Integrating Multiple Methods to Validate Key Genes Driving the Progression of Breast Ductal Carcinoma In Situ
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of DEGs
2.2. Search Tool for the Retrieval of Interacting Genes (https://cn.string-db.org/)
2.3. Gene Expression Level Validation
2.4. Bioinformatic Validation and Functional Annotation of Driver Genes
2.5. Cell Culture and siRNA Transfection
2.6. Migration and Invasion Assays
2.7. Cell Proliferation Assay
2.8. RNA Extraction and Real-Time Quantitative PCR
2.9. Statistical Analysis
3. Results
3.1. Identification of Candidate Genes for DCIS
3.2. Validation of the Expression of Candidate Genes in Breast Cancer
3.3. Exploring the Effect of Candidate Genes in the Progression of DCIS Lesions
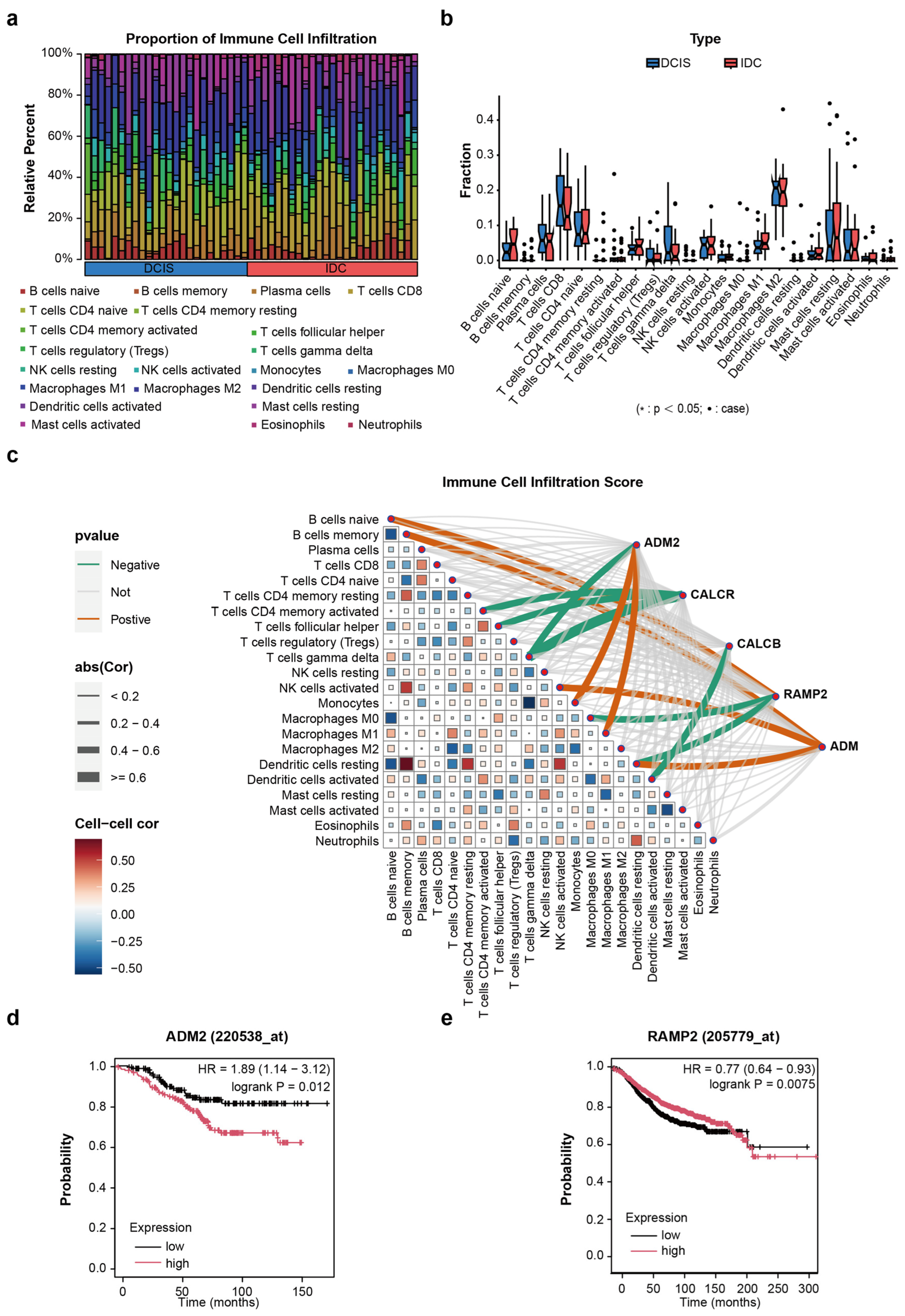
3.4. The Role of Candidate Genes and Immune Cells in the Progression of DCIS
3.5. The Relationship Between TPAGs and Patient OS
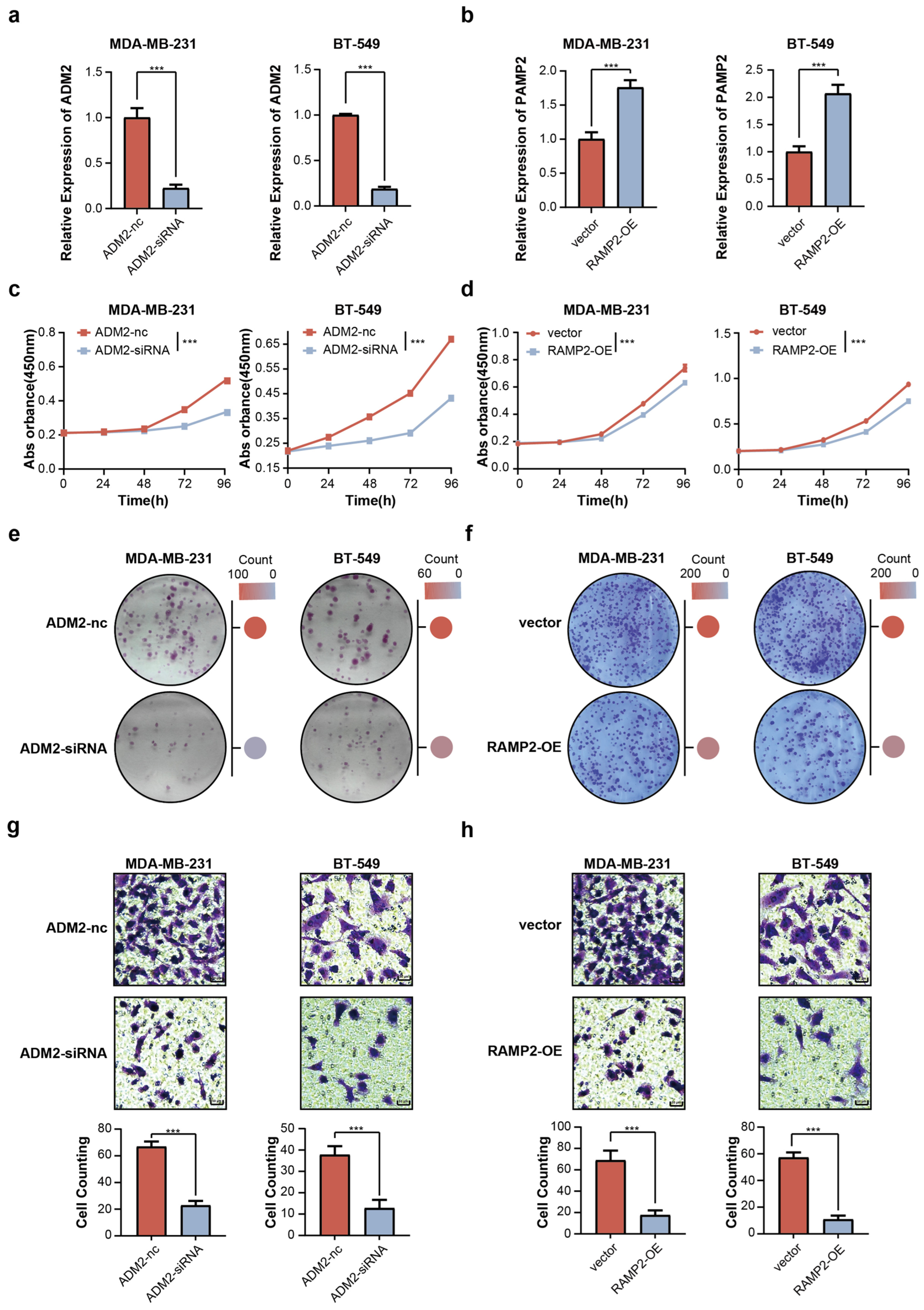
3.6. Influence of TPAGs on Malignant Behavior in Breast Cancer Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cowell, C.F.; Weigelt, B.; Sakr, R.A.; Ng, C.K.Y.; Hicks, J.; King, T.A.; Reis-Filho, J.S. Progression from ductal carcinoma in situ to invasive breast cancer: Revisited. Mol. Oncol. 2013, 7, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Hosein, A.N.; Brekken, R.A.; Maitra, A. Pancreatic cancer stroma: An update on therapeutic targeting strategies. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 487–505. [Google Scholar] [CrossRef] [PubMed]
- Bremnes, R.M.; Dønnem, T.; Al-Saad, S.; Al-Shibli, K.; Andersen, S.; Sirera, R.; Camps, C.; Marinez, I.; Busund, L.-T. The role of tumor stroma in cancer progression and prognosis: Emphasis on carcinoma-associated fibroblasts and non-small cell lung cancer. J. Thorac. Oncol. 2011, 6, 209–217. [Google Scholar] [CrossRef]
- Low, J.-Y.; Laiho, M. Caveolae-Associated Molecules, Tumor Stroma, and Cancer Drug Resistance: Current Findings and Future Perspectives. Cancers 2022, 14, 589. [Google Scholar] [CrossRef]
- Niwińska, A.; Olszewski, W.P. The role of stromal immune microenvironment in the progression of ductal carcinoma in situ (DCIS) to invasive breast cancer. Breast Cancer Res. 2021, 23, 118. [Google Scholar] [CrossRef]
- Strell, C.; Paulsson, J.; Jin, S.-B.; Tobin, N.P.; Mezheyeuski, A.; Roswall, P.; Mutgan, C.; Mitsios, N.; Johansson, H.; Wickberg, S.M.; et al. Impact of Epithelial-Stromal Interactions on Peritumoral Fibroblasts in Ductal Carcinoma in Situ. J. Natl. Cancer Inst. 2019, 111, 983–995. [Google Scholar] [CrossRef]
- Lisanti, M.P.; Tsirigos, A.; Pavlides, S.; Reeves, K.J.; Peiris-Pagès, M.; Chadwick, A.L.; Sanchez-Alvarez, R.; Lamb, R.; Howell, A.; Martinez-Outschoorn, U.E.; et al. JNK1 stress signaling is hyper-activated in high breast density and the tumor stroma: Connecting fibrosis, inflammation, and stemness for cancer prevention. Cell Cycle 2014, 13, 580–599. [Google Scholar] [CrossRef]
- Siziopikou, K.P. Ductal Carcinoma In Situ of the Breast: Current Concepts and Future Directions. Arch. Pathol. Lab. Med. 2013, 137, 462–466. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Győrffy, B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation 2024, 5, 100625. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–w102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, Y.; Liu, X.; Fu, L.; Gu, F.; Ma, Y. High Expression of Complement Component C7 Indicates Poor Prognosis of Breast Cancer and Is Insensitive to Taxane-Anthracycline Chemotherapy. Front. Oncol. 2021, 11, 724250. [Google Scholar] [CrossRef]
- Avalle, L.; Raggi, L.; Monteleone, E.; Savino, A.; Viavattene, D.; Statello, L.; Camperi, A.; Stabile, S.A.; Salemme, V.; De Marzo, N.; et al. STAT3 induces breast cancer growth via ANGPTL4, MMP13 and STC1 secretion by cancer associated fibroblasts. Oncogene 2022, 41, 1456–1467. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, M.; Li, J.; Xiong, Y.; Wang, J.; Jing, H.; Gu, Y. Overexpression of PSMC2 promotes the tumorigenesis and development of human breast cancer via regulating plasminogen activator urokinase (PLAU). Cell Death Dis. 2021, 12, 690. [Google Scholar] [CrossRef]
- Li, K.; Wu, J.L.; Qin, B.; Fan, Z.; Tang, Q.; Lu, W.; Zhang, H.; Xing, F.; Meng, M.; Zou, S.; et al. ILF3 is a substrate of SPOP for regulating serine biosynthesis in colorectal cancer. Cell Res. 2020, 30, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, Z.; Li, X.; Liu, B.; Liu, M.; Liu, L.; Chen, S.; Ren, M.; Wang, Y.; Yu, M.; et al. SHMT2 Desuccinylation by SIRT5 Drives Cancer Cell Proliferation. Cancer Res. 2018, 78, 372–386. [Google Scholar] [CrossRef]
- Chen, X.; Li, Z.; Liang, M.; Zhang, Z.; Zhu, D.; Lin, B.; Zhou, R.; Lu, Y. Identification of DDIT4 as a potential prognostic marker associated with chemotherapeutic and immunotherapeutic response in triple-negative breast cancer. World J. Surg. Oncol. 2023, 21, 194. [Google Scholar] [CrossRef]
- Wu, V.H.; Yung, B.S.; Faraji, F.; Saddawi-Konefka, R.; Wang, Z.; Wenzel, A.T.; Song, M.J.; Pagadala, M.S.; Clubb, L.M.; Chiou, J.; et al. The GPCR-Gα(s)-PKA signaling axis promotes T cell dysfunction and cancer immunotherapy failure. Nat. Immunol. 2023, 24, 1318–1330. [Google Scholar] [CrossRef]
- Dorsam, R.T.; Gutkind, J.S. G-protein-coupled receptors and cancer. Nat. Rev. Cancer 2007, 7, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tao, Y.; Chen, Y.; Wu, Y.; He, Y.; Yin, S.; Xu, S.; Yu, Y. Development of a metabolism-related signature for predicting prognosis, immune infiltration and immunotherapy response in breast cancer. Am. J. Cancer Res. 2022, 12, 5440–5461. [Google Scholar]
- Qian, B.Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef]
- Lips, E.H.; Kumar, T.; Megalios, A.; Visser, L.L.; Sheinman, M.; Fortunato, A.; Shah, V.; Hoogstraat, M.; Sei, E.; Mallo, D.; et al. Genomic analysis defines clonal relationships of ductal carcinoma in situ and recurrent invasive breast cancer. Nat. Genet. 2022, 54, 850–860. [Google Scholar] [CrossRef]
- Hayward, M.K.; Louise Jones, J.; Hall, A.; King, L.; Ironside, A.J.; Nelson, A.C.; Shelley Hwang, E.; Weaver, V.M. Derivation of a nuclear heterogeneity image index to grade DCIS. Comput. Struct. Biotechnol. J. 2020, 18, 4063–4070. [Google Scholar] [CrossRef]
- Wang, J.; Li, B.; Luo, M.; Huang, J.; Zhang, K.; Zheng, S.; Zhang, S.; Zhou, J. Progression from ductal carcinoma in situ to invasive breast cancer: Molecular features and clinical significance. Signal Transduct. Target. Ther. 2024, 9, 83. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Y.; Liu, Z.; Lin, C.; Meng, F.; Xu, L.; Zhang, X.; Zhang, C.; Zhang, P.; Gong, S.; et al. CircMYH9 drives colorectal cancer growth by regulating serine metabolism and redox homeostasis in a p53-dependent manner. Mol. Cancer 2021, 20, 114. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.C.; Maddocks, O.D.K. Serine and Functional Metabolites in Cancer. Trends Cell Biol. 2017, 27, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, J.; Sun, S.; Chen, X.; Zhang, H.; Li, B.; Sun, S. YAP/TAZ-mediated activation of serine metabolism and methylation regulation is critical for LKB1-deficient breast cancer progression. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef]
- Li, C.; Vides, A.; Kim, D.; Xue, J.Y.; Zhao, Y.; Lito, P. The G protein signaling regulator RGS3 enhances the GTPase activity of KRAS. Science 2021, 374, 197–201. [Google Scholar] [CrossRef]
- An, W.; Lin, H.; Ma, L.; Zhang, C.; Zheng, Y.; Cheng, Q.; Ma, C.; Wu, X.; Zhang, Z.; Zhong, Y.; et al. Progesterone activates GPR126 to promote breast cancer development via the Gi pathway. Proc. Natl. Acad. Sci. USA 2022, 119, e2117004119. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.M.; Ge, J.Y.; Chen, Y.F.; Liu, T.; Chen, L.; Liu, C.C.; Ma, D.; Chen, Y.Y.; Cai, Y.W.; Xu, Y.Y.; et al. Combined Single-Cell and Spatial Transcriptomics Reveal the Metabolic Evolvement of Breast Cancer during Early Dissemination. Adv. Sci. 2023, 10, e2205395. [Google Scholar] [CrossRef]
- Yang, J.; Yu, L.; Man, J.; Chen, H.; Zhou, L.; Zhao, L. Immune scoring model based on immune cell infiltration to predict prognosis in diffuse large B-cell lymphoma. Cancer 2023, 129, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Kotwal, A.; Gustafson, M.P.; Bornschlegl, S.; Dietz, A.B.; Delivanis, D.; Ryder, M. Circulating immunophenotypes are potentially prognostic in follicular cell-derived thyroid cancer. Front. Immunol. 2023, 14, 1325343. [Google Scholar] [CrossRef] [PubMed]
- Cassetta, L.; Fragkogianni, S.; Sims, A.H.; Swierczak, A.; Forrester, L.M.; Zhang, H.; Soong, D.Y.H.; Cotechini, T.; Anur, P.; Lin, E.Y.; et al. Human Tumor-Associated Macrophage and Monocyte Transcriptional Landscapes Reveal Cancer-Specific Reprogramming, Biomarkers, and Therapeutic Targets. Cancer Cell 2019, 35, 588–602.e510. [Google Scholar] [CrossRef]
- Jiang, P.; Gao, W.; Ma, T.; Wang, R.; Piao, Y.; Dong, X.; Wang, P.; Zhang, X.; Liu, Y.; Su, W.; et al. CD137 promotes bone metastasis of breast cancer by enhancing the migration and osteoclast differentiation of monocytes/macrophages. Theranostics 2019, 9, 2950–2966. [Google Scholar] [CrossRef]
- Wang, Q.; Sun, K.; Liu, R.; Song, Y.; Lv, Y.; Bi, P.; Yang, F.; Li, S.; Zhao, J.; Li, X.; et al. Single-cell transcriptome sequencing of B-cell heterogeneity and tertiary lymphoid structure predicts breast cancer prognosis and neoadjuvant therapy efficacy. Clin. Transl. Med. 2023, 13, e1346. [Google Scholar] [CrossRef]
- Roehrkasse, A.M.; Booe, J.M.; Lee, S.M.; Warner, M.L.; Pioszak, A.A. Structure-function analyses reveal a triple β-turn receptor-bound conformation of adrenomedullin 2/intermedin and enable peptide antagonist design. J. Biol. Chem. 2018, 293, 15840–15854. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, M.; Zheng, S.; Wen, Y.; Zhang, J.; Zhang, J.; Wang, H.; Mo, C.; Xu, S.; Chen, X. Integrating Multiple Methods to Validate Key Genes Driving the Progression of Breast Ductal Carcinoma In Situ. Curr. Issues Mol. Biol. 2025, 47, 864. https://doi.org/10.3390/cimb47100864
Zhong M, Zheng S, Wen Y, Zhang J, Zhang J, Wang H, Mo C, Xu S, Chen X. Integrating Multiple Methods to Validate Key Genes Driving the Progression of Breast Ductal Carcinoma In Situ. Current Issues in Molecular Biology. 2025; 47(10):864. https://doi.org/10.3390/cimb47100864
Chicago/Turabian StyleZhong, Minjie, Shengkai Zheng, Yahui Wen, Juansi Zhang, Jiahui Zhang, Hanwei Wang, Caiqin Mo, Sunwang Xu, and Xiangjin Chen. 2025. "Integrating Multiple Methods to Validate Key Genes Driving the Progression of Breast Ductal Carcinoma In Situ" Current Issues in Molecular Biology 47, no. 10: 864. https://doi.org/10.3390/cimb47100864
APA StyleZhong, M., Zheng, S., Wen, Y., Zhang, J., Zhang, J., Wang, H., Mo, C., Xu, S., & Chen, X. (2025). Integrating Multiple Methods to Validate Key Genes Driving the Progression of Breast Ductal Carcinoma In Situ. Current Issues in Molecular Biology, 47(10), 864. https://doi.org/10.3390/cimb47100864




