; Zou, T.; Ding, X.; Ma, L.; Zhang, Z. Grand Challenges in Genome Editing in Plants. Front. Genome Ed. 2020, 2, 2. [Google Scholar] [CrossRef] [PubMed]
- Gelinsky, E.; Hilbeck, A. European Court of Justice Ruling Regarding New Genetic Engineering Methods Scientifically Justified: A Commentary on the Biased Reporting About the Recent Ruling. Environ. Sci. Eur. 2018, 30, 52. [Google Scholar] [CrossRef]
- Hallerman, E.M.; Bredlau, J.P.; Camargo, L.S.A.; Dagli, M.L.Z.; Karembu, M.; Ngure, G.; Romero-Aldemita, R.; Rocha-Salavarrieta, P.J.; Tizard, M.; Walton, M.; et al. Towards Progressive Regulatory Approaches for Agricultural Applications of Animal Biotechnology. Transgenic Res. 2022, 31, 167–199. [Google Scholar] [CrossRef]
- ISAAA. Gene-Edited Animals for Agricultural Applications Database: Nile Tilapia (Oreochromis niloticus). 2025. Available online: https://www.isaaa.org/animalbiotechdatabase/specieslist/species/default.asp?SpeciesID=21&RegProcess=1 (accessed on 11 November 2025).
- Naylor, R.L.; Williams, S.L.; Strong, D.R. Aquaculture—A Gateway for Exotic Species. Science 2001, 294, 1655–1656. [Google Scholar] [CrossRef] [PubMed]
- Glover, K.A.; Solberg, M.F.; McGinnity, P.; Hindar, K.; Verspoor, E.; Coulson, M.W.; Skaala, Ø.; Svasand, T.; Ferguson, A. Half a Century of Genetic Interaction Between Farmed and Wild Atlantic Salmon: Status of Knowledge and Unanswered Questions. Fish Fish. 2017, 18, 890–927. [Google Scholar] [CrossRef]
- Blix, T.B.; Dalmo, R.A.; Wargelius, A.; Myhr, A.I. Genome Editing on Finfish: Current Status and Implications for Sustainability. Rev. Aquac. 2021, 13, 2344–2363. [Google Scholar] [CrossRef]
- Hallerman, E.M.; Dunham, R.; Houston, R.D.; Walton, M.; Wargelius, A.; Wray-Cahen, D. Towards Production of Genome-Edited Aquaculture Species. Rev. Aquac. 2023, 15, 404–408. [Google Scholar] [CrossRef]
- Robinson, N.A.; Østbye, T.K.K.; Kettunen, A.H.; Coates, A.; Barrett, L.T.; Robledo, D.; Dempster, T. A Guide to Assess the Use of Gene Editing in Aquaculture. Rev. Aquacult. 2024, 16, 775–784. [Google Scholar] [CrossRef]
- Dong, Z.; Ge, J.; Xu, Z.; Dong, X.; Cao, S.; Pan, J.; Zhao, Q. Generation of Myostatin B Knockout Yellow Catfish (Tachysurus fulvidraco) Using Transcription Activator-Like Effector Nucleases. Zebrafish 2014, 11, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Edvardsen, R.B.; Leininger, S.; Kleppe, L.; Skaftnesmo, K.O.; Wargelius, A. Targeted Mutagenesis in Atlantic Salmon (Salmo salar L.) Using the CRISPR/Cas9 System Induces Complete Knockout Individuals in the F0 Generation. PLoS ONE 2014, 9, e108622. [Google Scholar] [CrossRef]
- Square, T.; Romášek, M.; Jandzik, D.; Cattell, M.V.; Klymkowsky, M.; Medeiros, D.M. CRISPR/Cas9-Mediated Mutagenesis in the Sea Lamprey (Petromyzon marinus): A Powerful Tool for Understanding Ancestral Gene Functions in Vertebrates. Development 2015, 142, 4180–4187. [Google Scholar] [CrossRef]
- Zu, Y.; Zhang, X.; Ren, J.; Dong, X.; Zhu, Z.; Jia, L.; Zhang, Q.; Li, W. Biallelic Editing of a Lamprey Genome Using the CRISPR/Cas9 System. Sci. Rep. 2016, 6, 23496. [Google Scholar] [CrossRef]
- Chakrapani, V.; Patra, S.K.; Panda, R.P.; Rasal, K.D.; Jayasankar, P.; Barman, H.K. Establishing Targeted Carp TLR22 Gene Disruption via Homologous Recombination Using CRISPR/Cas9. Dev. Comp. Immunol. 2016, 61, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Li, Y.; Su, B.; Cheng, Q.; Ye, Z.; Perera, D.A.; Fobes, M.; Shang, M.; Dunham, R.A. Editing of the Luteinizing Hormone Gene to Sterilize Channel Catfish (Ictalurus punctatus) Using a Modified Zinc Finger Nuclease Technology with Electroporation. Mar. Biotechnol. 2016, 18, 255–263. [Google Scholar] [CrossRef]
- Gui, T.; Zhang, J.; Song, F.; Sun, Y.; Xie, S.; Yu, K.; Xiang, J. CRISPR/Cas9-Mediated Genome Editing and Mutagenesis of EcChi4 in Exopalaemon carinicauda. G3 2016, 6, 3757–3764. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, Y.; Wang, W.; Wang, Q.; Zhang, N.; Lin, F.; Wang, N.; Shao, C.; Dong, Z.; Li, Y.; et al. Genome Editing Reveals dmrt1 as an Essential Male Sex-Determining Gene in Chinese Tongue Sole (Cynoglossus semilaevis). Sci. Rep. 2017, 7, 42213. [Google Scholar] [CrossRef]
- Ma, J.; Fan, Y.; Zhou, Y.; Liu, W.; Jiang, N.; Zhang, J.; Zeng, L. Efficient Resistance to Grass Carp Reovirus Infection in JAM-A Knockout Cells Using CRISPR/Cas9. Fish Shellfish Immunol. 2018, 76, 206–215. [Google Scholar] [CrossRef]
- Klaassen, H.; Wang, Y.; Adamski, K.; Rohner, N.; Kowalko, J.E. CRISPR Mutagenesis Confirms the Role of oca2 in Melanin Pigmentation in Astyanax mexicanus. Dev. Biol. 2018, 441, 313–318. [Google Scholar] [CrossRef]
- Baloch, A.R.; Franěk, R.; Tichopád, T.; Fučíková, M.; Rodina, M.; Pšenička, M. Dnd1 Knockout in Sturgeons by CRISPR/Cas9 Generates Germ Cell-Free Host for Surrogate Production. Animals 2019, 9, 174. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Li, H.; Li, Q.; Xu, R.; Yue, C.; Du, S. Targeted Gene Disruption in Pacific Oyster Based on CRISPR/Cas9 Ribonucleoprotein Complexes. Mar. Biotechnol. 2019, 21, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Cao, X.; Gao, J. Production of a Mutant of Large-Scale Loach (Paramisgurnus dabryanus) with Skin Pigmentation Loss by Genome Editing with the CRISPR/Cas9 System. Transgenic Res. 2019, 28, 341–356. [Google Scholar] [CrossRef]
- Liu, Q.; Qi, Y.; Liang, Q.; Song, J.; Liu, J.; Li, W.; Shu, Y.; Tao, M.; Zhang, C.; Qin, Q.; et al. Targeted Disruption of Tyrosinase Causes Melanin Reduction in Carassius auratus cuvieri and Its Hybrid Progeny. Sci. China Life Sci. 2019, 62, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, K.; Kazeto, Y.; Ozaki, Y.; Yamaguchi, T.; Shimada, Y.; Ina, Y.; Soma, S.; Sakakura, Y.; Goto, R.; Matsubara, T.; et al. Targeted Mutagenesis of the Ryanodine Receptor by Platinum TALENs Causes Slow Swimming Behaviour in Pacific Bluefin Tuna (Thunnus orientalis). Sci. Rep. 2020, 9, 13871, Erratum in Sci. Rep. 2020, 10, 9351. [Google Scholar] [CrossRef]
- Sun, Y.; Zheng, G.D.; Nissa, M.; Chen, J.; Zou, S.M. Disruption of Mstna and Mstnb Genes through CRISPR/Cas9 Leads to Elevated Muscle Mass in Blunt Snout Bream (Megalobrama amblycephala). Aquaculture 2020, 528, 735597. [Google Scholar] [CrossRef]
- Maki, J.A.; Cavallin, J.E.; Lott, K.G.; Saari, T.W.; Ankley, G.T.; Villeneuve, D.L. A Method for CRISPR/Cas9 Mutation of Genes in Fathead Minnow (Pimephales promelas). Aquat. Toxicol. 2020, 222, 105464. [Google Scholar] [CrossRef]
- Tao, B.; Tan, J.; Chen, L.; Xu, Y.; Liao, X.; Li, Y.; Chen, J.; Song, Y.; Hu, W. CRISPR/Cas9 System-Based Myostatin-Targeted Disruption Promotes Somatic Growth and Adipogenesis in Loach (Misgurnus anguillicaudatus). Aquaculture 2021, 544, 737097. [Google Scholar] [CrossRef]
- Pandey, D.; Matsubara, T.; Saito, T.; Kazeto, Y.; Gen, K.; Sakuma, T.; Yamamoto, T.; Mekuchi, M.; Goto, R. TALEN-Mediated Gene Editing of slc24a5 (Solute Carrier Family 24, Member 5) in Kawakawa (Euthynnus affinis). J. Mar. Sci. Eng. 2021, 9, 1378. [Google Scholar] [CrossRef]
- Yan, M.; Li, B.; Wang, J.; Bai, Y.; Ke, Q.; Zhou, T.; Xu, P. Disruption of mstn Gene by CRISPR/Cas9 in Large Yellow Croaker (Larimichthys crocea). Mar. Biotechnol. 2022, 24, 681–689. [Google Scholar] [CrossRef]
- Clark, B.; Elkin, J.; Marconi, A.; Turner, G.F.; Smith, A.M.; Joyce, D.; Miska, E.A.; Juntti, S.A.; Santos, M.E. Oca2 Targeting Using CRISPR/Cas9 in the Malawi Cichlid Astatotilapia calliptera. R. Soc. Open Sci. 2022, 9, 220077. [Google Scholar] [CrossRef]
- Kawamura, W.; Hasegawa, N.; Yamauchi, A.; Kimura, T.; Yahagi, H.; Tani, R.; Morita, T.; Yazawa, R.; Yoshizaki, G. Production of Albino Chub Mackerel (Scomber japonicus) by slc45a2 Knockout and the Use of a Positive Phototaxis-Based Larviculture Technique to Overcome the Lethal Albino Phenotype. Aquaculture 2022, 560, 738490. [Google Scholar] [CrossRef]
- Molcho, J.; Manor, R.; Shamsian, M.; Sudarshan, G.; Ofir, R.; Parker, D.; Weil, S.; Wattad, H.; Hayun, E.; Levy, T.; et al. On Genome Editing in Embryos and Cells of the Freshwater Prawn (Macrobrachium rosenbergii). Aquaculture 2022, 558, 738391. [Google Scholar] [CrossRef]
- Ou, M.; Wang, F.; Li, K.; Wu, Y.; Huang, S.; Luo, Q.; Liu, H.; Zhang, X.; Fei, S.; Chen, K.; et al. Generation of Myostatin Gene-Edited Blotched Snakehead (Channa maculata) Using the CRISPR/Cas9 System. Aquaculture 2023, 563, 738988. [Google Scholar] [CrossRef]
- Kuang, Y.; Zheng, X.; Cao, D.; Sun, Z.; Tong, G.; Xu, H.; Yan, T.; Tang, S.; Chen, Z.; Zhang, T.; et al. Generation of a New Crucian Carp (Carassius auratus) Strain without Intermuscular Bones by Knocking Out bmp6. Aquaculture 2023, 569, 739407. [Google Scholar] [CrossRef]
- Gan, R.H.; Li, Z.; Wang, Z.W.; Li, X.Y.; Wang, Y.; Zhang, X.J.; Tong, J.F.; Wu, Y.; Xia, L.Y.; Gao, Z.X.; et al. Creation of Intermuscular Bone-Free Mutants in Amphitriploid Gibel Carp by Editing Two Duplicated runx2b Homeologs. Aquaculture 2023, 567, 739300. [Google Scholar] [CrossRef]
- Krug, J.; Perner, B.; Albertz, C.; Mörl, H.; Hopfenmüller, V.L.; Englert, C. Generation of a Transparent Killifish Line through Multiplex CRISPR/Cas9-Mediated Gene Inactivation. eLife 2023, 12, e81549. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, S.; Jiang, W.; Li, F.; Chi, M.; Cheng, S.; Liu, Y. CRISPR/Cas9-Mediated Mutation of Mstn Confers Growth Performance in Culter alburnus Juveniles. Aquac. Fish. 2024, 9, 900–907. [Google Scholar] [CrossRef]
- Yazawa, R.; Saitoh, K.; Yamauchi, A.; Eyüboğlu, O.; Ozawa, K.; Kawamura, W.; Morita, T.; Takeuchi, Y.; Yoshizaki, G. Reproductive Characteristics and Suitability of Sterile Dead End Knockout Nibe Croaker as a Recipient for Intraperitoneal Germ Cell Transplantation. Mar. Biotechnol. 2024, 26, 891–901. [Google Scholar] [CrossRef] [PubMed]
- Booncherd, K.; Sreebun, S.; Pasomboon, P.; Boonanuntanasarn, S. Effects of CRISPR/Cas9-Mediated dnd1 Knockout Impair Gonadal Development in Striped Catfish. Animals 2024, 18, 101039. [Google Scholar] [CrossRef] [PubMed]
- López-Porras, A.; Berg, R.S.; Burgerhout, E.; Hansen, Ø.J.; Györkei, Á.; Qiao, S.W.; Johansen, F.E. CRISPR-Cas9/Cas12a-Based Genome Editing in Atlantic Cod (Gadus morhua). Aquaculture 2024, 581, 740440. [Google Scholar] [CrossRef]
- Li, R.; Xu, Y.; Wu, F.; Peng, Z.; Chan, J.; Zhang, L. Easy-to-Use CRISPR-Cas9 Genome Editing in the Cultured Pacific Abalone (Haliotis discus hannai). CRISPR J. 2024, 7, 41–52. [Google Scholar] [CrossRef]
- Gao, T.; Wang, F.; Wu, Q.; Gan, L.; Jin, C.; Ma, L.; Wang, D.; Sun, L. Mutation of Genes Associated with Body Color, Growth, Intermuscular Bone, and Sex Differentiation in Onychostoma macrolepis Using CRISPR/Cas9. Fishes 2025, 10, 40. [Google Scholar] [CrossRef]
- Niu, S.; Li, X.; Feng, C.; Zhang, Z.; Sha, H.; Luo, X.; Zou, G.; Liang, H. Targeting and Editing the Second Exon of bmp6 Gene Results in a Silver Carp with Reduced Intramuscular Bones. Aquac. Rep. 2025, 40, 102586. [Google Scholar] [CrossRef]
- Chu, W.-K.; Huang, S.-C.; Chang, C.-F.; Lin, Y.-H.; Wu, J.-L.; Gong, H.-Y. Knockout of dead end 1 by CRISPR/Cas9 Leads to Loss of Germ Cells and Male-Biased Sex Development in Freshwater Angelfish (Pterophyllum scalare). Aquaculture 2025, 599, 742180. [Google Scholar] [CrossRef]
- Sun, L.; Gao, T.; Li, Z.; Yang, X.; Qin, Z.; Ye, M.; Li, Y.; Fei, F.; Wang, D.; Wang, F. Creation of Body Color Mutants by CRISPR/Cas9 Gene Editing in Largemouth Bass (Micropterus salmoides). Aquacult. Rep. 2025, 40, 102593. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable Base Editing of A•T to G•C in Genomic DNA without DNA Cleavage. Nature 2017, 551, 464–471, Erratum in Nature 2018, 559, E8. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raghuram, A.; et al. Search-and-Replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Brookes, G.; Smyth, S.J. Risk-Appropriate Regulations for Gene-Editing Technologies. GM Crops Food 2024, 15, 1–14. [Google Scholar] [CrossRef]
- FAO (Food and Agriculture Organization of the United Nations). Gene Editing and Food Safety—Technical Considerations and Potential Relevance to the Work of Codex Alimentarius; FAO: Rome, Italy, 2023. [Google Scholar] [CrossRef]
- Wu, Y.; Wu, T.; Yang, L.; Su, Y.; Zhao, C.; Li, L.; Cai, J.; Dai, X.; Wang, D.; Zhou, L. Generation of Fast Growth Nile Tilapia (Oreochromis niloticus) by Myostatin Gene Mutation. Aquaculture 2023, 562, 738762. [Google Scholar] [CrossRef]
- Shahi, N.; Kaleem, A.; Rasooli, S.A.; Mehrjou, M.; Seyyedsharifi, N.; Sultanban Haji, A.; Rahnama, M.; Soleimannejad, H.; Yoon, D.-H.; Kim, J.-H.; et al. Muscle Growth in Targeted Knockout Common Carp (Cyprinus carpio) mstn Gene with Low Off-Target Effects. Aquaculture 2022, 565, 738955. [Google Scholar] [CrossRef]
- ISAAA (International Service for the Acquisition of Agri-Biotech Applications). Japan: Gene-Edited Animals for Agricultural Applications Database; ISAAA: [online] 2024. Available online: https://www.isaaa.org/animalbiotechdatabase/countrylist/country/default.asp?CountryID=JP&RegProcess=1 (accessed on 3 November 2025).
- Kim, J.W.; Kim, J.; Cho, J.Y.; Shin, Y.; Son, H.; Sathiyamoorthy, S.; Kim, B.S.; Kim, Y.O.; Kang, B.C.; Kong, H.J. Association Between Muscle Growth and Transcription of a Mutant MSTN Gene in Olive Flounder (Paralichthys olivaceus). Mar. Biotechnol. 2024, 26, 599–608. [Google Scholar] [CrossRef]
- Buchanan, J.; Herbert, S.; Umazume, T.; Lauth, X. Genome Editing to Produce Monosex and Sterile Fish for Aquaculture. In Proceedings of the Aquaculture Europe 2021, Funchal, Portugal, 4–7 October 2021; European Aquaculture Society: Funchal, Portugal, 2021. Available online: https://aquaeas.org/Program/PaperDetail/39097 (accessed on 3 November 2025).
- Ohga, H.; Shibata, K.; Sakanoue, R.; Ogawa, T.; Kitano, H.; Kai, S.; Ohta, K.; Nagano, N.; Nagasako, T.; Uchida, S.; et al. Development of a Chub Mackerel with Less-Aggressive Fry Stage by Genome Editing of Arginine Vasotocin Receptor V1a2. Sci. Rep. 2023, 13, 3190. [Google Scholar] [CrossRef]
- Segev-Hadar, A.; Slosman, T.; Rozen, A.; Sherman, A.; Cnaani, A.; Biran, J. Genome Editing Using the CRISPR-Cas9 System to Generate a Solid-Red Germline of Nile Tilapia (Oreochromis niloticus). CRISPR J. 2021, 4, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, J.; Du, J.; Si, Z.; Yang, H.; Xu, X.; Wang, C. ASIP Disruption via CRISPR/Cas9 System Induces Black Patches Dispersion in Oujiang Color Common Carp. Aquaculture 2019, 498, 230–235. [Google Scholar] [CrossRef]
- Pavelin, J.; Jin, Y.H.; Gratacap, R.L.; Taggart, J.B.; Hamilton, A.; Verner-Jeffreys, D.W.; Paley, R.K.; Rubin, C.J.; Bishop, S.C.; Bron, J.E.; et al. The nedd8 Activating Enzyme Gene Underlies Genetic Resistance to Infectious Pancreatic Necrosis Virus in Atlantic Salmon. Genomics 2021, 113, 3842–3850. [Google Scholar] [CrossRef]
- Liu, X.; Cai, X.; Zhang, D.; Xu, C.; Xiao, W. Zebrafish foxo3b Negatively Regulates Antiviral Response through Suppressing the Transactivity of irf3 and irf7. J. Immunol. 2016, 197, 4736–4749. [Google Scholar] [CrossRef]
- Qu, Z.L.; Gong, X.Y.; An, L.L.; Sun, H.Y.; Guo, W.H.; Luan, H.Y.; Wu, M.Y.; Dan, C.; Gui, J.F.; Zhang, Y.B. Genome Editing of FTR42 Improves Zebrafish Survival Against Virus Infection by Enhancing IFN Immunity. iScience 2024, 27, 109497. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, L.; Wu, X.; Tao, M.; Chen, Y.; Chang, M. CRISPR/Cas9 Mutagenesis Reveals an Essential Role of PI4KB in Promoting Growth and Resisting Hemorrhagic Disease Caused by GCRV-II Infection in Juvenile Grass Carp. Water Biol. Secur. 2025, 4, 100323. [Google Scholar] [CrossRef]
- Wang, J.; Su, B.; Xing, D.; Bruce, T.J.; Li, S.; Bern, L.; Shang, M.; Johnson, A.; Simora, R.M.C.; Coogan, M.; et al. Generation of Eco-Friendly and Disease-Resistant Channel Catfish (Ictalurus punctatus) Harboring the Alligator Cathelicidin Gene via CRISPR/Cas9 Engineering. Engineering 2024, 39, 273–286. [Google Scholar] [CrossRef]
- Dionglay, C. Japan’s Three Genome-Edited Food Products Reach Consumers. Science Speaks—ISAAA. International Service for the Acquisition of Agri-biotech Applications. 2022. Available online: https://www.isaaa.org/blog/entry/default.asp?BlogDate=1/19/2022 (accessed on 21 June 2023).
- Washio, Y.; Ohama, M.; Kishimoto, K.; Kinoshita, M.; Kato, K. Growth Performance and Edible Ratio of Myostatin-Knockout Young Red Sea Bream (Pagrus major) Produced by Genome Editing with CRISPR/Cas9. Aquac. Sci. 2021, 69, 101–112. [Google Scholar] [CrossRef]
- Shigi, R.; Seo, Y. Consumer Acceptance of Genome-Edited Foods in Japan. Sustainability 2023, 15, 9662. [Google Scholar] [CrossRef]
- ISAAA (International Service for the Acquisition of Agri-Biotech Applications). Gene-Edited Tilapia Not Classified as GMO in Argentina. Crop Biotech Update. 16 January 2019. Available online: https://www.isaaa.org/kc/cropbiotechupdate/article/default.asp?ID=17132 (accessed on 3 November 2025).
- ISAAA (International Service for the Acquisition of Agri-Biotech Applications). Brazilian Fish and CAT Introduce First Gene-Edited Tilapia in Brazil. Crop Biotech Update. 19 February 2025. Available online: https://www.isaaa.org/kc/cropbiotechupdate/article/default.asp?ID=21216 (accessed on 3 November 2025).
- Genetic Literacy Project. Argentina: Animals—Gene-Edited Tilapia Regulatory Status. 2023. Available online: https://crispr-gene-editing-regs-tracker.geneticliteracyproject.org/argentina-animals/ (accessed on 3 November 2025).
- Camargo, L.S.A. Brazil—Non-GMO Decisions for Genome-Edited Animals [Presentation]. In Proceedings of the 4th International Workshop on Regulatory Approaches for Agricultural Applications of Animal Biotechnologies, São Paulo, Brazil, 12–16 September 2022; International Service for the Acquisition of Agri-biotech Applications (ISAAA). Available online: https://www.isaaa.org/kc/proceedings/animalbiotechnology/2022-09-12-4th-intl-workshop/session07/pdf/43.%20Camargo%20-%20Brazil%20Non-GMO%20decisions.pdf (accessed on 22 October 2025).
- Vieira, L.R.; Freitas, N.C.; Justen, F.; Miranda, V.J.; Garcia, B.O.; Nepomuceno, A.L.; Fuganti-Pagliarini, R.; Soares Felipe, M.S.; Mertz-Henning, L.M.; Kobayashi, A.K.; et al. In CRISPR Technology in Plant Genome Editing: Biotechnology Applied to Agriculture; Molinari, H.B.C., Vieira, L.R., Silva, N.V.e., Prado, G.S., Lopes Filho, J.H., Eds.; Regulatory Framework of Genome Editing in Brazil and Worldwide. Embrapa Agroenergy: Brasília, Brazil, 2021; pp. 169–195. ISBN 978-65-87380-10-0. [Google Scholar]
- CAT (Center for Aquaculture Technologies). Brazilian Fish Announce the First Commercial-Scale Genetically Edited Tilapia for Improved Performance in Brazil. 2025. Available online: https://aquatechcenter.com/news/brazilian-fish-announce-the-first-commercial-scale-genetically-edited-tilapia-for-improved-performance-in-brazil/ (accessed on 22 October 2025).
- Puthumana, J.; Chandrababu, A.; Sarasan, M.; Joseph, V.; Singh, I.S.B. Genetic Improvement in Edible Fish: Status, Constraints, and Prospects on CRISPR-Based Genome Engineering. 3 Biotech 2024, 14, 44. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Huang, J.; Jia, J.; Lou, Q.; Shi, C.; Yasheng, M.; Zhao, Y.; Yuan, Q.; Tang, K.; Liu, X.; et al. The Food Safety Assessment of All-Female Common Carp (Cyprinus carpio) (cyp17a1+/−; XX Genotype) Generated Using Genome Editing Technology. Food Chem. Toxicol. 2023, 181, 114103. [Google Scholar] [CrossRef]
- Blix, T.B.; Myhr, A.I. A Sustainability Assessment Framework for Genome-Edited Salmon. Aquaculture 2023, 562, 738803. [Google Scholar] [CrossRef]
- Roy, S.; Kumar, V.; Behera, B.K.; Parhi, J.; Mohapatra, S.; Chakraborty, T.; Das, B.K. CRISPR/Cas Genome Editing—Can It Become a Game Changer in Future Fisheries Sector? Front. Mar. Sci. 2022, 9, 924475. [Google Scholar] [CrossRef]
- Gratacap, R.L.; Wargelius, A.; Edvardsen, R.B.; Houston, R.D. Potential of Genome Editing to Improve Aquaculture Breeding and Production. Trends Genet. 2019, 35, 672–684. [Google Scholar] [CrossRef]
- Kondo, K.; Taguchi, C. Japanese Regulatory Framework and Approach for Genome-Edited Foods Based on Latest Scientific Findings. Food Saf. 2022, 10, 113–128. [Google Scholar] [CrossRef]
- MAFF (Ministry of Agriculture, Forestry and Fisheries, Japan). ゲノム編集技術の利用により得られた生物の使用等に係る確認結果 ─ 可食部増量マダイ(E189-E90系統) [Confirmation Results on the Use of Organisms Obtained by Genome Editing Technology—Edible Part-Increased Red Seabream (E189-E90 Strain)]. MAFF: Tokyo, Japan, 2021. Available online: https://www.maff.go.jp/j/syouan/nouan/carta/tetuduki/attach/pdf/nbt_tetuzuki-10.pdf (accessed on 11 July 2025).
- MAFF (Ministry of Agriculture, Forestry and Fisheries, Japan). ノム編集技術の利用により得られた生物の使用等に係る確認結果: 高成長トラフグ(4D-4D系統) [Confirmation Results on the Use of Organisms Obtained Through Genome Editing Technology: High-Growth Tiger Puffer (4D-4D Strain)]. MAFF: Tokyo, Japan, 2021. Available online: https://www.maff.go.jp/j/syouan/nouan/carta/tetuduki/attach/pdf/nbt_tetuzuki-12.pdf (accessed on 11 July 2025).
- MAFF (Ministry of Agriculture, Forestry and Fisheries, Japan). ゲノム編集技術の利用により得られた生物の使用等に係る確認結果: 高成長ヒラメ(8D系統) [Confirmation Results on the Use of Organisms Obtained Through Genome Editing Technology: High-Growth Olive Flounder (8D Strain)]. MAFF: Tokyo, Japan, 2023. Available online: https://www.maff.go.jp/j/syouan/nouan/carta/tetuduki/attach/pdf/nbt_tetuzuki-16.pdf (accessed on 11 July 2025).
- MAFF (Ministry of Agriculture, Forestry and Fisheries, Japan). ゲノム編集技術を用いて得られた生物等の利用に関する情報提供書:可食筋肉量を増加させたマダイ(系統 E189-E90) [Information Provision Document on the Use of Organisms Obtained through Genome Editing Technology: Edible Muscle-Enhanced Red Seabream (E189-E90 Strain)]. Document Code: 様式第1(第3の1の(1)の①関係). 2021. Available online: https://www.maff.go.jp/j/syouan/nouan/carta/tetuduki/attach/pdf/nbt_tetuzuki-9.pdf (accessed on 11 July 2025).
- MAFF (Ministry of Agriculture, Forestry and Fisheries, Japan). ゲノム編集技術の利用により得られた生物の使用等に関する情報提供: 高成長トラフグ(4D-4D系統) [Information Provision Document on the Use of Organisms Obtained Through Genome Editing Technology: High-Growth Tiger Puffer (4D-4D Strain)]. Tokyo, Japan, 2021. Available online: https://www.maff.go.jp/j/syouan/nouan/carta/tetuduki/attach/pdf/nbt_tetuzuki-30.pdf (accessed on 11 July 2025).
- MAFF (Ministry of Agriculture, Forestry and Fisheries, Japan). ゲノム編集技術の利用により得られた生物の使用等に関する情報提供: 高成長ヒラメ(8D系統) [Information Provision Document on the Use of Organisms Obtained Through Genome Editing Technology: High-Growth Olive flounder (8D Strain)]. Tokyo, Japan, 2023. Available online: https://www.maff.go.jp/j/syouan/nouan/carta/tetuduki/attach/pdf/nbt_tetuzuki-17.pdf (accessed on 11 July 2025).
- MAFF (Ministry of Agriculture, Forestry and Fisheries, Japan). ゲノム編集技術の利用により得られた生物に関する生物多様性影響等検討会 議事概要(E189-E90系統) [Summary of the Expert Review Meeting on the Use of Organisms Obtained by Genome-Editing Technology and Their Potential Biodiversity Impacts (Increased Edible Portion of Red Seabream (E189-E90 Strain)]. MAFF, Tokyo, Japan, 2021. Available online: https://www.maff.go.jp/j/syouan/nouan/carta/tetuduki/attach/pdf/kentokai-10.pdf (accessed on 11 July 2025).
- MAFF (Ministry of Agriculture, Forestry and Fisheries, Japan). ゲノム編集技術の利用により得られた生物に関する生物多様性影響等検討会 議事概要: 高成長トラフグ(4D-4D系統) [Summary of the Expert Review Meeting on the Use of Organisms Obtained by Genome-Editing Technology and Their Potential Biodiversity Impacts: High-Growth Tiger Puffer (4D-4D Strain)]. MAFF, Tokyo, Japan, 2021. Available online: https://www.maff.go.jp/j/syouan/nouan/carta/tetuduki/attach/pdf/kentokai-4.pdf (accessed on 11 July 2025).
- MAFF (Ministry of Agriculture, Forestry and Fisheries, Japan). ゲノム編集技術の利用により得られた生物に関する生物多様性影響等検討会: 高成長ヒラメ(8D系統) [Summary of the Expert Review Meeting on the Use of Organisms Obtained by Genome-Editing Technology and Their Potential Biodiversity Impacts: High-Growth Olive Flounder (8D Strain)]. MAFF, Tokyo, Japan, 2023. Available online: https://www.maff.go.jp/j/syouan/nouan/carta/tetuduki/attach/pdf/kentokai-8.pdf (accessed on 11 July 2025).
- MHLW (Ministry of Health, Labour and Welfare, Japan). 別紙3–1(公表様式:食品): 名称:可食部増量マダイ(E189-E90 系統) [Appendix 3-1 (Publication Format: Food): Name: Red Sea Bream with Increased Edible portion (E189-E90 Strain)]. 2021. Available online: https://www.mhlw.go.jp/content/11120000/000833887.pdf (accessed on 11 July 2025).
- Japan Institute of Design Promotion (Good Design Award—G-Mark). Water Product Variety Development Platform—Good Design Award 2022 Winner (Award No. 22G181428). Recipient: Regional Fish Co., Ltd. 2022. Available online: https://www.g-mark.org/gallery/winners/9821 (accessed on 2 November 2025).
- MHLW (Ministry of Health, Labour and Welfare, Japan). 別紙3–1(公表様式:食品): 名称:高成長トラフグ(4D-4D 系統) [Appendix 3-1 (Publication Format: Food): Name: High-Growth Tiger Pufferfish (4D-4D Strain)] [PDF]. 2021. Available online: https://www.mhlw.go.jp/content/11120000/000849318.pdf (accessed on 11 July 2025).
- Yasuda, S. 「ゲノム編集食品は未来の食卓をどう変えるのか」 (“How Will Genome-Edited Foods Change the Future Dining Table?”)—From a Lecture by Setsuko Yasuda, Food Policy Center Vision 21. 長周新聞 [Chosyu Journal]. 11 October 2023. Available online: https://www.chosyu-journal.jp/shakai/27785 (accessed on 2 November 2025).
- MHLW (Ministry of Health, Labour and Welfare, Japan). 別紙3–1(公表様式:食品): 高成長ヒラメ(8D 系統) (Attachment 3-1 (Publication Format: Food): High-Growth Flounder (8D Strain)) [PDF]. Tokyo, Japan, 2023. Available online: https://www.mhlw.go.jp/content/11120000/001160416.pdf (accessed on 11 July 2025).
- Ramboll Japan KK. 3rd Gene Edited Fish Notified in Japan. SCC Japan: News & Guidance. 27 December 2023. Available online: https://scc-japan.jp/en/3rd-gene-edited-fish-notified-japan-2/ (accessed on 3 November 2025).
- AquaBounty. Intrexon and AquaBounty Receive Regulatory Exemption Development of Gene-Edited Tilapia. Investor News. 18 December 2018. Available online: https://investors.aquabounty.com/news-releases/news-release-details/intrexon-and-aquabounty-receive-regulatory-exemption-development (accessed on 25 October 2025).
- CTNBio (National Technical Commission on Biosafety). Normative Resolution No. 16/2018: Procedures for Analysis of Products from New Breeding Techniques (NBTs); English Summary Available via United States Department of Agriculture Foreign Agricultural Service, Agricultural Biotechnology Annual Report (BR2022-0064); CTNBio: Brasília, Brazil, 2018. Available online: https://apps.fas.usda.gov/newgainapi/api/Report/DownloadReportByFileName?fileName=Agricultural%20Biotechnology%20Annual_Brasilia_Brazil_BR2022-0064.pdf (accessed on 11 November 2025).
- Aquaculture Magazine. Brazilian Fish Announce the First Commercial-Scale Genetically Edited Tilapia for Improved Performance in Brazil. 2025. Available online: https://www.aquaculturemag.com/2025/02/21/brazilian-fish-announce-the-first-commercial-scale-genetically-edited-tilapia-for-improved-performance-in-brazil/ (accessed on 22 October 2025).
- Shukla-Jones, A.; Friedrichs, S.; Winickoff, D.E. Gene Editing in an International Context: Scientific, Economic and Social Issues Across Sectors; OECD Science, Technology and Industry Working Papers 2018, No. 2018/04; OECD Publishing: Paris, France, 2018. [Google Scholar] [CrossRef]
- Lim, D.; Yoon, J.; Lee, J.; Kim, H.; Park, S.; Cho, S. Global Trends in Regulatory Frameworks for Animal Genome Editing. J. Anim. Breed. Genet. 2023, 38, 247–259. [Google Scholar] [CrossRef]
- Tsuda, M.; Watanabe, K.N.; Ohsawa, R. Regulatory Status of Genome-Edited Organisms under the Japanese Cartagena Act. Front. Bioeng. Biotechnol. 2019, 7, 387. [Google Scholar] [CrossRef] [PubMed]
- MHLW (Ministry of Health, Labour and Welfare, Japan). Precautionary Notes on the Handling of Fishes Obtained via Genome Editing Technology; MHLW: Tokyo, Japan, 2021; English Summary Available in USDA Foreign Agricultural Service, GAIN Report No. JA2021-0156, 2021. Available online: https://apps.fas.usda.gov/newgainapi/api/Report/DownloadReportByFileName?fileName=MHLW+Publishes+Considerations+for+Genome+Edited+Fish_Tokyo_Japan_09-22-2021 (accessed on 28 October 2025).
- USDA (U.S. Department of Agriculture). Modernizing the Regulatory Framework for Agricultural Biotechnology Products. Federal Register, 14 June 2019; 84 FR 27899. Available online: https://www.federalregister.gov/documents/2019/06/14/2019-12802/modernizing-the-regulatory-framework-for-agricultural-biotechnology-products (accessed on 28 October 2025).
- FDA (U.S. Food and Drug Administration). Guidance for Industry #187: Heritable Intentional Genomic Alterations in Animals: Risk-Based Approach; FDA: Silver Spring, MD, USA, 2024. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/cvm-gfi-187a-heritable-intentional-genomic-alterations-animals-risk-based-approach (accessed on 28 October 2025).
- European Commission. Proposal for a Regulation on Plants Obtained by Certain New Genomic Techniques and Their Food and Feed, and Amending Regulation (EU) 2017/625. Brussels, Belgium; 5 July 2023. (COM(2023) 411 Final). Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A52023PC0411 (accessed on 28 October 2025).
- MARA (Ministry of Agriculture and Rural Affairs of the People’s Republic of China). Guidelines for the Safety Evaluation of Gene-Edited Plants (Trial) and Administrative Measures for Livestock and Poultry Genetic Resources. Beijing, China, 2022. Available online: https://www.moa.gov.cn/ztzl/zjyqwgz/sbzn/202201/t20220124_6387561.htm (accessed on 28 October 2025).
- Whelan, A.I.; Lema, M.A. Regulatory Framework for Gene Editing and Other New Breeding Techniques (NBTs) in Argentina. GM Crops Food 2015, 6, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Organisation for Economic Co-Operation and Development (OECD). Harmonisation of Regulatory Oversight in Biotechnology; OECD Publishing: Paris, France, 2018. [Google Scholar] [CrossRef]
- Sato, A.; Watanabe, D.; Saito, Y. Growing Knowledge Impact of Gene-Editing Technology on Public Acceptance: A Longitudinal Analysis in Japan. GM Crops Food 2024, 15, 411–428. [Google Scholar] [CrossRef]
- Woźniak, E.; Tyczewska, A.; Twardowski, T. Public Opinion on Biotechnology and Genetic Engineering in the European Union: Polish Consumer Study. BioTechnologia 2022, 103, 185–201. [Google Scholar] [CrossRef]
- Bearth, A.; Otten, C.D.; Cohen, A.S. Consumers’ Perceptions and Acceptance of Genome Editing in Agriculture: Insights from the United States of America and Switzerland. Food Res. Int. 2024, 178, 113982. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Ezaki, K.; Ito, K. Exploring the Landscape of Public Attitudes towards Gene-Edited Foods in Japan. J. Soc. Breed. Sci. 2024, 74, 11–21. [Google Scholar] [CrossRef]
- Taguchi, C.; Shibata, N.; Soga, K.; Yoshiba, S.; Narushima, J.; Sugino, M.; Kondo, K. Providing Appropriate Information to Consumers Boosts the Acceptability of Genome-Edited Foods in Japan. GM Crops Food 2023, 14, 1–14. [Google Scholar] [CrossRef]
- Martin-Collado, D.; Byrne, T.J.; Crowley, J.J.; Kirk, T.; Ripoll, G.; Whitelaw, C.B.A. Gene-Edited Meat: Disentangling Consumers’ Attitudes and Potential Purchase Behavior. Front. Nutr. 2022, 9, 856491. [Google Scholar] [CrossRef]
- Robbins, M.; Calabrese, C.; Featherstone, J.D.; Barnett, G.A. Understanding Knowledge and Perceptions of Genome Editing Technologies: A Textual Analysis of Major Agricultural Stakeholder Groups. J. Sci. Commun. 2021, 20, A07. [Google Scholar] [CrossRef]
- Gutási, A.; Hammer, S.E.; El-Matbouli, M.; Saleh, M. Recent Applications of Gene Editing in Fish Species and Aquatic Medicine. Animals 2023, 13, 1250. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Sumana, S.L.; Abdullateef, M.M.; Falayi, O.C.; Shui, Y.; Zhang, C.; Zhu, J.; Su, S. CRISPR/Cas9 Technology for Enhancing Desirable Traits of Fish Species in Aquaculture. Int. J. Mol. Sci. 2024, 25, 9299. [Google Scholar] [CrossRef] [PubMed]
- Dara, M.; Carbonara, P.; La Corte, C.; Parrinello, D.; Cammarata, M.; Parisi, M.G. Fish Welfare in Aquaculture: Physiological and Immunological Activities for Diets, Social and Spatial Stress on Mediterranean Aqua Cultured Species. Fishes 2023, 8, 414. [Google Scholar] [CrossRef]
- Sneddon, L.U.; Wolfenden, D.C.C.; Leach, M.C. In Fish Physiology—Volume 35: Wild-Caught Fish; Schreck, C.B., Tort, L., Farrell, A.P., Brauner, C.J., Eds.; Stress Management and Welfare. Elsevier: Amsterdam, The Netherlands, 2016; pp. 313–347. [Google Scholar] [CrossRef]
- Huntingford, F.A.; Adams, C.; Braithwaite, V.A.; Kadri, S.; Pottinger, T.G.; Sandøe, P.; Turnbull, J.F. Current Issues in Fish Welfare. J. Fish Biol. 2006, 68, 332–372. [Google Scholar] [CrossRef]
- Ashley, P.J. Fish Welfare: Current Issues in Aquaculture. Appl. Anim. Behav. Sci. 2007, 104, 199–235. [Google Scholar] [CrossRef]
- Percie du Sert, N.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; Emerson, M.; et al. Reporting Animal Research: Explanation and Elaboration for the ARRIVE Guidelines 2.0. PLoS Biol. 2020, 18, e3000411. [Google Scholar] [CrossRef]
- Leggatt, R.A.; Biagi, C.A.; Sakhrani, D.; Dominelli, R.; Eliason, E.J.; Farrell, A.P.; Devlin, R.H. Fitness Component Assessments of Wild-Type and Growth Hormone Transgenic Coho Salmon Reared in Seawater Mesocosms. Aquaculture 2017, 473, 31–42. [Google Scholar] [CrossRef]
- Nassar, M.; Auffan, M.; Santaella, C.; Masion, A.; Rose, J. Robustness of Indoor Aquatic Mesocosm Experimentations and Data Reusability to Assess the Environmental Risks of Nanomaterials. Front. Environ. Sci. 2021, 9, 625201. [Google Scholar] [CrossRef]
- Holman, L.E.; Garcia de la Serrana, D.; Onoufriou, A.; Hillestad, B.; Johnston, I.A. A Workflow Used to Design Low-Density SNP Panels for Parentage Assignment and Traceability in Aquaculture Species and Its Validation in Atlantic Salmon. Aquaculture 2017, 476, 59–64. [Google Scholar] [CrossRef]
- Huang, S.; Yoshitake, K.; Watabe, S.; Asakawa, S. Environmental DNA Study on Aquatic Ecosystem Monitoring and Management: Recent Advances and Prospects. J. Environ. Manag. 2022, 323, 116310. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for Scientific Data Management and Stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Y.; Luo, Z.; Lin, X.; Liu, M.; Wu, F.; Shao, H.; Zhang, W. Advances in Off-Target Detection for CRISPR-Based Genome Editing. Hum. Gene Ther. 2023, 34, 112–128. [Google Scholar] [CrossRef]
- Höijer, I.; Emmanouilidou, A.; Östlund, R.; van Schendel, R.; Bozorgpana, S.; Tijsterman, M.; Feuk, L.; Gyllensten, U.; den Hoed, M.; Ameur, A. CRISPR-Cas9 Induces Large Structural Variants at On-Target and Off-Target Sites In Vivo That Segregate Across Generations. Nat. Commun. 2022, 13, 627. [Google Scholar] [CrossRef]
- Watson, C.J.; Monstad-Rios, A.T.; Bhimani, R.M.; Gistelinck, C.; Willaert, A.; Coucke, P.; Hsu, Y.-H.; Kwon, R.Y. Phenomics-Based Quantification of CRISPR-Induced Mosaicism in Zebrafish. Cell Syst. 2020, 10, 275–286.e5. [Google Scholar] [CrossRef]
- Okoli, A.S.; Blix, T.B.; Myhr, A.I.; Xu, W.; Zhu, X. Sustainable Use of CRISPR/Cas in Fish Aquaculture: The Biosafety Perspective. Transgenic Res. 2022, 31, 1–21. [Google Scholar] [CrossRef]
| SL | Species | Year | Target Gene(s) | Effect | Ref. |
|---|---|---|---|---|---|
| 1. | Rainbow trout (Oncorhynchus mykiss) | 2013 | sdY | Induced male-to-female sex reversal | [8] |
| 2. | Yellow catfish (Pelteobagrus fulvidraco) | 2014 | mstnb | Proof-of-concept study | [31] |
| 3. | Nile tilapia (Oreochromis niloticus) | 2014 | nanos, dmrt1, foxl2 | Produced sex reversal/sterility | [2] |
| 4. | Atlantic salmon (Salmo salar) | 2014 | tyr, slc45a2 | Induced loss of pigmentation | [32] |
| 5. | Sea lamprey (Petromyzon marinus) | 2015 | tyr, fgf8/17/18 | Induced loss of pigmentation | [33] |
| 6. | Common carp (Cyprinus carpio) | 2016 | sp7, mstn | Increased muscle mass | [9] |
| 7. | Northeast Chinese lamprey (Lethenteron morii) | 2016 | gol, kctd10 | Induced loss of pigmentation | [34] |
| 8. | Rohu carp (Labeo rohita) | 2016 | tlr-22 | Decreased immunity | [35] |
| 9. | Channel catfish (Ictalurus punctatus) | 2016 | lh | Produced sterile offspring | [36] |
| 10. | Ridgetail white prawn (Exopalaemon carinicauda) | 2016 | chi4 | Heritable editing | [37] |
| 11. | Chinese tongue sole (Cynoglossus semilaevis) | 2017 | dmrt1 | Disrupted spermatogenesis | [38] |
| 12. | Red seabream (Pagrus major) | 2018 | mstn | Increased muscle mass | [11] |
| 13. | Grass carp (Ctenopharyngodon idella) | 2018 | jam-a | Reduced viral replication | [39] |
| 14. | Mexican tetra (Astyanax mexicanus) | 2018 | oca2 | Produced albino offspring | [40] |
| 15. | Olive flounder (Paralichthys olivaceus) | 2019 | mstn | Increased growth | [12] |
| 16. | Tiger puffer (Takifugu rubripes) | 2019 | mstn | Improved growth by 1.95-fold | [13] |
| 17. | Sterlet (Acipenser ruthenus) | 2019 | dnd1 | Reduced primordial germ cell | [41] |
| 18. | Pacific oyster (Crassostrea gigas) | 2019 | mstn | Induced muscle dysfunction | [42] |
| 19. | Large-scale loach (Paramisgurnus dabryanus) | 2019 | tyr | Induced loss of pigmentation | [43] |
| 20. | White crucian carp (Carassius auratus cuvieri) | 2019 | tyr | Induced loss of pigmentation | [44] |
| 21. | Pacific bluefin tuna (Thunnus orientalis) | 2019 | RyR1b | Slow swimming behavior | [45] |
| 22. | Blunt snout bream (Megalobrama amblycephala) | 2020 | mstna, mstnb | Improved growth | [46] |
| 23. | Fathead minnow (Pimephales promelas) | 2020 | tyr | Induced loss of pigmentation | [47] |
| 24. | Loach (Misgurnus anguillicaudatus) | 2021 | mstn | Increased growth | [48] |
| 25. | Mackerel tuna (Euthynnus affinis) | 2021 | slc24a5 | Reduced melanin pigments | [49] |
| 26. | Large yellow croaker (Larimichthys crocea) | 2022 | mstn | Produced F0 mutant founders | [50] |
| 27. | Malawi cichlid (Astatotilapia calliptera) | 2022 | oca2 | Reduced melanin production | [51] |
| 28. | Chub mackerel (Scomber japonicus) | 2022 | slc45a2 | Produced albino offspring | [52] |
| 29. | Freshwater prawn (Macrobrachium rosenbergii) | 2022 | pax6 | Developed distinct eye phenotype | [53] |
| 30. | Blotched snakehead (Channa maculata) | 2023 | mstn | Produced F0 mutant founders | [54] |
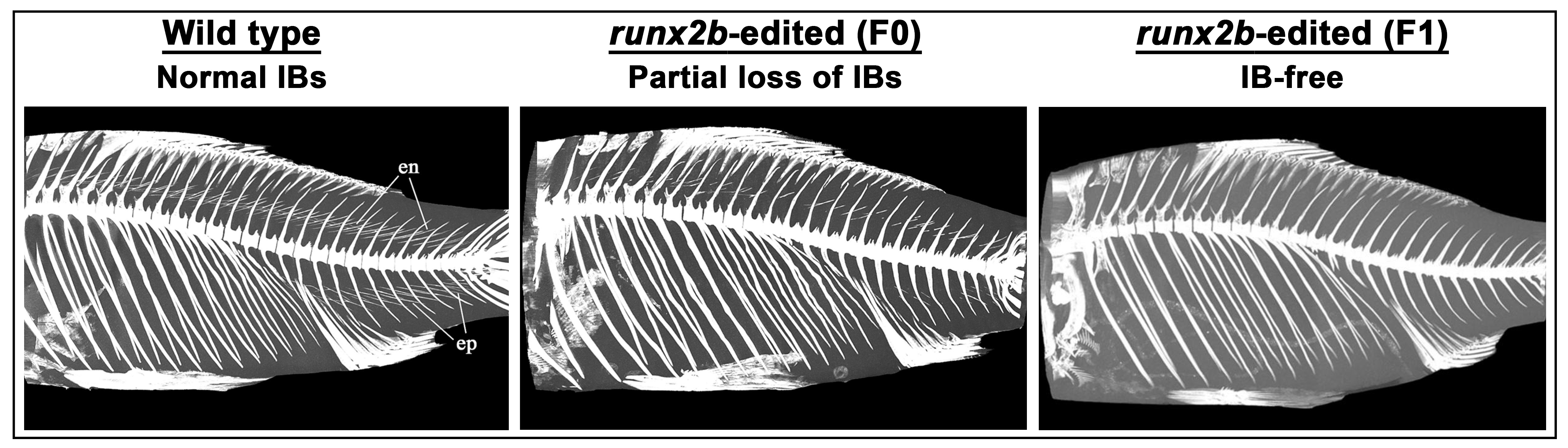
| 31. | Crucian carp (Carassius auratus) | 2023 | bmp6 | Eliminated IBs in F2 mutants | [55] |
| 32. | Gibel carp (Carassius gibelio) | 2023 | runx2b | Eliminated IBs in F3 mutants | [56] |
| 33. | African killifish (Nothobranchius furzeri) | 2023 | mitfa, ltk, csf1ra | Generate transparent fish | [57] |
| 34. | Topmouth culter (Culter alburnus) | 2024 | mstn | Increased muscle growth | [58] |
| 35. | Nibe croaker (Nibea mitsukurii) | 2024 | dnd | Eliminated germ cells | [59] |
| 36. | Striped catfish (Pangasianodon hypophthalmus) | 2024 | dnd1 | Reduced primordial germ cells | [60] |
| 37. | Atlantic cod (Gadus morhua) | 2024 | slc45a2 | Developed albino-like larvae | [61] |
| 38. | Pacific abalone (Haliotis discus hannai) | 2024 | β-tubulin | Affected cilia development | [62] |
| 39. | Large shoveljaw fish (Onychostoma macrolepis) | 2025 | mstnb, tyr, bmp6 | Improved growth, reduced IBs | [63] |
| 40. | Silver carp (Hypophthalmichthys molitrix) | 2025 | bmp6 | Reduced IBs by 30% in F0 mutants | [64] |
| 41. | Freshwater angelfish (Pterophyllum scalare) | 2025 | dnd1 | Produced a male-biased sex ratio | [65] |
| 42. | Largemouth bass (Micropterus salmoides) | 2025 | tyrb, csf1ra | tyrb mutation induced albinism | [66] |
| Country/Study | Trait | Trial System Type | Control | Welfare/Health Metric | Generation Tracked | Data Access | Key Gaps |
|---|---|---|---|---|---|---|---|
| Japan: Red seabream (mstn-KO) | Growth | Land-based aquaculture facility | Wild-type/sibling line | Mortality, growth rate, feed use efficiency | F2 | Government and regulatory reports (partial public data) | Limited ecological interaction data |
| Japan: Tiger puffer (lepr-KO) | Appetite, growth | Land-based aquaculture facility | Non-edited control | Growth, feed conversion, health screening | F1 | Regulatory dossiers and government summaries (partial public data) | Limited multi-generation data |
| Japan: Olive flounder (lepr-KO) | Appetite, growth | Land-based aquaculture facility | Non-edited control | Growth, feed conversion, health screening | F1 | Regulatory dossiers and government summaries (partial public data) | Limited multi-generation data |
| Brazil/Argentina: Nile tilapia (mstn KO) | Growth | Earthen pond | Conventional tilapia strains | Survival, growth, fertility checks | F1/F2 | Company and regulatory documents, press release | Lack of peer-reviewed, open data |
| Species | Brand and Strain Name | Country | Institute | Year | Edited Gene | Trait |
|---|---|---|---|---|---|---|
| Red seabream (Pagrus major) | 22nd century red seabream (4D strain) | Japan | Regional Fish Institute Ltd., Kyoto University, Kindai University | 2021 | mstn | Growth |
| Tiger puffer (Takifugu rubripes) | 22nd century fugu (13D strain) | Japan | Regional Fish Institute Ltd., Kyoto University | 2021 | lepr | Appetite, growth |
| Olive flounder (Paralichthys olivaceus) | 22nd century flounder (8D strain) | Japan | Regional Fish Institute Ltd., Kyoto University | 2023 | lepr | Appetite, growth |
| Nile tilapia (Oreochromis niloticus) | FLT-01 | Argentina | AquaBounty | 2018 | mstn | Growth |
| - | Brazil | CAT, Brazilian Fish | 2025 | mstn | Growth |
| Country/Region | Regulatory Model | Legal Basis/Key Agencies | Scope and Criteria | Labeling Policy | Commercial Status |
|---|---|---|---|---|---|
| Japan | Product-based (SDN-1 exemption) | MHLW, MAFF, MOE (2019 framework) | Organisms without foreign DNA are considered outside GMO law; developer notification required | Mandatory labeling: QR code-based traceability | Three fish species approved (mstn-edited red seabream, lepr-edited tiger puffer, lepr-edited olive flounder) |
| Argentina | Product-based | Resolution No. 173/2015 (CONABIA) | Organisms are considered non-GMO in the absence of foreign DNA | Not required if classified as non-GMO | mstn-edited Nile tilapia authorized (2021) |
| Brazil | Product-based | CTNBio Normative Resolution No. 16/2018 | Case-by-case determination of non-GMO status | Not required if classified as non-GMO | Pilot production of mstn-edited tilapia (2025) |
| USA | Product-based, risk-tiered | USDA and FDA (2020 modernization framework) | USDA exempts low-risk edits; FDA retains oversight for food safety and animal health | Voluntary disclosure | Research-stage GED fish; no commercial release |
| China | Developing hybrid model | MARA (2022 Guidelines) | Case-by-case biosafety evaluation and semi-field testing | To be determined | Research trials (carp, catfish); no market approvals yet |
| Canada | Process-neutral/precautionary | Health Canada (2021 Guidance) | All organisms with novel traits require premarket assessment | Case-by-case | No commercial approvals to date |
| European Union | Process-based (GMO Directive) | Directive 2001/18/EC and ECJ ruling (2018); EC, EFSA | All GED organisms regulated as GMOs | Mandatory GMO labeling | Field testing and commercialization are possible, but require full GMO authorization under Directive 2001/18/EC and Regulation (EC) No. 1829/2003 |
| Category | Reporting Elements | Minimum Requirement/Indicator | Rationale |
|---|---|---|---|
| Molecular characterization | Target gene, edit type, and verification method | Specify locus, CRISPR target, and edit category | Ensures edit traceability |
| Verification of on-target edit and allele state | Deep sequencing; confirm homozygosity, heterozygosity, or mosaicism | Confirms molecular integrity | |
| Screening for off-target mutations | Apply unbiased methods (GUIDE-seq, CIRCLE-seq, WGS) | Demonstrates genomic precision | |
| Confirmation of absence of exogenous DNA | Use PCR or WGS to detect foreign fragments | Determines SDN-1 classification | |
| Validation of heritable transmission | Multi-generational verification (F0–F2) | Confirms germline stability | |
| Performance evaluation | Quantification of production metrics | Report growth rate, feed conversion ratio, survival, and reproduction | Determines aquaculture viability |
| Inclusion of matched control group | Parallel rearing under identical environmental conditions | Isolates genomic effects | |
| Multi-environment trial design | At least two sites or seasons | Tests robustness and scalability | |
| Welfare and health | Assessment of physiological stress | Measure cortisol, HSPs, oxidative stress, and immune responses | Evaluates health resilience |
| Behavioral monitoring | Record activity, aggression, feeding rate, and social interaction | Detects welfare compromise | |
| Morphological integrity | Conduct radiographic or deformity analysis; document mortality | Identifies unintended effects | |
| Ethics and oversight | Institutional animal care approval and ARRIVE 2.0 compliance | Ensures ethical compliance | |
| Biosafety and ecological | Escape probability and containment assessment | Reports on physical and biological confinement measures | Prevents environmental release |
| Simulated ecological exposure | Use mesocosm or semi-natural setup experimental setups | Evaluates ecological risk | |
| Biological confinement | Apply sterility strategies (e.g., dnd-knockout, triploidy, hormonal induction) | Minimizes gene flow and reproductive risk | |
| Environmental monitoring | Conduct eDNA surveillance | Enables early detection of escapees | |
| Apply molecular or phenotypic tagging | Apply SNP barcoding or CRISPR-based markers | Enables traceability | |
| Data Transparency and reporting | Deposition of raw data and metadata | Submit to NCBI SRA, Dryad; ensure FAIR compliance | Enables data reuse |
| Reporting of statistical methods and null results | Include model details and all outcomes in publication | Reduces reporting bias | |
| Inclusion of socio-economic metrics | Record production cost, market response, and perception metrics | Links science to sustainability |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kho, K.H.; Sukhan, Z.P.; Cho, Y.; Cho, D.; Choi, C.Y. Genome-Edited Fish in the Field. Curr. Issues Mol. Biol. 2025, 47, 1013. https://doi.org/10.3390/cimb47121013
Kho KH, Sukhan ZP, Cho Y, Cho D, Choi CY. Genome-Edited Fish in the Field. Current Issues in Molecular Biology. 2025; 47(12):1013. https://doi.org/10.3390/cimb47121013
Chicago/Turabian StyleKho, Kang Hee, Zahid Parvez Sukhan, Yusin Cho, Doohyun Cho, and Cheol Young Choi. 2025. "Genome-Edited Fish in the Field" Current Issues in Molecular Biology 47, no. 12: 1013. https://doi.org/10.3390/cimb47121013
APA StyleKho, K. H., Sukhan, Z. P., Cho, Y., Cho, D., & Choi, C. Y. (2025). Genome-Edited Fish in the Field. Current Issues in Molecular Biology, 47(12), 1013. https://doi.org/10.3390/cimb47121013







