CAMF-DTI: Enhancing Drug–Target Interaction Prediction via Coordinate Attention and Multi-Scale Feature Fusion
Abstract
1. Introduction
- To tackle the lack of directional information in spatial features, coordinate attention is introduced to jointly encode spatial position and sequence directionality, improving the localization of key interaction regions.
- To address the limitation of single-scale receptive fields, a multi-scale fusion module is designed to extract structural features at different scales via parallel branches, enabling unified representation of local and global information.
- To overcome insufficient modeling of drug–target interactions, a cross-attention mechanism is employed to capture dynamic dependencies between drugs and targets during representation learning.
- Experimental results on multiple benchmark datasets demonstrate that CAMF-DTI outperforms existing state-of-the-art methods across AUROC, AUPRC, Accuracy, F1 score, and MCC metrics, indicating superior predictive performance and strong generalization ability.
2. Materials and Methods
2.1. The Architecture of CAMF-DTI
2.2. Drug Encoder
2.2.1. Drug Feature Construction
2.2.2. GCN
2.2.3. Multi-Scale Feature Fusion
2.3. Protein Encoder
2.3.1. Protein Feature Construction
2.3.2. Coordinate Attention Mechanism
2.4. Feature Fusion Stage
2.4.1. Interaction Feature Extraction Module
2.4.2. Prediction Module
3. Results and Discussion
3.1. Evaluation Metrics
3.2. Datasets
3.3. Baseline Methods
- CPI-GNN [30]: Uses GNNs for drug graphs and CNNs for protein sequences, with a single-sided attention mechanism to model the effect of protein subsequences on drugs.
- BACPI [31]: Combines CNNs and dual attention to extract and integrate compound–protein features, emphasizing local interaction sites.
- CPGL [32]: Applies GAT and LSTM for robust and generalizable compound–protein feature extraction, followed by a fully connected layer.
- BINDTI [33]: Encodes drugs via molecular graphs and proteins via ACMix, then fuses their features using a bidirectional intention network.
- FOTF-CPI [34]: Enhances Transformer with optimal fragmentation and attention fusion to improve interaction prediction.
- CAT-CPI [35]: Employs CNNs and Transformers to encode proteins and captures drug–target interaction via cross-attention.
- DO-GMA [36]: Extracts representations using CNNs and GCNs and integrates features via gated and multi-head attention with bilinear fusion.
3.4. Results
3.4.1. Analysis of Performance
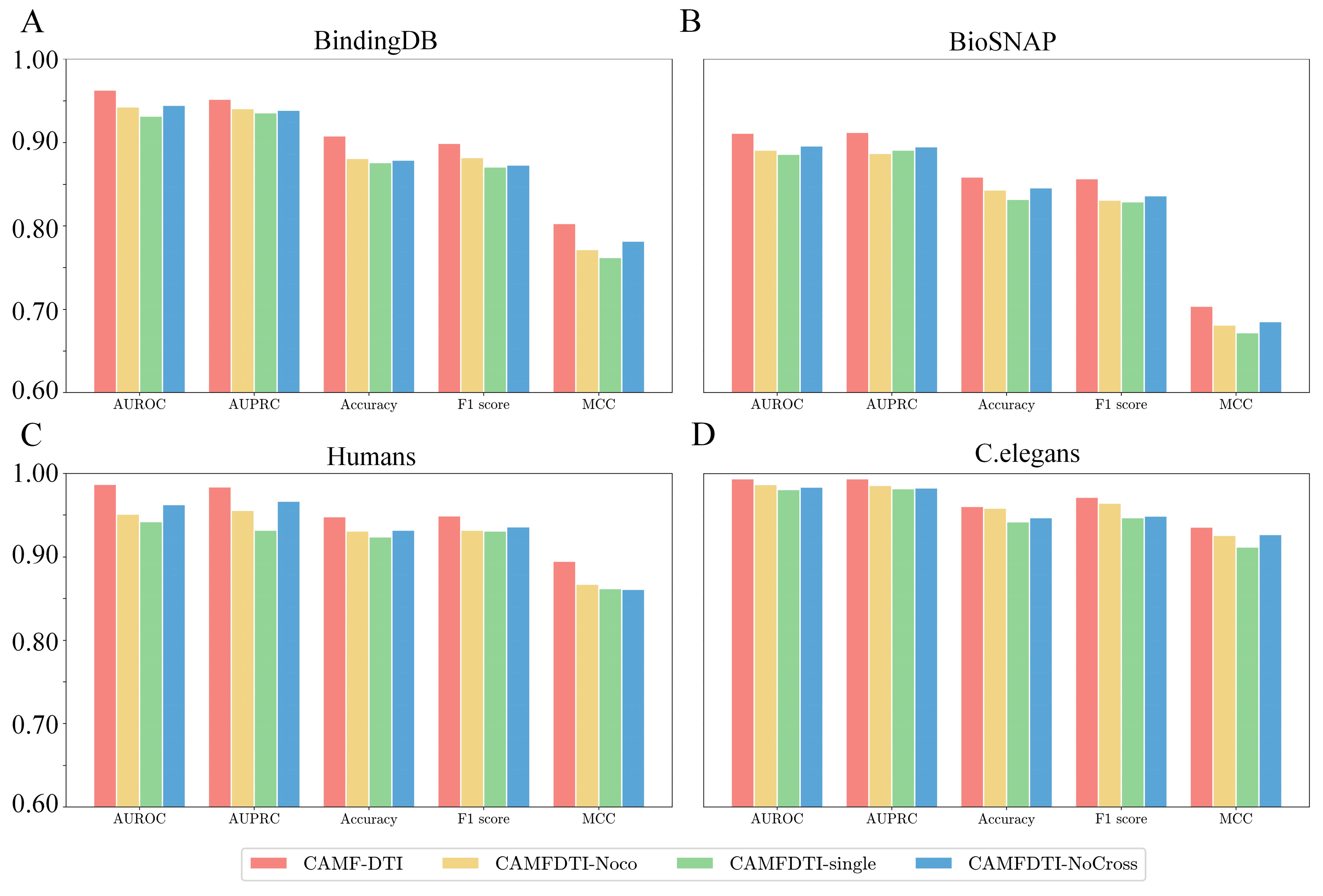
3.4.2. Ablation Experiments
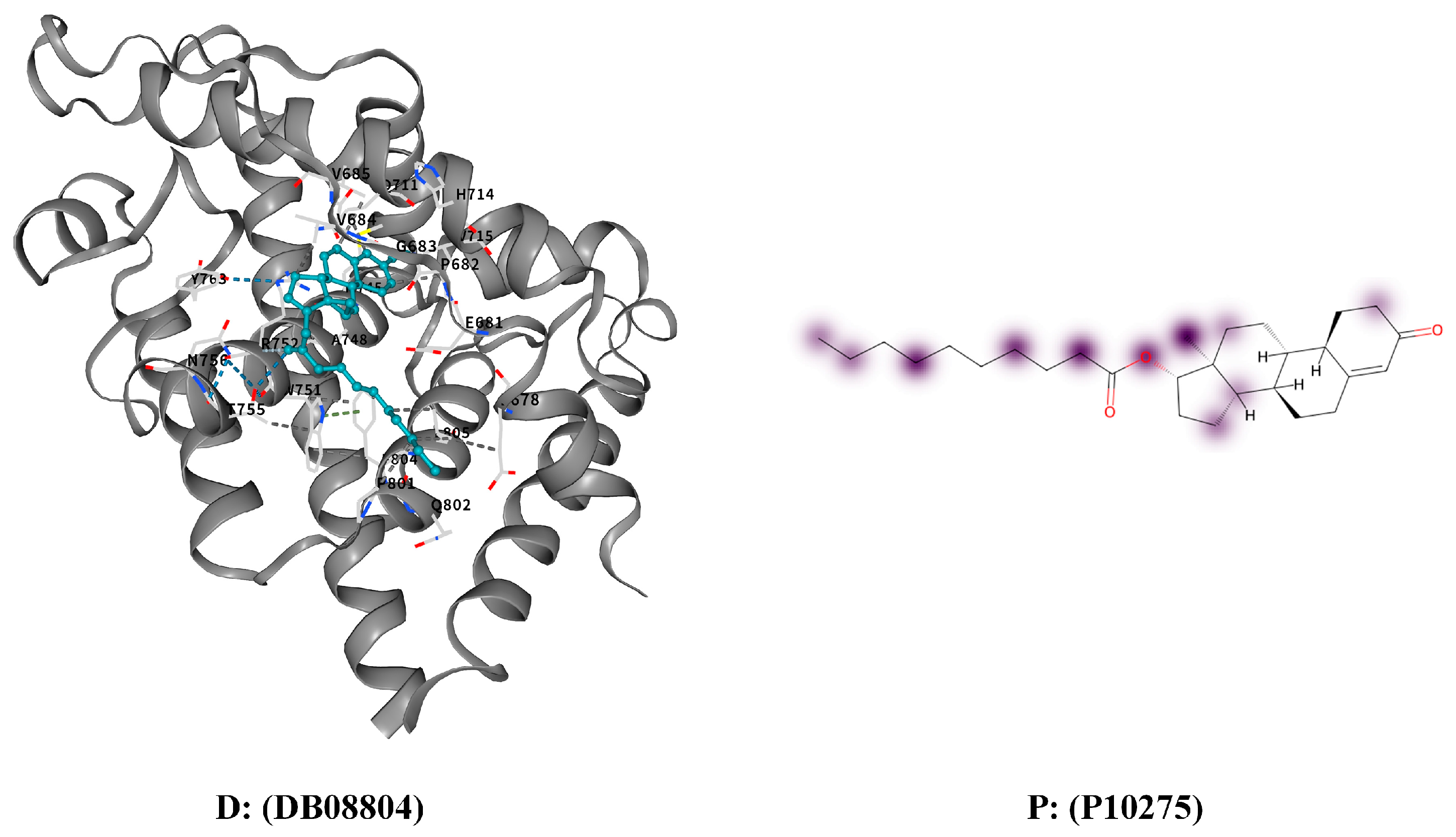
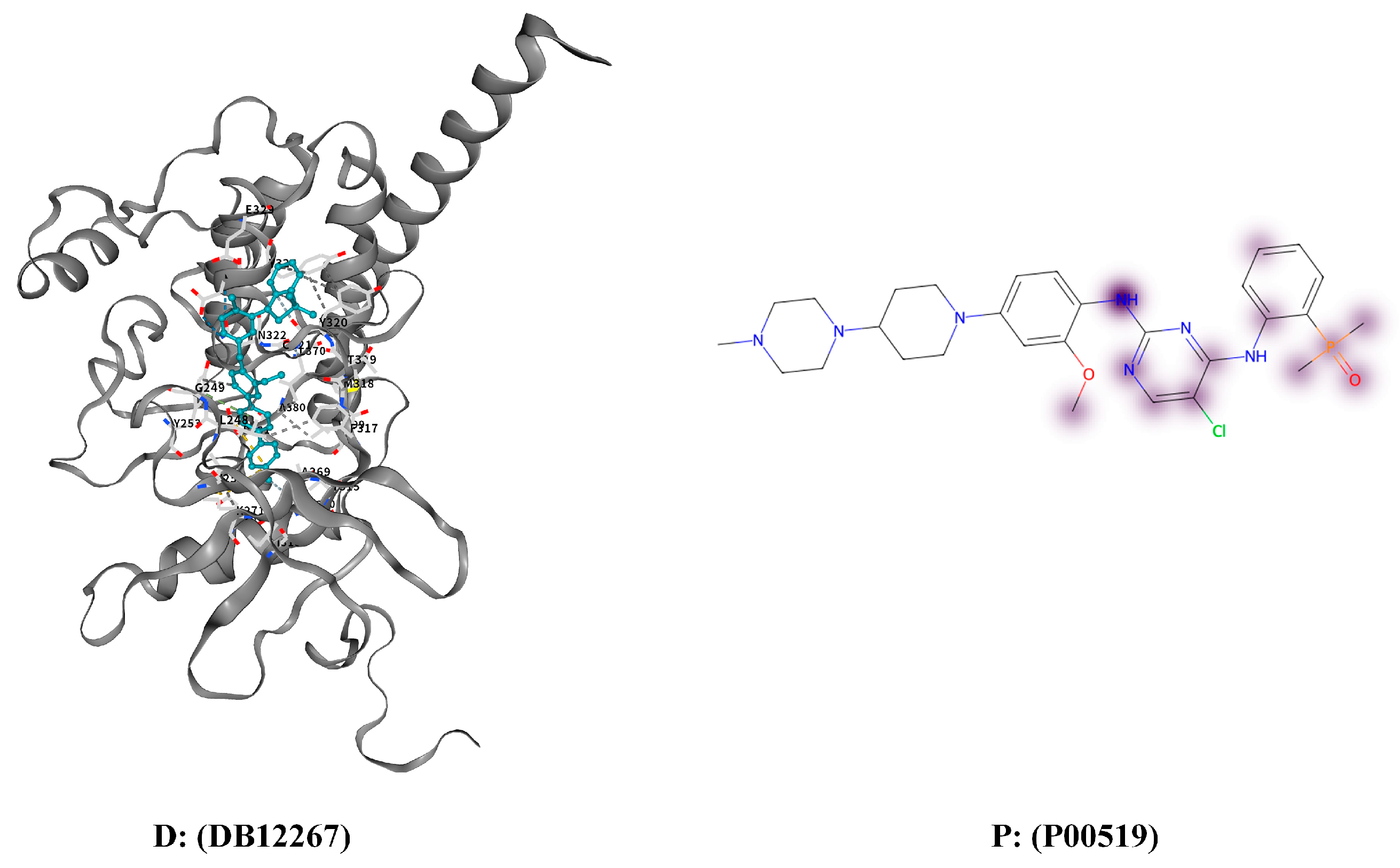
3.4.3. Interpretation and Case Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, M.; Guo, Z.; Wu, Y.; Guo, P.; Shi, Y.; Hu, S.; Wan, W.; Hu, S. ViDTA: Enhanced Drug-Target Affinity Prediction via Virtual Graph Nodes and Attention-based Feature Fusion. In Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Lisbon, Portugal, 3–6 December 2024. [Google Scholar]
- Luo, Z.; Wu, W.; Sun, Q.; Wang, J. Accurate and transferable drug–target interaction prediction with DrugLAMP. Bioinformatics 2024, 40, btae693. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Wu, C.; Zhan, L.; Wang, A.; Cheng, C.; Zhao, J.; Zhang, W.; Chen, J.; Li, P. Drug–target interaction prediction by integrating heterogeneous information with mutual attention network. BMC Bioinform. 2024, 25, 361. [Google Scholar] [CrossRef]
- Meng, Z.; Meng, Z.; Yuan, K.; Ounis, I. FusionDTI: Fine-grained Binding Discovery with Token-level Fusion for Drug–Target Interaction. In Proceedings of the 2025 Conference on Empirical Methods in Natural Language Processing, Suzhou, China, 4–9 November 2025. [Google Scholar] [CrossRef]
- Liu, B.; Wu, S.; Wang, J.; Deng, X.; Zhou, A. HiGraphDTI: Hierarchical Graph Representation Learning for Drug–Target Interaction Prediction. In Joint European Conference on Machine Learning and Knowledge Discovery in Databases; Springer: Cham, Switzerland, 2024. [Google Scholar]
- Li, C.; Mi, J.; Wang, H.; Liu, Z.; Gao, J.; Wan, J. MGMA-DTI: Drug target interaction prediction using multi-order gated convolution and multi-attention fusion. Comput. Biol. Chem. 2025, 118, 108449. [Google Scholar] [CrossRef]
- Honda, S.; Shi, S.; Ueda, H.R. Smiles transformer: Pre-trained molecular fingerprint for low data drug discovery. arXiv 2019, arXiv:1911.04738. [Google Scholar] [CrossRef]
- Ozturk, H.; Ozgur, A.; Ozkirimli, E. DeepDTA: Deep drug–target binding affinity prediction. Bioinformatics 2018, 34, i821–i829. [Google Scholar] [CrossRef]
- Öztürk, H.; Ozkirimli, E.; Özgür, A. WideDTA: Prediction of Drug-Target Binding Affinity. arXiv 2019, arXiv:1902.04166. [Google Scholar]
- Chen, L.; Tan, X.; Wang, D.; Zhong, F.; Liu, X.; Yang, T.; Luo, X.; Chen, K.; Jiang, H.; Zheng, M. TransformerCPI: Improving compound–protein interaction prediction by sequence-based deep learning with self-attention mechanism. Bioinformatics 2020, 36, 4406–4414. [Google Scholar] [CrossRef]
- Huang, K.; Xiao, C.; Glass, L.M.; Sun, J. MolTrans: Molecular interaction transformer for drug–target interaction prediction. Bioinformatics 2021, 37, 830–836. [Google Scholar] [CrossRef]
- Nguyen, T.; Le, H.; Quinn, T.P.; Nguyen, T.; Le, T.D.; Venkatesh, S. GraphDTA: Predicting Drug–Target Binding Affinity with Graph Neural Networks. Bioinformatics 2021, 37, 1140–1147. [Google Scholar] [CrossRef]
- Yang, Z.; Zhong, W.; Zhao, L.; Chen, C.Y.-C. MGraphDTA: Deep Multiscale Graph Neural Network for Explainable Drug–Target Binding Affinity Prediction. Chem. Sci. 2022, 13, 816–833. [Google Scholar] [CrossRef]
- Wan, F.; Hong, L.; Xiao, A.; Jiang, T.; Zeng, J. NeoDTI: Neural integration of neighbor information from a heterogeneous network for discovering new drug–target interactions. Bioinformatics 2019, 35, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Du, G.; Hu, J.; Cai, Y. Graph-DTI: A New Model for Drug-Target Interaction Prediction Based on Heterogenous Network Graph Embedding. Curr. Comput.-Aided Drug Des. 2024, 20, 1013–1024. [Google Scholar] [CrossRef]
- Stark, A.; Pinsler, R.; Corso, G.; Fout, A.; Zitnik, M. EquiBind: Geometric deep learning for drug binding structure prediction. In Proceedings of the International Conference on Machine Learning (ICML), Baltimore, MD, USA, 17–23 July 2022; pp. 20503–20521. [Google Scholar]
- Wang, C.; Liu, Y.; Song, S.; Cao, K.; Liu, X.; Sharma, G.; Guo, M. DHAG-DTA: Dynamic Hierarchical Affinity Graph Model for Drug–Target Binding Affinity Prediction. IEEE Trans. Comput. Biol. Bioinform. 2025, 22, 697–709. [Google Scholar] [CrossRef]
- Liu, T.; Lin, Y.; Wen, X.; Jorissen, R.N.; Gilson, M.K. BindingDB: A web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 2007, 35, D198–D201. [Google Scholar] [CrossRef]
- Davis, M.I.; Hunt, J.P.; Herrgard, S.; Ciceri, P.; Wodicka, L.M.; Pallares, G.; Hocker, M.; Treiber, D.K.; Zarrinkar, P.P. Comprehensive analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2011, 29, 1046–1051. [Google Scholar] [CrossRef]
- Tang, J.; Szwajda, A.; Shakyawar, S.; Xu, T.; Hintsanen, P.; Wennerberg, K.; Aittokallio, T. Making sense of large-scale kinase inhibitor bioactivity data sets: A comparative and integrative analysis. J. Chem. Inf. Model. 2014, 54, 735–743. [Google Scholar] [CrossRef]
- Yao, Q.; Chen, Z.; Cao, Y.; Hu, H. Enhancing Drug-Target Interaction Prediction with Graph Representation Learning and Knowledge-Based Regularization. Front. Bioinform. 2025, 5, 1649337. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, X.; Pan, Z.; Rodríguez Patón, A.; Wang, S.; Song, T.; Gu, Y. Multi-TransDTI: Transformer for Drug–Target Interaction Prediction Based on Simple Universal Dictionaries with Multi-View Strategy. Biomolecules 2022, 12, 644. [Google Scholar] [CrossRef]
- Geng, G.; Wang, L.; Xu, Y.; Wang, T.; Ma, W.; Duan, H.; Zhang, J.; Mao, A. MGDDI: A multi-scale graph neural networks for drug–drug interaction prediction. Methods 2024, 228, 22–29. [Google Scholar] [CrossRef]
- Alon, U.; Yahav, E. On the Bottleneck of Graph Neural Networks and its Practical Implications. arXiv 2020, arXiv:2006.05205. [Google Scholar]
- Ouyang, D.; He, S.; Zhang, G.; Luo, M.; Guo, H.; Zhan, J.; Huang, Z. Efficient Multi-Scale Attention Module with Cross-Spatial Learning. In Proceedings of the ICASSP 2023—IEEE International Conference on Acoustics, Speech and Signal Processing, Rhodes Island, Greece, 4–10 June 2023; pp. 1–5. [Google Scholar] [CrossRef]
- Hou, Q.; Zheng, M.; Lu, M.; Wu, E. Coordinate Attention for Efficient Mobile Network Design. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 13713–13722. [Google Scholar] [CrossRef]
- McDermott, M.; Zhang, H.; Hansen, L.; Angelotti, G.; Gallifant, J. A closer look at AUROC and AUPRC under class imbalance. Adv. Neural Inf. Process. Syst. 2024, 37, 44102–44163. [Google Scholar]
- Metrics and Scoring: Quantifying the Quality of Predictions. Available online: https://scikit-learn.org/stable/modules/model_evaluation.html (accessed on 12 October 2025).
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef]
- Tsubaki, M.; Tomii, K.; Sese, J. Compound–Protein Interaction Prediction with End-to-End Learning of Neural Networks for Graphs and Sequences. Bioinformatics 2019, 35, 309–318. [Google Scholar] [CrossRef]
- Li, M.; Lu, Z.; Wu, Y.; Li, Y.H. BACPI: A bi-directional attention neural network for compound–protein interaction and binding affinity prediction. Bioinformatics 2022, 38, 1995–2002. [Google Scholar] [CrossRef]
- Zhao, M.; Yuan, M.; Yang, Y.; Xu, S.X. CPGL: Prediction of Compound-Protein Interaction by Integrating Graph Attention Network with Long Short-Term Memory Neural Network. IEEE/ACM Trans. Comput. Biol. Bioinform. 2023, 20, 1935–1942. [Google Scholar] [CrossRef]
- Peng, L.; Liu, X.; Yang, L.; Liu, L.; Bai, Z.; Chen, M.; Lu, X.; Nie, L. BINDTI: A Bi-Directional Intention Network for Drug–Target Interaction Identification Based on Attention Mechanisms. IEEE J. Biomed. Health Inform. 2025, 29, 1602–1612. [Google Scholar] [CrossRef]
- Yin, Z.; Chen, Y.; Hao, Y.; Pandiyan, S.; Shao, J.; Wang, L. FOTF-CPI: A compound-protein interaction prediction transformer based on the fusion of optimal transport fragments. iScience 2023, 27, 108756. [Google Scholar] [CrossRef]
- Qian, Y.; Wu, J.; Zhang, Q. CAT-CPI: Combining CNN and transformer to learn compound image features for predicting compound-protein interactions. Front. Mol. Biosci. 2022, 9, 963912. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Mao, J.; Huang, G.; Han, G.; Liu, X.; Liao, W.; Tian, G.; Yang, J. DO-GMA: An End-to-End Drug–Target Interaction Identification Framework with a Depthwise Overparameterized Convolutional Network and the Gated Multihead Attention Mechanism. J. Chem. Inf. Model. 2025, 65, 1318–1337. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Watton, P.N.; Neu, C.P.; Pierce, D.M. A chemo-mechano-biological modeling framework for cartilage evolving in health, disease, injury, and treatment. Comput. Methods Programs Biomed. 2023, 231, 107419. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Proceedings of the Advances in Neural Information Processing Systems (NeurIPS), Long Beach, CA, USA, 4–9 December 2017; Volume 30, pp. 5998–6008. [Google Scholar]

| Dataset | Drug | Protein | Interaction | Positive | Negative |
|---|---|---|---|---|---|
| binding DB | 14,643 | 2623 | 49,199 | 20,674 | 28,525 |
| BioSNAP | 4510 | 2181 | 27,464 | 13,830 | 13,634 |
| C.elegans | 2726 | 2001 | 6278 | 3364 | 3364 |
| Human | 2726 | 2001 | 6278 | 3364 | 3364 |
| Dataset | Baseline | AUROC | AUPRC | Accuracy | F1-Score | MCC |
|---|---|---|---|---|---|---|
| binding DB | CPI-GNN | 0.559 | 0.470 | 0.561 | 0.336 | 0.053 |
| BACPI | 0.954 | 0.941 | 0.892 | 0.871 | 0.779 | |
| GPGL | 0.934 | 0.914 | 0.869 | 0.844 | 0.731 | |
| bindTI | 0.960 | 0.946 | 0.907 | 0.909 | 0.812 | |
| FOTF-CPI | 0.953 | 0.937 | 0.894 | 0.873 | 0.782 | |
| CAT-DTI | 0.960 | 0.947 | 0.900 | 0.884 | 0.797 | |
| DO-GMA | 0.962 | 0.950 | 0.901 | 0.905 | 0.801 | |
| CAMF-DTI | 0.963 | 0.952 | 0.908 | 0.899 | 0.803 | |
| BioSNAP | CPI-GNN | 0.720 | 0.719 | 0.654 | 0.652 | 0.309 |
| BACPI | 0.888 | 0.895 | 0.808 | 0.810 | 0.616 | |
| GPGL | 0.886 | 0.893 | 0.813 | 0.813 | 0.621 | |
| bindTI | 0.903 | 0.903 | 0.832 | 0.840 | 0.673 | |
| FOTF-CPI | 0.899 | 0.902 | 0.827 | 0.829 | 0.654 | |
| CAT-DTI | 0.902 | 0.907 | 0.836 | 0.835 | 0.664 | |
| DO-GMA | 0.923 | 0.926 | 0.851 | 0.854 | 0.704 | |
| CAMF-DTI | 0.915 | 0.912 | 0.859 | 0.857 | 0.706 | |
| C.elegans | CPI-GNN | 0.986 | 0.986 | 0.949 | 0.918 | 0.829 |
| BACPI | 0.986 | 0.986 | 0.949 | 0.948 | 0.933 | |
| GPGL | 0.986 | 0.986 | 0.928 | 0.928 | 0.853 | |
| bindTI | 0.982 | 0.983 | 0.966 | 0.966 | 0.932 | |
| FOTF-CPI | 0.990 | 0.990 | 0.966 | 0.966 | 0.932 | |
| CAT-DTI | 0.983 | 0.986 | 0.967 | 0.964 | 0.932 | |
| DO-GMA | 0.993 | 0.993 | 0.974 | 0.973 | 0.948 | |
| CAMF-DTI | 0.987 | 0.984 | 0.948 | 0.949 | 0.895 | |
| Human | CPI-GNN | 0.967 | 0.966 | 0.907 | 0.906 | 0.834 |
| BACPI | 0.967 | 0.967 | 0.905 | 0.907 | 0.835 | |
| GPGL | 0.968 | 0.967 | 0.902 | 0.904 | 0.832 | |
| bindTI | 0.981 | 0.976 | 0.940 | 0.938 | 0.879 | |
| FOTF-CPI | 0.983 | 0.980 | 0.941 | 0.932 | 0.881 | |
| CAT-DTI | 0.982 | 0.969 | 0.942 | 0.944 | 0.886 | |
| DO-GMA | 0.986 | 0.984 | 0.950 | 0.951 | 0.900 | |
| CAMF-DTI | 0.987 | 0.984 | 0.948 | 0.949 | 0.895 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mi, J.; Li, C.; Jiang, D.; Wan, J. CAMF-DTI: Enhancing Drug–Target Interaction Prediction via Coordinate Attention and Multi-Scale Feature Fusion. Curr. Issues Mol. Biol. 2025, 47, 964. https://doi.org/10.3390/cimb47110964
Mi J, Li C, Jiang D, Wan J. CAMF-DTI: Enhancing Drug–Target Interaction Prediction via Coordinate Attention and Multi-Scale Feature Fusion. Current Issues in Molecular Biology. 2025; 47(11):964. https://doi.org/10.3390/cimb47110964
Chicago/Turabian StyleMi, Jia, Chang Li, Daguang Jiang, and Jing Wan. 2025. "CAMF-DTI: Enhancing Drug–Target Interaction Prediction via Coordinate Attention and Multi-Scale Feature Fusion" Current Issues in Molecular Biology 47, no. 11: 964. https://doi.org/10.3390/cimb47110964
APA StyleMi, J., Li, C., Jiang, D., & Wan, J. (2025). CAMF-DTI: Enhancing Drug–Target Interaction Prediction via Coordinate Attention and Multi-Scale Feature Fusion. Current Issues in Molecular Biology, 47(11), 964. https://doi.org/10.3390/cimb47110964





