Morphological and Molecular Characterization of Apple Scab (Venturia inaequalis) in Kazakhstan and Kyrgyzstan
Abstract
1. Introduction
2. Materials and Methods
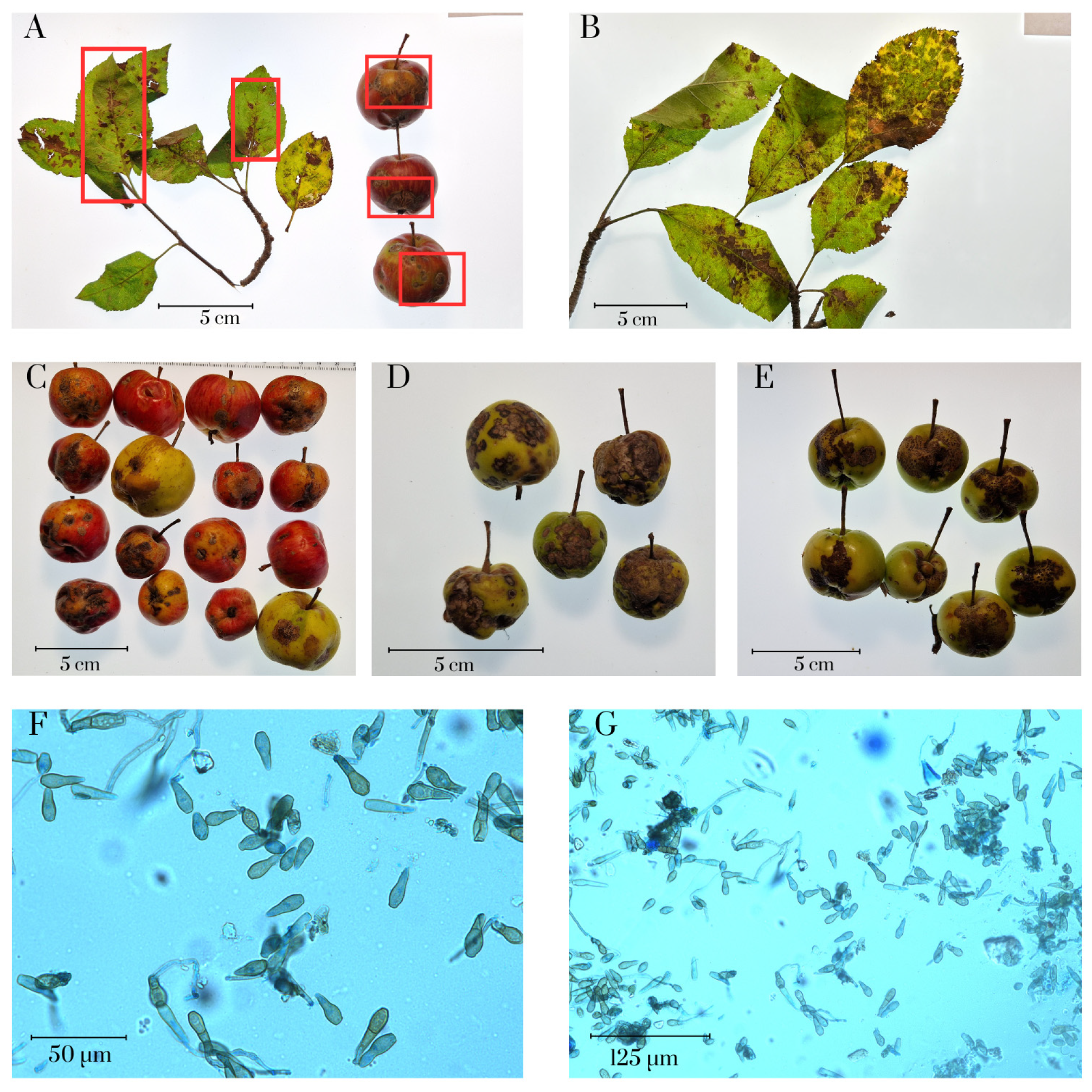
2.1. Collection and Morphology of Venturia inaequalis Isolates
2.2. Real-Time PCR Identification
2.3. Targeted DNA Sequencing Using the Oxford Nanopore Technologies (ONT) Platform
2.4. Data Analysis
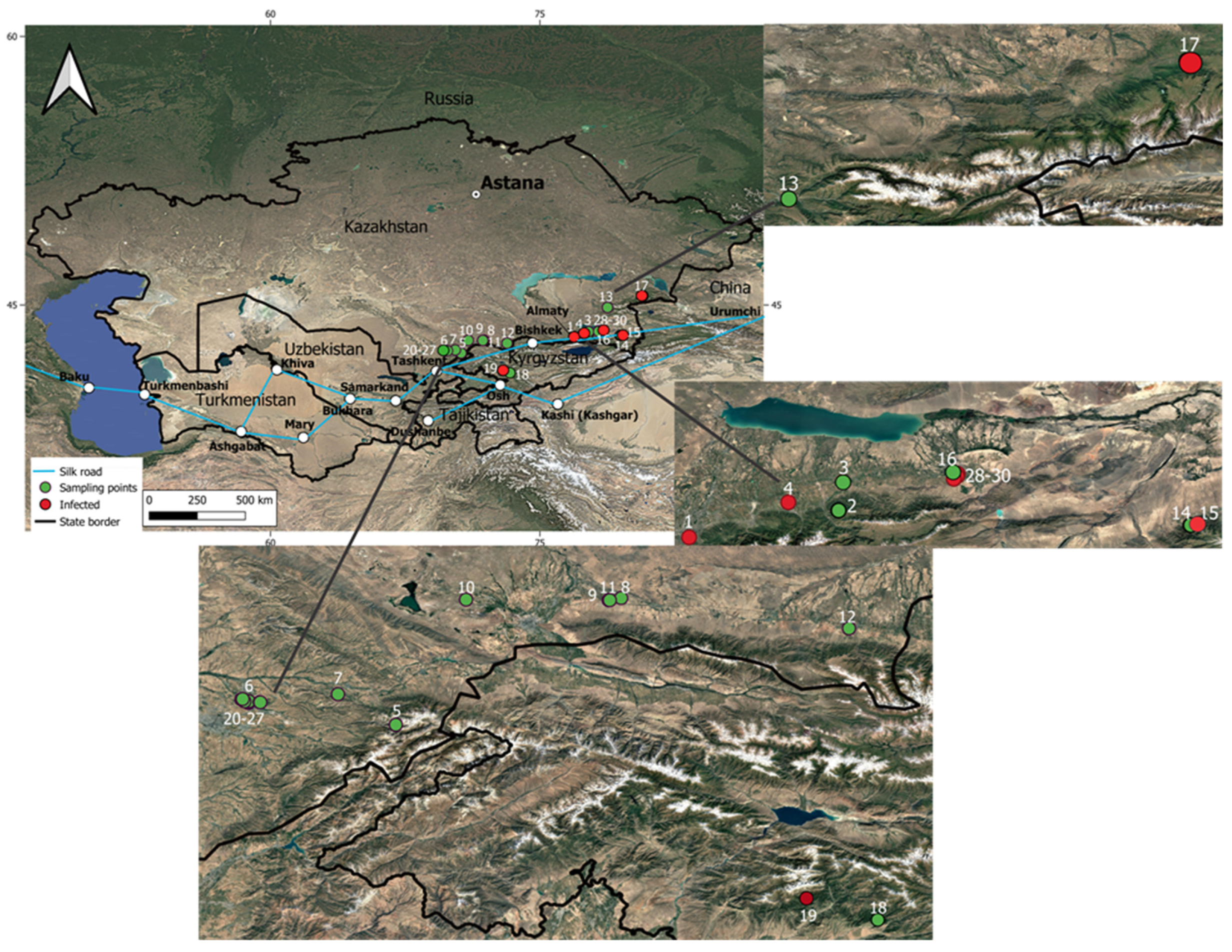
2.5. Mapping the Circulation of Fungal Pathogens
3. Results and Discussion
3.1. Morphological Analysis
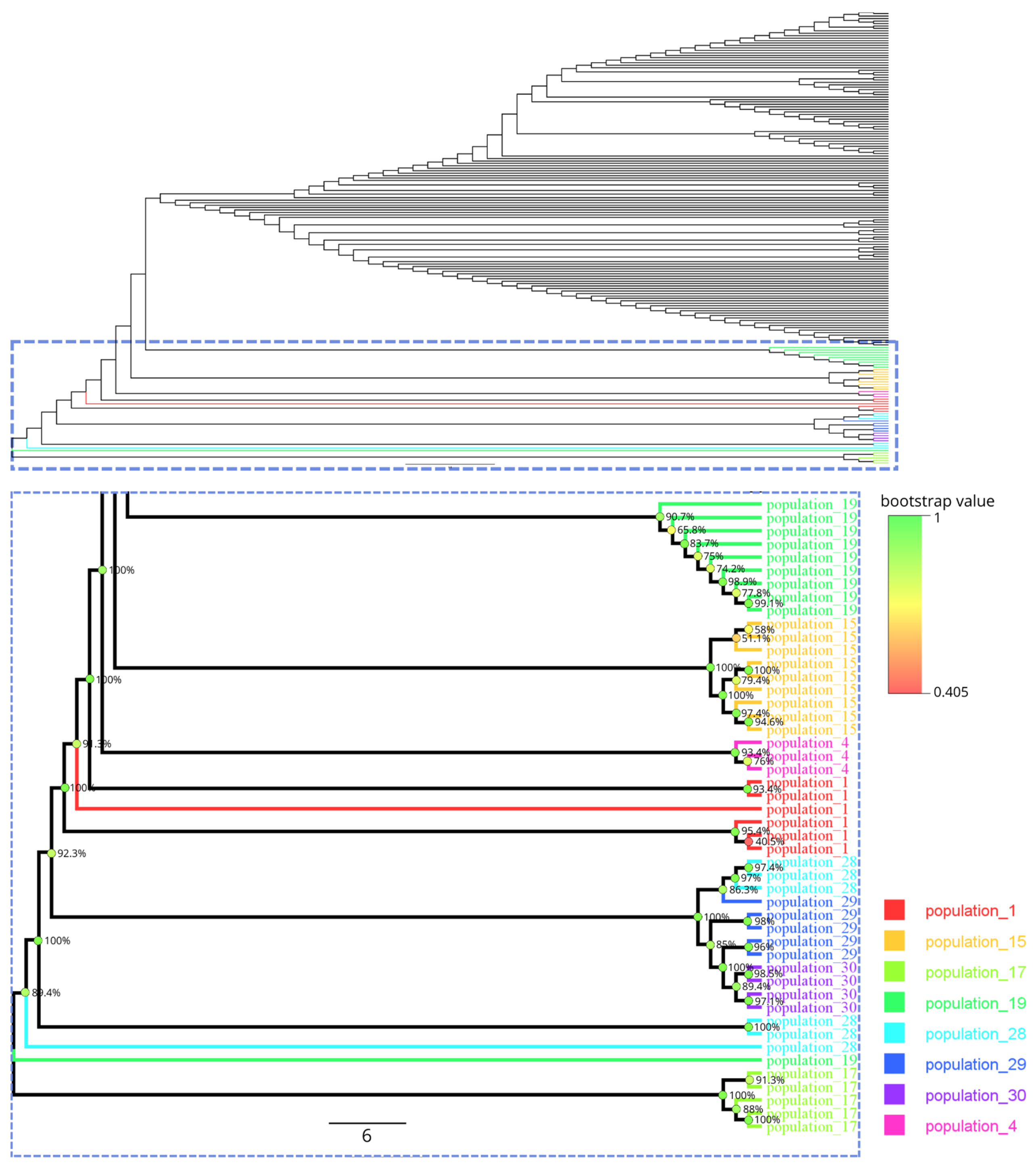
3.2. Molecular Identification of Venturia inaequalis and Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Spotts, R.A. Photosynthesis, Transpiration, and Water Potential of Apple Leaves Infected by Venturia Inaequalis. Phytopathology 1979, 69, 717. [Google Scholar] [CrossRef]
- Naqvi, S.A.M.H. (Ed.) Diseases of Fruits and Vegetables: Volume II: Diagnosis and Management; Springer: Dordrecht, The Netherlands, 2004; ISBN 978-1-4020-1823-7. [Google Scholar]
- Antal, G.; Szabó, S.; Szarvas, P.; Holb, I.J. Yield and Cost–Benefit Analyses for Apple Scab Sanitation Practices in Integrated and Organic Apple Management Systems. Plants People Planet 2024, 6, 470–489. [Google Scholar] [CrossRef]
- Švara, A.; De Storme, N.; Carpentier, S.; Keulemans, W.; De Coninck, B. Phenotyping, Genetics, and “-Omics” Approaches to Unravel and Introgress Enhanced Resistance against Apple Scab (Venturia inaequalis) in Apple Cultivars (Malus × domestica). Hortic. Res. 2024, 11, uhae002. [Google Scholar] [CrossRef]
- Chane, T.; Boyraz, N. Critical Review on Apple Scab (Venturia Inaequalis) Biology, Epidemiology, Economic Importance, Management and Defense Mechanisms to the Causal Agent. J. Plant Physiol. Pathol. 2017, 5, 2. [Google Scholar] [CrossRef]
- Bowen, J.K.; Mesarich, C.H.; Bus, V.G.M.; Beresford, R.M.; Plummer, K.M.; Templeton, M.D. Venturia Inaequalis: The Causal Agent of Apple Scab. Mol. Plant Pathol. 2011, 12, 105–122. [Google Scholar] [CrossRef]
- González-Domínguez, E.; Armengol, J.; Rossi, V. Biology and Epidemiology of Venturia Species Affecting Fruit Crops: A Review. Front. Plant Sci. 2017, 8, 1496. [Google Scholar] [CrossRef]
- Gladieux, P.; Zhang, X.-G.; Afoufa-Bastien, D.; Valdebenito Sanhueza, R.-M.; Sbaghi, M.; Le Cam, B. On the Origin and Spread of the Scab Disease of Apple: Out of Central Asia. PLoS ONE 2008, 3, e1455. [Google Scholar] [CrossRef]
- Lê Van, A.; Gladieux, P.; Lemaire, C.; Cornille, A.; Giraud, T.; Durel, C.-E.; Caffier, V.; Le Cam, B. Evolution of Pathogenicity Traits in the Apple Scab Fungal Pathogen in Response to the Domestication of Its Host. Evol. Appl. 2012, 5, 694–704. [Google Scholar] [CrossRef]
- Gladieux, P.; Zhang, X.-G.; Róldan-Ruiz, I.; Caffier, V.; Leroy, T.; Devaux, M.; Van Glabeke, S.; Coart, E.; Le Cam, B. Evolution of the Population Structure of Venturia Inaequalis, the Apple Scab Fungus, Associated with the Domestication of Its Host. Mol. Ecol. 2010, 19, 658–674. [Google Scholar] [CrossRef]
- Tegtmeier, R.; Švara, A.; Gritsenko, D.; Khan, A. Malus Sieversii: A Historical, Genetic, and Conservational Perspective of the Primary Progenitor Species of Domesticated Apples. Hortic. Res. 2025, 12, uhae244. [Google Scholar] [CrossRef] [PubMed]
- Kanat, G.; Suleimanova, G.; Irkitbay, A.; Madenova, A.; Aitymbet, Z. Fungal Diseases of Apple Trees in Kazakhstan. In Proceedings of the The XIII International Science Conference «Perspectives of Development of Science and Practice», Prague, Czech Republic, 14–17 December 2021; 626p. pp. 20–25. [Google Scholar]
- Madenova, A.; Aitymbet, Z.; Bolat, M.; Kaldybayeva, D.; Galymbek, K.; Kuan, A.; Kabylbekova, B.; Irkitbay, A.; Yeszhanov, T.; Bakirov, S.; et al. Screening of Apple Cultivars for Scab Resistance in Kazakhstan. Horticulturae 2024, 10, 184. [Google Scholar] [CrossRef]
- Urazova, M.; Satenova, A.M.; Askarova, M.; Tuyakova, A.; Abilkhadirov, A.; Shaikhin, S. Biocontrol Activity of Metschnikowia Pulcherrima Strains Isolated from Local Varieties of Apples in Kazakhstan. Int. J. Agric. Biosci. 2025, 14, 265–275. [Google Scholar] [CrossRef]
- Papp, D.; Gao, L.; Thapa, R.; Olmstead, D.; Khan, A. Field Apple Scab Susceptibility of a Diverse Malus Germplasm Collection Identifies Potential Sources of Resistance for Apple Breeding. CABI Agric. Biosci. 2020, 1, 16. [Google Scholar] [CrossRef]
- Li, X.; Li, H.; Yu, Z.; Gao, L.; Yang, J. Investigating the Sensitivity of Venturia Inaequalis Isolates to Difenoconazole and Pyraclostrobin in Apple Orchards in China. Eur. J. Plant Pathol. 2021, 161, 207–217. [Google Scholar] [CrossRef]
- Prencipe, S.; Sillo, F.; Garibaldi, A.; Gullino, M.L.; Spadaro, D. Development of a Sensitive TaqMan qPCR Assay for Detection and Quantification of Venturia Inaequalis in Apple Leaves and Fruit and in Air Samples. Plant Dis. 2020, 104, 2851–2859. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chaisiri, C.; Luo, M.; Fan, F.; Wang, Y.-F.; Yin, L.-F.; Yin, W.-X.; Luo, C.-X. Genetic Diversity of Venturia Carpophila Populations from Different Hosts and Geographic Regions in China. Front. Microbiol. 2022, 13, 985691. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; The UGENE Team. Unipro UGENE: A Unified Bioinformatics Toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, C.N. Statistical Analysis of Real-Time PCR Data. BMC Bioinform. 2006, 7, 85. [Google Scholar] [CrossRef]
- About the Silk Roads | Silk Roads Programme. Available online: https://en.unesco.org/silkroad/about-silk-roads (accessed on 26 November 2025).
- Apple Scab Fruit Fact Sheet | CALS. Available online: https://cals.cornell.edu/integrated-pest-management/outreach-education/fact-sheets/apple-scab-fruit-fact-sheet (accessed on 17 November 2025).
- Apple Scab Disease. Available online: https://www.ages.at/en/plant/plant-health/pests-from-a-to-z/apple-scab-disease (accessed on 17 November 2025).
- Jha, G.; Thakur, K.; Thakur, P. The Venturia Apple Pathosystem: Pathogenicity Mechanisms and Plant Defense Responses. BioMed Res. Int. 2009, 2009, 680160. [Google Scholar] [CrossRef] [PubMed]
- Gouit, S.; Chiadmi, S.; Goura, K.; Legrifi, I.; El Jarroudi, M.; Belabess, Z.; Tahiri, A.; Lazraq, A.; Baala, M.; Lahlali, R. Assessing Venturia Inaequalis Response to Common Fungicides in Morocco. J. Fungi 2025, 11, 493. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, C.F.A.; Steiner, U.; Dehne, H.-W.; Oerke, E.-C. Localized Adhesion of Nongerminated Venturia Inaequalis Conidia to Leaves and Artificial Surfaces. Phytopathology 2008, 98, 760–768. [Google Scholar] [CrossRef] [PubMed]
| Value | Infection Severity |
|---|---|
| 2 | Less than 1% of leaves and fruits affected; symptoms are noticeable only upon close inspection |
| 3 | Up to 5% of affected organs, with no noticeable impact on the tree |
| 4 | Intermediate |
| 5 | Affected leaves and fruits are widely distributed, involving a significant portion of the tree, with approximately 25% infection severity |
| 6 | Intermediate |
| 7 | Severe infection: approximately 50% of organs heavily affected |
| Primer Design | Name | 5′-3′ Sequence | Source |
|---|---|---|---|
| Real-time PCR analysis | F1 | F: CACTTCCCCGCTATTCACGT | [17] |
| R11 | R: GCAATCGTTAGCATCGTCATAGTG | ||
| Ven1 | [FAM]CTCAAGGCAGCCCAACTTTCTCCGGT[BHQ1] | ||
| ONT sequencing | ITS4 | F: TCCTCCGCTTATTGATATGC | [18] |
| ITS5 | R: GGAAGTAAAAGTCGTAACAAGG |
| Garden Definition | Region | No. on Map | Sampling Location | Coordinates |
|---|---|---|---|---|
| Cultivated apple orchards | Almaty Region | 1 | Institute of Plant Biology and Biotechnology | 43.2268, 76.91625 |
| 2 | v. Shelek | N/A | ||
| 3 | v. Malovodnoye | 43.51056, 77.70278 | ||
| 4 | v. Bolek | 43.40639, 77.42306 | ||
| Turkestan Region | 5 | Bekzhan Farm | 42.31667, 70.61667 | |
| 6 | Shymkent, “Dala Fruit” | 42.44023, 69.7849 | ||
| 7 | Tyulkubas District, “Almaly Sai” | 42.49, 70.29 | ||
| Zhambyl Region | 8 | Taraz, “Ecofruit” | 43.03519, 71.88858 | |
| 9 | Taraz, “Alina Apple” | 43.02272, 71.81756 | ||
| 10 | Taraz, “Auli Ata” | 43.02607, 71.0131 | ||
| 11 | Taraz, “Grand apple” | 43.02285, 71.82382 | ||
| 12 | v. Merke | 42.86333, 73.17417 | ||
| Zhetysu Region | 13 | Tekeli | 44.86306, 78.76417 | |
| Wild apple tree populations | Almaty Region | 14 | Sumbé | 43.29306, 79.48417 |
| 15 | Ketpentau | 43.30528, 79.75139 | ||
| 16 | Ile-Alatau State National Natural Park | 43.36424, 77.68041 | ||
| Zhetysu Region | 17 | Zhongar-Alatau State National Natural Park | 45.51746, 80.72224 | |
| Kyrgyz Republic | 18 | v. Kara-Alma | 41.210000953, 73.336669017 | |
| 19 | Arslan Bob | 41.333334292, 72.933335690 | ||
| Abandoned cultivated apple orchards | Turkestan Region | 20 | Garden 1 | 42.44863821, 69.78857 |
| 21 | Garden 2 | 42.44779721, 69.79035526 | ||
| 22 | Garden 3 | 42.44968945, 69.78657079 | ||
| 23 | Garden 4 | 42.44874333, 69.76197175 | ||
| 24 | Garden 5 | 42.46041211, 69.74557239 | ||
| 25 | Garden 6 | 42.46230434, 69.75187984 | ||
| 26 | Garden 7 | 42.44522167, 69.84775302 | ||
| 27 | Garden 8 | 42.44401275, 69.85153749 | ||
| Almaty Region | 28 | Garden 11 | 43.55170546, 78.28706375 | |
| 29 | Garden 12 | 43.55202083, 78.28900855 | ||
| 30 | Garden 13 | 43.54077255, 78.28506639 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostyukova, V.; Pozharskiy, A.; Khusnitdinova, M.; Nizamdinova, G.; Gritsenko, D. Morphological and Molecular Characterization of Apple Scab (Venturia inaequalis) in Kazakhstan and Kyrgyzstan. Curr. Issues Mol. Biol. 2025, 47, 1011. https://doi.org/10.3390/cimb47121011
Kostyukova V, Pozharskiy A, Khusnitdinova M, Nizamdinova G, Gritsenko D. Morphological and Molecular Characterization of Apple Scab (Venturia inaequalis) in Kazakhstan and Kyrgyzstan. Current Issues in Molecular Biology. 2025; 47(12):1011. https://doi.org/10.3390/cimb47121011
Chicago/Turabian StyleKostyukova, Valeriya, Alexandr Pozharskiy, Marina Khusnitdinova, Gulnaz Nizamdinova, and Dilyara Gritsenko. 2025. "Morphological and Molecular Characterization of Apple Scab (Venturia inaequalis) in Kazakhstan and Kyrgyzstan" Current Issues in Molecular Biology 47, no. 12: 1011. https://doi.org/10.3390/cimb47121011
APA StyleKostyukova, V., Pozharskiy, A., Khusnitdinova, M., Nizamdinova, G., & Gritsenko, D. (2025). Morphological and Molecular Characterization of Apple Scab (Venturia inaequalis) in Kazakhstan and Kyrgyzstan. Current Issues in Molecular Biology, 47(12), 1011. https://doi.org/10.3390/cimb47121011






