Epigenomic Transcriptome Regulation of Growth and Development and Stress Response in Cucurbitaceae Plants: The Role of RNA Methylation
Abstract
1. Introduction
2. The Functional Role and Mechanism of RNA Methylation in the Cucurbitaceae
3. Advances in Techniques and Methods for RNA Methylation Research and Their Application Prospects in Cucurbitaceae Crops
3.1. MeRIP-Seq (m6A-Seq)
3.2. RNA-BisSeq
3.3. Nanopore Direct RNA Sequencing
4. The Application of Epitranscriptomics in Cucurbitaceae Breeding and Genetic Improvement
4.1. Identification of RNA Methylation Marks and Their Potential in MAS
4.1.1. Identification of RNA Methylation Marks
4.1.2. From Mark Identification to MAS Application
4.2. Frontiers in Biotechnological Breeding
4.2.1. Precise Targeting with CRISPR Gene Editing Technology
4.2.2. Fine-Tuning of “Writer” Enzymes
4.3. Concept Validation and Prospects in Model Plants
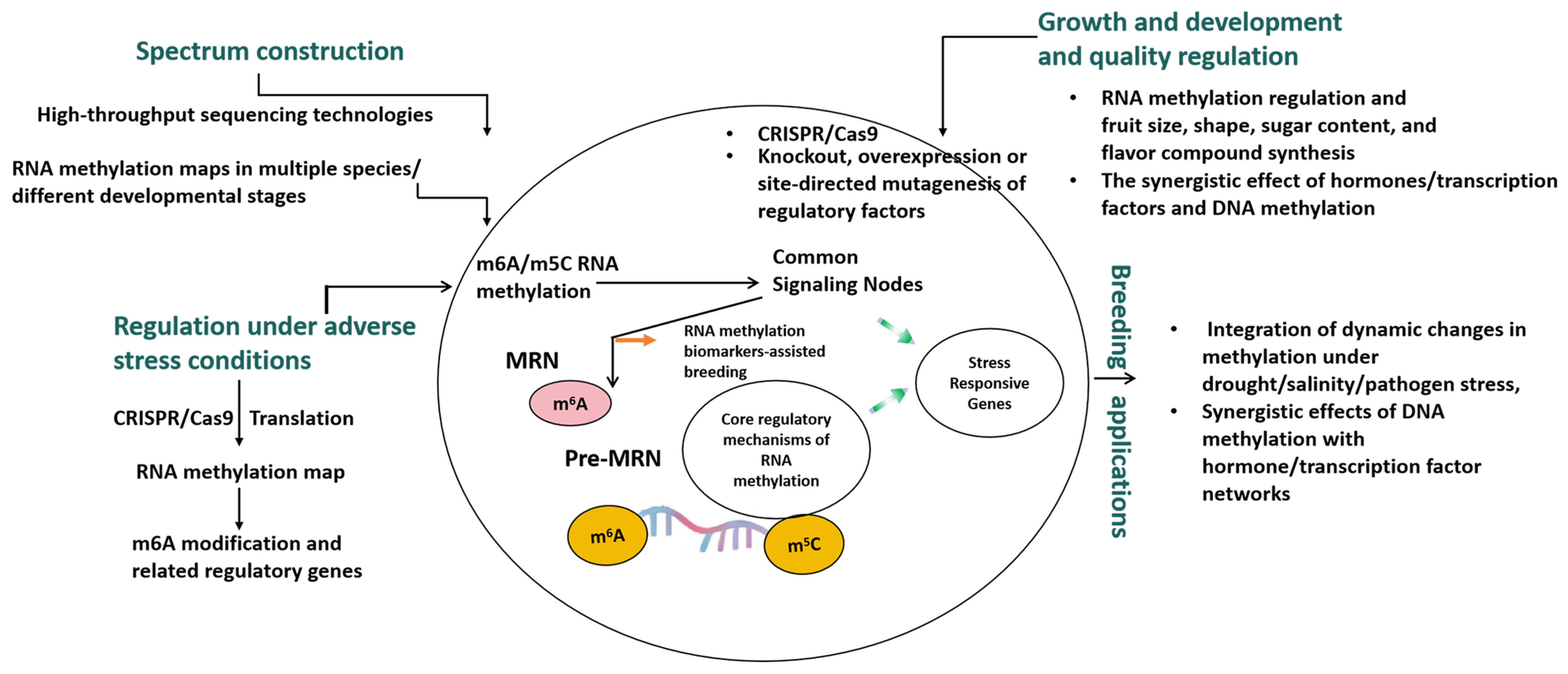
5. Key Issues and Limitations of the Study
6. Future Research Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chmielowska-Bąk, J.; Arasimowicz-Jelonek, M.; Deckert, J. In search of the mRNA modification landscape in plants. BMC Plant Biol. 2019, 19, 421. [Google Scholar] [CrossRef]
- Takahashi, Y.; Valencia, M.M.; Yu, Y.; Ouchi, Y.; Takahashi, K.; Shokhirev, M.N.; Lande, K.; Williams, A.E.; Fresia, C.; Kurita, M.; et al. Transgenerational inheritance of acquired epigenetic signatures at CpG islands in mice. Cell 2023, 186, 715–731.e19. [Google Scholar] [CrossRef]
- Zhang, H.M.; Lang, Z.B.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Zhang, P.; Yu, Y.; Cao, L.; Zhu, G.; Wu, W.; Guo, Y.; Yin, G.; Jia, M. Research Progress of m6A Methylation Modification Response to Plant Biotic and Abiotic Stresses. Acta Hortic. Sin. 2023, 50, 1841–1853. [Google Scholar]
- He, L.; Huang, H.; Bradai, M.; Zhao, C.; You, Y.; Ma, J.; Zhao, L.; Lozano-Durán, R.; Zhu, J.-K. DNA Methylation-Free Arabidopsis Reveals Crucial Roles of DNA Methylation in Regulating Gene Expression and Development. Nat. Commun. 2022, 13, 1355. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.Z.; Min, L.; Wang, M.J.; Wang, C.; Zhao, Y.; Li, Y.; Fang, Q.; Wu, Y.; Xie, S.; Ding, Y.; et al. Disrupted genome methylation in response to high temperature has distinct affects on microspore abortion and anther indehiscence. Plant Cell 2018, 30, 1387–1403. [Google Scholar] [CrossRef]
- Liu, G.; Wang, J.; Hou, X. Transcriptome-wide N6-methyladenosine (m6A) methylome profiling of heat stress in Pak-choi (Brassica rapa ssp. chinensis). Plants 2020, 9, 1080. [Google Scholar] [CrossRef]
- Zhang, K.; Zhuang, X.; Dong, Z.; Xu, K.; Chen, X.; Liu, F.; He, Z. The dynamics of N6 -methyladenine RNA modification in interactions between rice and plant viruses. Genome Biol. 2021, 22, 189. [Google Scholar] [CrossRef]
- Chen, H.S.; Wang, F.; Chen, J.G. Epigenetic Mechanisms in Depression: Implications for Pathogenesis and Treatment. Curr. Opin. Neurobiol. 2024, 85, 102854. [Google Scholar] [CrossRef]
- Reddy, D.; Wickman, J.R.; Ajit, S.K. Epigenetic Regulation in Opioid Induced Hyperalgesia. Neurobiol. Pain 2023, 14, 100146. [Google Scholar] [CrossRef]
- Long, S.; Yan, Y.; Xu, H.; Wang, L.; Jiang, J.; Xu, Z.; Liu, R.; Zhou, Q.; Huang, X.; Chen, J.; et al. Insights into the Regulatory Role of RNA Methylation Modifications in Glioma. J. Transl. Med. 2023, 21, 810. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Lin, H.; Miao, L.; Guo, H.; Chen, Y.; Zhuo, Z.; He, J. Functions, Mechanisms, and Therapeutic Implications of METTL14 in Human Cancer. J. Hematol. Oncol. 2022, 15, 13. [Google Scholar] [CrossRef] [PubMed]
- Moshitch-Moshkovitz, S.; Sevilla-Sharon, M.; Ashwal-Fluss, R.; Glick-Saar, E.; Rechavi, G.; Dominissini, D. mRNA m6A Detection. Nat. Rev. Methods Primers 2024, 4, 87. [Google Scholar] [CrossRef]
- Zhang, B.; Jiang, H.; Dong, Z.; Sun, A.; Ge, J. The Critical Roles of m6A Modification in Metabolic Abnormality and Cardiovascular Diseases. Genes Dis. 2021, 8, 746–758. [Google Scholar] [CrossRef]
- Aziz, M.A. Multiomics Approach towards Characterization of Tumor Cell Plasticity and Its Significance in Precision and Personalized Medicine. Cancer Metastasis Rev. 2024, 43, 1549–1559. [Google Scholar] [CrossRef]
- Mansoor, S.; Karunathilake, E.M.B.M.; Tuan, T.T.; Chung, Y.S. Genomics, Phenomics, and Machine Learning in Transforming Plant Research: Advancements and Challenges. Hortic. Plant J. 2025, 11, 486–503. [Google Scholar] [CrossRef]
- Ali, M.; Yang, T.; He, H.; Zhang, Y. Plant Biotechnology Research with Single-Cell Transcriptome: Recent Advancements and Prospects. Plant Cell Rep. 2024, 43, 18. [Google Scholar] [CrossRef]
- Li, D.; Sheng, Y.; Niu, H.; Li, Z. Gene Interactions Regulating Sex Determination in Cucurbits. Front. Plant Sci. 2019, 10, 1231. [Google Scholar] [CrossRef]
- Argyris, J.M.; Pujol, M.; Martín-Hernández, A.M.; Garcia-Mas, J. Combined Use of Genetic and Genomics Resources to Understand Virus Resistance and Fruit Quality Traits in Melon. Physiol. Plant. 2015, 155, 4–11. [Google Scholar] [CrossRef]
- Ma, L.; Wang, Q.; Zheng, Y.; Guo, J.; Yuan, S.; Fu, A.; Bai, C.; Zhao, X.; Zheng, S.; Wen, C.; et al. Cucurbitaceae Genome Evolution, Gene Function, and Molecular Breeding. Hortic. Res. 2022, 9, uhab057. [Google Scholar] [CrossRef]
- Wang, X.Y.; Zhang, D.; Cui, N.; Yu, Y.; Yu, G.C.; Fan, H.Y. Transcriptome and miRNA analyses of the response to Corynespora cassiicola in cucumber. Sci. Rep. 2018, 8, 7798. [Google Scholar] [CrossRef] [PubMed]
- Shnaider, Y.; Elad, Y.; Rav-David, D.; Pashkovsky, E.; Leibman, D.; Kravchik, M.; Shtarkman-Cohen, M.; Gal-On, A.; Spiegelman, Z. Development of Powdery Mildew Resistance in Cucumber Using CRISPR/Cas9-Mediated Mutagenesis of CsaMLO8. Phytopathology 2023, 113, 786–790. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wang, N.; Li, Y.; Wang, H.; Zhang, W.; Wang, H.; Chai, S. Recent Progress in Genetic Transformation and Gene Editing Technology in Cucurbit Crops. Agronomy 2023, 13, 755. [Google Scholar] [CrossRef]
- Xin, T.; Tian, H.; Ma, Y.; Wang, S.; Yang, L.; Li, X.; Zhang, M.; Chen, C.; Wang, H.; Li, H.; et al. Targeted Creation of New Mutants with Compact Plant Architecture Using CRISPR/Cas9 Genome Editing by an Optimized Genetic Transformation Procedure in Cucurbit Plants. Hortic. Res. 2022, 9, uhab086. [Google Scholar] [CrossRef]
- Li, X.; Lin, S.; Xiang, C.; Liu, W.; Zhang, X.; Wang, C.; Lu, X.; Liu, M.; Wang, T.; Liu, Z.; et al. CUCUME: An RNA Methylation Database Integrating Systemic mRNAs Signals, GWAS and QTL Genetic Regulation and Epigenetics in Different Tissues of Cucurbitaceae. Comput. Struct. Biotechnol. J. 2023, 21, 837–846. [Google Scholar] [CrossRef]
- Ferraz, R.; Coimbra, S.; Correia, S.; Canhoto, J. RNA Methyltransferases in Plants: Breakthroughs in Function and Evolution. Plant Physiol. Biochem. 2023, 194, 449–460. [Google Scholar] [CrossRef]
- Wang, M.K.; Gao, C.C.; Yang, Y.G. Emerging Roles of RNA Methylation in Development. Acc. Chem. Res. 2023, 56, 3417–3427. [Google Scholar] [CrossRef]
- Zhang, C.; Guo, X.; Zhang, Y.; Pang, H.; Chen, Q. Identification and Functional Exploration of the ALKBH Gene Family in Oriental Melon Fruit Ripening. Int. J. Mol. Sci. 2025, 26, 4254. [Google Scholar] [CrossRef]
- He, Y.; Li, Y.; Yao, Y.; Zhang, H.; Wang, Y.; Gao, J.; Fan, M. Overexpression of Watermelon m6A Methyltransferase CIMTB Enhances Drought Tolerance in Tobacco by Mitigating Oxidative Stress and Photosynthesis Inhibition and Modulating Stress-Responsive Gene Expression. Plant Physiol. Biochem. 2021, 168, 340–352. [Google Scholar] [CrossRef]
- Li, X.; Wang, C.; Chen, Y.; Liu, W.; Zhang, M.; Wang, N.; Xiang, C.; Gao, L.; Dong, Y.; Zhang, W. m5C and m6A Modifications Regulate the Mobility of Pumpkin CHOLINE KINASE 1 mRNA under Chilling Stress. Plant Physiol. 2024, 197, kiae511. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, F.; Li, J.; Sun, X.; Zhang, X.; Wang, H.; Fan, P.; Lai, L.; Li, Z.; Sui, T. Programmable RNA 5-Methylcytosine (m5C) Modification of Cellular RNAs by dCasRx Conjugated Methyltransferase and Demethylase. Nucleic Acids Res. 2024, 52, 2776–2791. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Jia, Y.; Pan, Y.; Lin, H.; Lv, S.; Nawaz, M.; Song, B.; Nie, X. Genome-Wide Identification of m6A-Related Gene Family and the Involvement of TdFIP37 in Salt Stress in Wild Emmer Wheat. Plant Cell Rep. 2024, 43, 254. [Google Scholar] [CrossRef]
- Kong, Y.; Mead, E.A.; Fang, G. Navigating the Pitfalls of Mapping DNA and RNA Modifications. Nat. Rev. Genet. 2023, 24, 363–381. [Google Scholar] [CrossRef]
- Feng, Y.; Gao, F. Bsgenova: An Accurate, Robust, and Fast Genotype Caller for Bisulfite-Sequencing Data. BMC Bioinform. 2024, 25, 206. [Google Scholar] [CrossRef]
- Wang, M.; Xie, N.B.; Chen, K.K.; Ji, T.T.; Xiong, J.; Guo, X.; Yu, S.Y.; Tang, F.; Xie, C.; Feng, Y.Q.; et al. Engineered APOBEC3C Sequencing Enables Bisulfite-Free and Direct Detection of DNA Methylation at a Single-Base Resolution. Anal. Chem. 2023, 95, 1556–1565. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, S.W.; Zhang, Y.C.; Liu, H.; Zhang, L.; Chen, R.; Huang, Y.; Meng, J. Decomposition of RNA Methylome Reveals Co-Methylation Patterns Induced by Latent Enzymatic Regulators of the Epitranscriptome. Mol. BioSyst. 2015, 11, 262–274. [Google Scholar] [CrossRef]
- Zhang, L.S.; Dai, Q.; He, C. Base-Resolution Sequencing Methods for Whole-Transcriptome Quantification of mRNA Modifications. Acc. Chem. Res. 2023, 57, 47–58. [Google Scholar] [CrossRef]
- Johnson, Z.; Xu, X.; Pacholec, C.; Xie, H. Systematic Evaluation of Parameters in RNA Bisulfite Sequencing Data Generation and Analysis. NAR Genom. Bioinform. 2022, 4, lqac045. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Song, B.; Wei, Z.; Huang, D.; Zhang, Y.; Su, J.; de Magalhães, J.P.; Rigden, D.J.; Meng, J.; Chen, K. m5C-Atlas: A Comprehensive Database for Decoding and Annotating the 5-Methylcytosine (m5C) Epitranscriptome. Nucleic Acids Res. 2021, 50, D196–D203. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Feng, J.; Zeng, C.; Liu, J.; Fu, Z.; Wang, D.; Wang, Y.; Zhang, L.; Li, J.; Jiang, A.; et al. NSUN2-Mediated M5C Methylation of IRF3 mRNA Negatively Regulates Type I Interferon Responses during Various Viral Infections. Emerg. Microbes Infect. 2023, 12, 2178238. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Liu, H.; Tan, W.; Chen, Y.; Dong, J.; Bai, S.; Wu, Z.; Zeng, Y. Dynamic Regulation and Key Roles of Ribonucleic Acid Methylation. Front. Cell. Neurosci. 2022, 16, 1058083. [Google Scholar] [CrossRef]
- Chachar, S.; Chachar, M.; Riaz, A.; Shaikh, A.A.; Li, X.; Li, X.; Guan, C.; Zhang, P. Epigenetic Modification for Horticultural Plant Improvement Comes of Age. Sci. Hortic. 2022, 292, 110633. [Google Scholar] [CrossRef]
- Rehman, A.; Tian, C.; He, S.; Li, H.; Lu, S.; Du, X.; Peng, Z. Transcriptome Dynamics of Gossypium Purpurascens in Response to Abiotic Stresses by Iso-Seq and RNA-Seq Data. Sci. Data 2024, 11, 477, Erratum in: Plant Cell Rep. 2012, 31, 1379. [Google Scholar] [CrossRef]
- Liu, Y.; Shen, Y.; Wang, H.; Zhang, Y.; Zhu, X. m5Cpred-XS: A New Method for Predicting RNA m5C Sites Based on XGBoost and SHAP. Front. Genet. 2022, 13, 853258. [Google Scholar] [CrossRef]
- Molho, D.; Ding, J.; Tang, W.; Li, Z.; Wen, H.; Wang, Y.; Venegas, J.; Jin, W.; Liu, R.; Su, R.; et al. Deep Learning in Single-Cell Analysis. ACM Trans. Intell. Syst. Technol. 2024, 15, 1–62. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, H.; Kohnen, M.V.; Prasad, K.V.S.K.; Gu, L.; Reddy, A.S.N. Analysis of Transcriptome and Epitranscriptome in Plants Using PacBio Iso-Seq and Nanopore-Based Direct RNA Sequencing. Front. Genet. 2019, 10, 253. [Google Scholar] [CrossRef]
- Tang, J.; Zheng, F.; Liu, X.; Li, Y.; Guo, Z.; Lin, X.; Zhou, J.; Zhang, Y.; Yu, G.; Hu, H.; et al. Cobalt Induces Neurodegeneration through FTO-Triggered Autophagy Impairment by Targeting TSC1 in an m6A-YTHDF2-Dependent Manner. J. Hazard. Mater. 2023, 453, 131354. [Google Scholar] [CrossRef]
- Kamali, S.; Singh, A. Genomic and Transcriptomic Approaches to Developing Abiotic Stress-Resilient Crops. Agronomy 2023, 13, 2903. [Google Scholar] [CrossRef]
- Raza, A.; Chen, H.; Zhang, C.; Zhuang, Y.; Sharif, Y.; Cai, T.; Yang, Q.; Soni, P.; Pandey, M.K.; Varshney, R.K.; et al. Designing Future Peanut: The Power of Genomics-Assisted Breeding. Theor. Appl. Genet. 2024, 137, 66. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Stewart, C.N. Functional Markers for Precision Plant Breeding. Int. J. Mol. Sci. 2020, 21, 4792. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wei, X.; Gao, G.; Jiang, H. Development and Evaluation of Improved Lines Based on an Elite Rice Variety 9311 for Overcoming Hybrid Sterility in Rice. Mol. Breed. 2020, 40, 102. [Google Scholar] [CrossRef]
- Guan, Y.X.; Wang, B.; Feng, Y.; Li, P. Development and Application of Marker-Assisted Reverse Breeding Using Hybrid Maize Germplasm. J. Integr. Agric. 2015, 14, 2538–2546. [Google Scholar] [CrossRef][Green Version]
- Zyprian, E.; Ochßner, I.; Schwander, F.; Šimon, S.; Hausmann, L.; Bonow-Rex, M.; Moreno-Sanz, P.; Grando, M.S.; Wiedemann-Merdinoglu, S.; Merdinoglu, D.; et al. Quantitative Trait Loci Affecting Pathogen Resistance and Ripening of Grapevines. Mol. Genet. Genom. 2016, 291, 1573–1594. [Google Scholar] [CrossRef]
- Abdul Aziz, M.; Masmoudi, K. Molecular Breakthroughs in Modern Plant Breeding Techniques. Hortic. Plant J. 2025, 11, 15–41. [Google Scholar] [CrossRef]
- Muha-Ud-Din, G.; Ali, F.; Hameed, A.; Naqvi, S.A.H.; Nizamani, M.M.; Jabran, M.; Sarfraz, S.; Yong, W. CRISPR/Cas9-Based Genome Editing: A Revolutionary Approach for Crop Improvement and Global Food Security. Physiol. Mol. Plant Pathol. 2024, 129, 102191. [Google Scholar] [CrossRef]
- Mohamed, H.I.; Khan, A.; Basit, A. CRISPR-Cas9 System Mediated Genome Editing Technology: An Ultimate Tool to Enhance Abiotic Stress in Crop Plants. J. Soil Sci. Plant Nutr. 2024, 24, 1799–1822. [Google Scholar] [CrossRef]
- Li, T.; Hu, J.; Sun, Y.; Li, B.; Zhang, D.; Li, W.; Liu, J.; Li, D.; Gao, C.; Zhang, Y.; et al. Highly Efficient Heritable Genome Editing in Wheat Using an RNA Virus and Bypassing Tissue Culture. Mol. Plant 2021, 14, 1787–1798. [Google Scholar] [CrossRef] [PubMed]
- Geng, R.; Fan, X.; Sarwar, R.; Wang, Y.; Dong, K.; Tan, X.L. CRISPR Mutant Rapid Identification in B. Napus: RNA-Seq Functional Profiling and Breeding Technology Application. Front. Plant Sci. 2025, 16, 1572020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, Z.; Feng, L.; Zhi, Z.; Liu, Y.; Zhang, M.; Yue, H.; Zhu, G.-P.; Gao, F. The Transcriptional Regulatory Mechanisms Exploration of Jujube Biological Traits through Multi-Omics Analysis. Forests 2024, 15, 395. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, L.; Wang, Y.; Zhang, W.; Zhang, X.; Dong, J.; Ying, J.; Wang, L.; Ma, Y.; Liu, L. Integration of Transcriptome and DNA Methylome Analysis Reveals the Molecular Mechanism of Taproot Yield Heterosis in Radish. Hortic. Plant J. 2025, 11, 645–660. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X.; Guo, X.; Wang, X.-J.; Zhang, X. Arabidopsis AGO3 Predominantly Recruits 24-Nt Small RNAs to Regulate Epigenetic Silencing. Nat. Plants 2016, 2, 16049. [Google Scholar] [CrossRef]
- Xin, T.; Zhang, Z.; Zhang, Y.; Li, X.; Wang, S.; Wang, G.; Li, H.; Wang, B.; Zhang, M.; Li, W.; et al. Recessive Epistasis of a Synonymous Mutation Confers Cucumber Domestication through Epitranscriptomic Regulation. Cell 2025, 188, 4517–4529.e15. [Google Scholar] [CrossRef]
- Tek, M.I.; Calis, O.; Fidan, H.; Shah, M.D.; Celik, S.; Wani, S.H. CRISPR/Cas9 based mlo-mediated resistance against Podosphaera xanthii in cucumber (Cucumis sativus L.). Front. Plant Sci. 2022, 13, 1081506. [Google Scholar] [CrossRef]
- Fang, R.; Chen, X.; Shen, J.; Wang, B. Targeted mRNA Demethylation in Arabidopsis Using Plant m6A Editor. Plant Methods 2023, 19, 81. [Google Scholar] [CrossRef]
- Furci, L.; Berthelier, J.; Saze, H. RNA N6-Adenine Methylation Dynamics Impact Hyaloperonospora arabidopsidis Resistance in Arabidopsis. Plant Physiol. 2024, 196, 745–753. [Google Scholar] [CrossRef]
- Zheng, H.; Sun, X.; Li, J.; Song, Y.; Song, J.; Wang, F.; Liu, L.; Zhang, X.; Sui, N. Analysis of N6-Methyladenosine Reveals a New Important Mechanism Regulating the Salt Tolerance of Sweet Sorghum. Plant Sci. 2021, 304, 110801. [Google Scholar] [CrossRef]
- Shinde, H.; Dudhate, A.; Kadam, U.S.; Hong, J.C. RNA Methylation in Plants: An Overview. Front. Plant Sci. 2023, 14, 1132959. [Google Scholar] [CrossRef]
- Zhou, L.; Tang, R.; Li, X.; Tian, S.; Li, B.; Qin, G. N6-Methyladenosine RNA Modification Regulates Strawberry Fruit Ripening in an ABA-Dependent Manner. Genome Biol. 2021, 22, 168. [Google Scholar] [CrossRef]
- Hooghvorst, I.; Nogués, S. Opportunities and Challenges in Doubled Haploids and Haploid Inducer-Mediated Genome-Editing Systems in Cucurbits. Agronomy 2020, 10, 1441. [Google Scholar] [CrossRef]
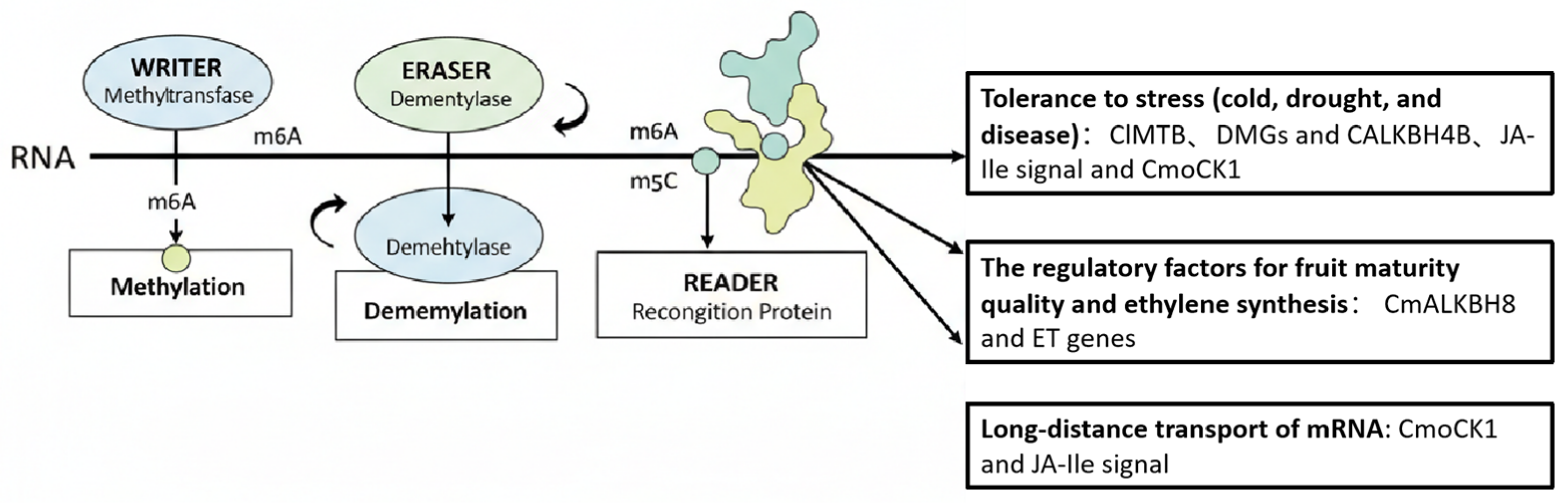
| Species | Published/Available Data on RNA Methylation | Regulatory Factor | Physiological Processes | Reference |
|---|---|---|---|---|
| Cucumbers and pumpkins (Cucumis sativus L.) and (Cucurbita moschata D.) | m6A modification is related to RNA mobility and has breeding potential | Genome-wide mRNA (including m5C and m6A modification sites) | RNA mobility, crop breeding | [25] |
| Oriental melon (Cucumis melo var. Makuwa) | m6A methylation positively regulates fruit ripening and ethylene synthesis. | CmALKBH8, ethylene genes | Fruit ripening, ethylene biosynthesis | [28] |
| Watermelon (Citrullus lanatus) | m6A methylation enhances drought resistance through ClMTB. | CIMTB | Drought stress response | [29] |
| Cucumbers and Pumpkin (Cucumis sativus L.) and (Cucurbita moschata D.) | m6A and m5C directly regulate the long-distance mobility of CmoCK1 mRNA at low temperatures. | CmoCK1, JA-Ile signal | Cold tolerance, long-distance mRNA transport, stress response | [30] |
| Watermelon (Citrullus lanatus) | Decreased m6A levels activate early immunity. | DMGs, CALKBH4B | Early virus immunity | [31] |
| Melon (Cucumis melo L.) | Transgenic activation of RNA silencing mechanism confers antiviral ability. siRNA, methylated | siRNA, methylated DNA | RNA silencing-mediated high plant disease resistance | [32] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, G.; Wang, Z.; Jia, L.; Huang, H. Epigenomic Transcriptome Regulation of Growth and Development and Stress Response in Cucurbitaceae Plants: The Role of RNA Methylation. Curr. Issues Mol. Biol. 2025, 47, 938. https://doi.org/10.3390/cimb47110938
Yu G, Wang Z, Jia L, Huang H. Epigenomic Transcriptome Regulation of Growth and Development and Stress Response in Cucurbitaceae Plants: The Role of RNA Methylation. Current Issues in Molecular Biology. 2025; 47(11):938. https://doi.org/10.3390/cimb47110938
Chicago/Turabian StyleYu, Guangchao, Zhipeng Wang, Lian Jia, and Hua Huang. 2025. "Epigenomic Transcriptome Regulation of Growth and Development and Stress Response in Cucurbitaceae Plants: The Role of RNA Methylation" Current Issues in Molecular Biology 47, no. 11: 938. https://doi.org/10.3390/cimb47110938
APA StyleYu, G., Wang, Z., Jia, L., & Huang, H. (2025). Epigenomic Transcriptome Regulation of Growth and Development and Stress Response in Cucurbitaceae Plants: The Role of RNA Methylation. Current Issues in Molecular Biology, 47(11), 938. https://doi.org/10.3390/cimb47110938





