Abstract
The plant Actinostemma tenerum is endemic to East Asia and has been used as a traditional medicinal herb for over 1400 years. Investigating the chloroplast genome characteristics and codon usage bias (CUB) is essential for advancing research on molecular markers and genetic diversity in A. tenerum. In this study, we sequenced the complete chloroplast genome of A. tenerum, revealing a length of 160,579 bp, with a GC content of 36.5%. The genome comprised 132 coding genes, including 87 protein-coding genes (CDSs), 8 rRNA genes, and 37 tRNA genes. Analysis of the 51 selected CDSs showed average GC1, GC2, and GC3 values of 46.95%, 39.52%, and 28.11%, respectively. The effective number of codons (ENC) ranged from 35.34% to 56.23%, with an average of 45.57%, indicating a weak CUB. Nucleotide composition analysis revealed unequal distribution of A, T, C, and G, with codon preference biased towards A or U. Neutrality plots, ENC-plots, and PR2-bias plots indicated that natural selection predominantly influences on CUB. A total of 18 optimal codons were identified. This study contributes genetic insights into A. tenerum and enhances our understanding of codon usage patterns in plant chloroplast genomes.
1. Introduction
Chloroplasts are essential semi-autonomous organelles responsible for photosynthesis in plants, possessing their own genome [1]. The chloroplast genome is typically organized into a circular configuration, comprising a large single copy region (LSC), a small single copy region (SSC), and two inverted repeat sequences (IRs) [2,3,4]. The chloroplast genome holds significant application values for plant identification, assessment of genetic diversity, and study of gene expression [5,6,7,8]. With the decreasing cost of high-throughput sequencing, an increasing number of plant chloroplast genomes have been sequenced and annotated [9,10]. Comprehensive investigations of plant chloroplast genomes play a vital role in species taxonomy, phylogeny and conservation, while simultaneously shedding light on the evolutionary processes of plant lineages.
Codons, which link nucleotides to proteins, can encode a single amino acid through multiple synonymous codons, a phenomenon known as codon usage bias (CUB) [11]. CUB, prevalent in plants, reflects an adaptive selection mechanism for specific nucleotide combinations during transcription and translation [12]. This bias is influenced by multiple factors, including gene expression levels, GC content, natural selection, mutational pressure, and selective pressure, which collectively shape the patterns of codon preferences in plants [13,14,15]. Understanding CUB is crucial for revealing evolutionary relationships among species and optimizing gene expression efficiency.
Actinostemma tenerum Griff. (1845), a monotypic genus of the Cucurbitaceae, has long been utilized as a traditional medicine herb for approximately 1400 years. This species is endemic to East Asia and distributes across China, Indochina Peninsula, Korea and Japan. A. tenerum is an annual herb characterized by a tufted growth habit and creeping rhizome, with its entire plant being used for medicinal purposes and having been cultivated as an ornamental species in gardens [16]. Based on the morphological data, Actinostemma lobatum (Maxim.) Maxim. ex Franch. & Sav. was treated as synonymous with A. tenerum (https://www.plantplus.cn/ (accessed on 8 August 2024)). Phytochemical studies confirmed that this species contains saponins, polyphenols, lipids, and other compounds, especially 19 types of saponin were identified, which are similar to ginsenoside [17,18,19,20,21,22,23,24,25]. Additionally, the seeds of this species consisted of 11 fatty oil compounds by GC-MS analysis, with highest content of unsaturated fatty acid [24]. Further studies have proved that this species possesses several biological activities, such as anti-tumor, anti-bacterial, anti-inflammatory, and anti-thrombotic [26,27,28].
In this study, we utilized high-throughput sequencing technology to obtain the chloroplast genome of A. tenerum for the first time. Through assembly and annotation, we analyzed the chloroplast genome’s structure and examined its codon usage bias. Furthermore, we investigated the factors influencing the codon preference, aiming to provide a reference for organism evolution and help to understand the patterns of codons in chloroplast genomes in Cucurbitaceae.
2. Materials and Methods
2.1. Sampling, DNA Extraction and Sequencing
The samples were collected from Anshan City, Liaoning Province, China (41°6′26″ N, 122°59′20″ E). A voucher specimen (accession no. ZJS_2024075) was deposited in the specimen room of Anshan Normal University (https://www.asnc.edu.cn/ (accessed on 28 August 2024), Contact: Ji-Si Zhang, E-mail: zhangjisi@asnc.edu.cn). Genomic DNA was extracted from silica gel-dried leaves using the modified CTAB method [29], and an Illumina paired-end (PE) library was prepared and sequenced at Personalbio Biotechnology Co., Ltd., Shanghai, China.
2.2. Chloroplast Genome Assembly and Annotation
Raw sequencing reads were quality-filtered using Trimmomatic v.0.39 to remove adapter sequences and low-quality bases [30]. GetOrganelle v.1.5 was then employed to assemble the clean reads [31]. The chloroplast genome of A. tenerum was annotated using GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html (accessed on 2 January 2025)) and Geneious v.9.0.5 (http://www.geneious.com/ (accessed on 2 January 2025)) with Gynostemma pubescens (NC_036142) as the reference [32]. The annotated complete chloroplast genome of A. tenerum was deposited in GenBank under the accession number PV938953.
2.3. Calculation of Parameters Related to Codon Usage Bias
To minimize analytical errors, coding sequences shorter than 300 bp and repetitive coding sequences were excluded, and, thus, there were 51 CDSs selected for subsequent analysis [33]. CodonW v.1.4.2 and the Codon Usage Statistics Program (CUSP) tool from the European Molecular Biology Open Software Suite (EMBOSS) website (https://bioinformatics.nl/cgi-bin/emboss/cusp (accessed on 20 January 2025)) were employed to calculate the effective number of codons (ENC), relative synonymous codon usage (RSCU), and GC content at the 1st, 2nd, and 3rd positions of the codons (namely GC1, GC2, GC3, respectively) in the 51 CDSs of A. tenerum. Correlation analysis and significance testing were performed using SPSS v.29.0.
2.4. Neutrality Plot Analysis
A neutrality plot was constructed using GC3 and GC12 (the mean of GC1 and GC2). The plot was annotated with the regression equation and the coefficient of determination (R2) to assess the relationship between these variables. A regression coefficient nearing 1 signifies a predominant influence of mutational pressure on the gene, while a coefficient nearing 0 indicates that natural selection is the primary determinant affecting the gene [34,35].
2.5. ENC-Plot Analysis
An effective number of codons plot (ENC-plot) can reflect the extent to which codon preference is influenced by mutation and natural selection. A scatter plot correlating GC3 content with ENC values was constructed, alongside a fitted standard curve described by the equation ENCexp = 2 + GC3s + 29/[GC3s2 + (1 − GC3s)2]. The standard curve serves as a reference boundary: ENC points above this curve suggest that mutational processes influence codon bias, indicating reduced impact of natural selection. Conversely, substantial deviations of points from the curve imply a stronger influence of natural selection pressure on codon bias [36].
2.6. PR2-Bias Plot Analysis
The ratios A3/(A3 + T3) and G3/(G3 + C3) were determined from the 51 CDSs of the A. tenerum chloroplast genome. A scatter plot was created with G3/(G3 + C3) and A3/(A3 + T3), with the center point marking A = T and C = G. Each point reflects the extent and direction of base deviation. Points in the upper half suggest a higher frequency of third-position codon A over T, while points in the left half indicate a higher frequency of C over G. If base mutations were the sole influence, base usage would be uniform, resulting in an even scatter distribution. Otherwise, it suggests codon preference is influenced by both mutation and natural selection [37,38].
2.7. Identification of Optimal Codons
Optimal codons are characterized by both high expression and frequency. The 51 CDS sequences of the A. tenerum chloroplast genome were ranked by ENC, and the top and bottom 10% were selected to form high-expression (ycf3, ycf4, atpE, ycf2, rpl22) and low-expression (rps8, rps14, petD, rpl16, ndhJ) gene libraries, respectively. CodonW 1.4.2 was used to calculate RSCU and ΔRSCU (ΔRSCU = RSCUhigh − RSCUlow) for these libraries. Codons with RSCU > 1 were deemed high-frequency, while those with ΔRSCU ≥ 0.08 were considered high-expression. Codons meeting both criteria were identified as optimal [33,36].
3. Results
3.1. Chloroplast Genome Characteristics of A. tenerum
A total of 6 Gb of 150 bp paired-end raw reads were utilized for the assembly of the chloroplast genome of A. tenerum. The chloroplast genome of A. tenerum spanned 160,579 bp, with a GC content of 36.5%. It displayed the typical quadripartite circular structure (Figure 1). The LSC region, encompassing 89,766 bp, harbored genes associated with photosynthesis and gene expression regulation. The SSC region with 18,553 bp predominantly contained genes involved in chloroplast function. Each IR region included 26,130 bp and maintained the stability of the chloroplast genome.
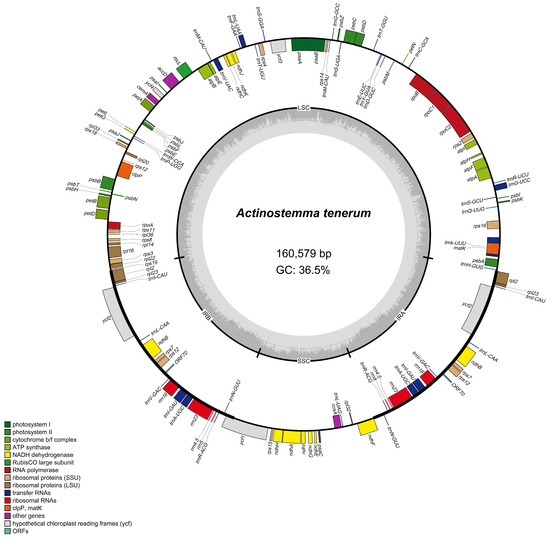
Figure 1.
Circular map of the A. tenerum chloroplast genome. Genes transcribed counterclockwise were inside the circle, while those transcribed clockwise were outside. Colors differentiate gene functions. The inner circle’s dashed area showed GC content in dark grey and AT content in light grey. LSC: large single-copy region; IR: inverted repeat; SSC: small single-copy region.
The chloroplast genome of A. tenerum comprised a total of 132 genes, consisting of 87 CDSs, 37 tRNA genes, and eight rRNA genes (Table S1). Among these annotated genes, 15 genes contained a single intron, while three genes (rps12, clpP, ycf3) harbored two introns (Table S1). These genes were classified into four main groups: 76 genes involved in self-replication, 46 genes related to photosynthesis, five genes with various functional roles, and five genes with uncharacterized functions. Notably, within the 46 photosynthesis-related genes, 18 genes encoded subunits of NADH dehydrogenase.
3.2. Codon Usage Patterns
Analysis of 51 CDSs selected from A. tenerum revealed a range of GC content from 31.23% (ndhF) to 46.04% (rps11) (Table S2). Notably, significant differences in GC content were observed among the three codon positions (Figure 2), with average GC1, GC2, and GC3 values of 46.95%, 39.52%, and 28.11%, respectively (Table S2). Nucleotide composition analysis demonstrated unequal distribution of A, T, C, and G, with a preference for codons ending in A or U. The ENC for most chloroplast genes exceeded 35, ranging from 35.34 to 56.22 (Table S2). Specifically, six genes (ndhJ, petD, psbA, rpl16, rps14 and rps8) exhibited ENC values below 40, while four genes (atpE, ycf2, ycf3 and ycf4) had ENC values exceeding 50. The ENC values of the remaining genes fell within 40 to 50. These findings suggested a relatively weak codon bias in the chloroplast genome of A. tenerum.
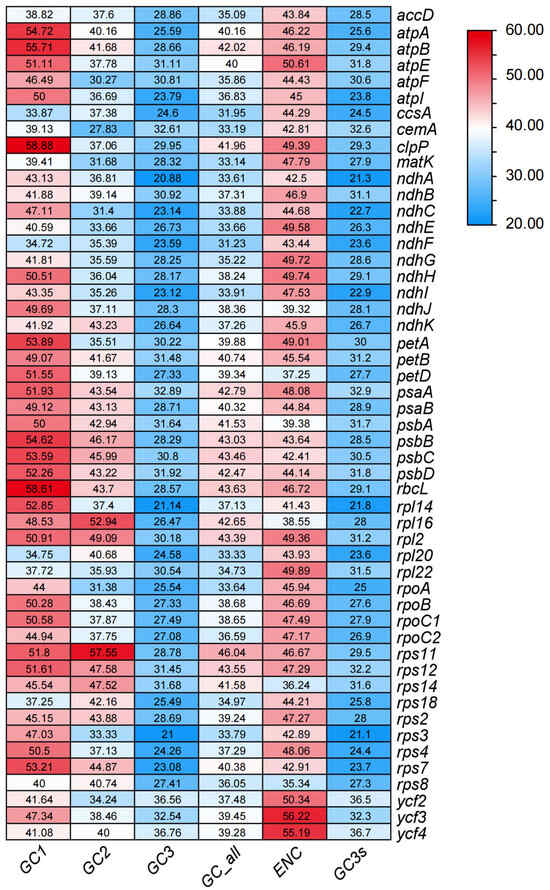
Figure 2.
The heatmap of ENC in A. tenerum.
Correlation analysis of codon parameters revealed that GC_all exhibited highly significant correlations with GC1, GC2, and GC3 (Table 1). GC1 and GC2 showed an extremely significant correlation, while neither correlated with GC3. These results suggested that the base compositions at the first and second codon positions were similar, but distinct from the third position. ENC values were statistically correlated with GC3, but not with GC1 and GC2. No correlations were observed between the number of codons and any parameters, indicating that gene sequence length does not influence GC content at different positions or the ENC value.

Table 1.
Correlation analysis of codon parameters in A. tenerum chloroplast genes.
With the exception of the codons encoding Methionine and Tryptophan, 30 out of the remaining codons had RSCU values greater than 1 (Figure 3, Table S3). Notably, all these codons terminated with either A or U, except for UUG. This finding further denoted that the chloroplast genome of A. tenerum has a preference for codons ending with A or T.
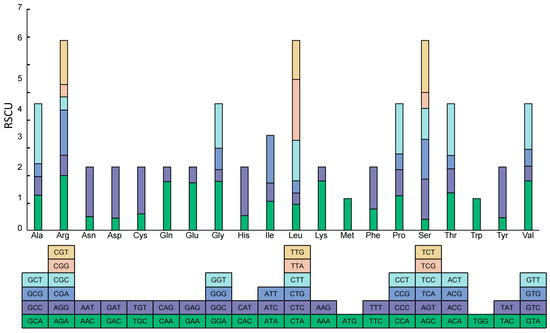
Figure 3.
Codon content of 20 amino acids in all protein-coding genes of the A. tenerum chloroplast genome.
3.3. Neutrality Plot Analysis
In the chloroplast genome of A. tenerum, the GC12 values of 51 CDSs varied from 33.5% to 54.7%, while the GC3 values ranged from 21.0% to 36.8%. These findings indicated that there is a certain difference in the proportions of GC12 and GC3 in the chloroplast genome of A. tenerum. The regression analysis yielded a coefficient of 0.2027 for GC12 with respect to GC3, accompanied by an R2 value of 0.0219, suggesting an absence of a significant correlation between GC3 and GC12 (Figure 4A). These results implied that CUB in A. tenerum may be marginally influenced by mutational pressures, while the interplay of natural selection and other factors likely holds substantial importance.
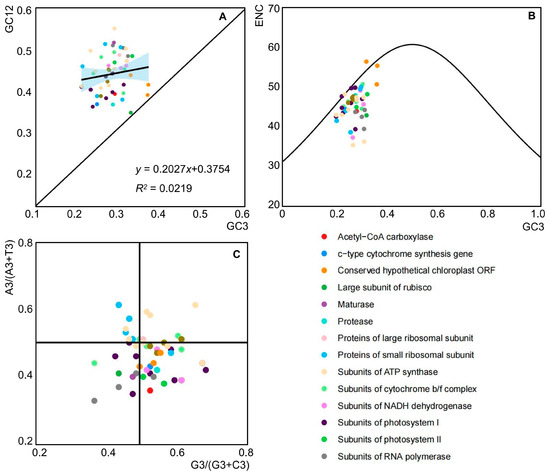
Figure 4.
Plots of the causes of codon preference in the chloroplast genome of A. tenerum. (A) Neutrality plot analysis; (B) ENC plot analysis; (C) PR2-plot analysis.
3.4. ENC-Plot Analysis
The ENC-plot analysis revealed that the majority of genes exhibited a distribution below the standard curve (the theoretical distribution of the genes’ ENC values), with only a minority clustering near it (Figure 4B). Analysis of the ENC ratio frequency distribution highlighted discrepancies between observed and expected ENC values (Table 2). Notably, eight genes (15.7%) fell within the range of −0.05 to 0.05, indicative of higher mutation pressure than selection pressure influencing codon bias. The remaining genes (84.3%) fell into other intervals, indicating a predominant impact of selection pressure. These findings inferred that a significant influence of natural selection on codon usage bias in the chloroplast genome of A. tenerum.

Table 2.
Distribution of ENC ratio frequency.
3.5. Parity-Rule 2 (PR2) Bias Plot Analysis
The PR2-plot analysis highlights the differential usage of the bases A, T, G, and C at the third of codons. With selective pressure, mutations at these positions should occur randomly, leading to similar base frequencies. However, as shown in Figure 4C, the chloroplast genes of A. tenerum exhibited an uneven distribution across the four quadrants, with the majority concentrated in the lower half. Specifically, the G3/(G3 + C3) values of 31 genes exceeded 0.5, while the A3/(A3 + T3) values of 35 genes were less than 0.5, indicating a preference for T over A and G over C in the third position base usage (Table S4). These findings suggested that natural selection primarily influenced the third base during the process of evolution.
3.6. Identification of Optimal Codons
The optimal codons for the CDSs of A. tenerum were presented in Table 3. Among the 30 high-frequency codons with an RSCU value exceeding 1, 29 of them terminated with either A or U (Table S3). Additionally, 26 codons exhibited high expression, with a ΔRSCU value of at least 0.08. By analyzing the intersection of these two sets, there were 18 optimal codons identified in the chloroplast genome of A. tenerum, including GCA, AGA, CAA, GAA, GGA, GGU, CAU, AUA, AUU, UUA, UUG, AAA, UUU, UCU, ACA, UAU, GUA and GUU. Notably, 17 of these optimal codons ended with A or U. These results also suggested a preference for codons ending with A or U in CDSs of the chloroplast genome of A. tenerum.

Table 3.
Optimal codons in chloroplast genome of A. tenerum.
4. Discussion
4.1. The Chloroplast Genome Characteristics of A. tenerum
In this study, the complete chloroplast genome of A. tenerum was firstly sequenced and annotated. The chloroplast genome had a size of 160,579 bp with a GC content of 36.5% (Figure 1), and displayed the typical quadripartite structure with other angiosperms [4,9]. It consisted of the largest LSC region (89,766 bp), SSC region (18,553 bp), and two IR regions (26,130 bp) (Table S1). This result is consistent with the other Cucurbitaceae plants [39,40,41,42]. For instance, Zhang et al. [39] detected a comparative chloroplast genome characteristic in the Cucurbitaceae, and revealed that the ten genera exhibited a conserved quadripartite structure with similar region lengths, comprising an LSC region spanning 86,642 to 88,374 bp, an SSC region spanning 17,897 to 18,653 bp, and two IR regions ranging from 25,193 to 26,242 bp. Similarly, Jiang et al. [42] demonstrated that the chloroplast genome sizes of 11 Trichosanthes ranged from 156,413 to 157,556 bp with a uniform GC content of 37%. Specifically, within the Trichosanthes, the LSC region spanned 85,642 to 88,374 bp, the SSC region ranged from 17,897 to 18,653 bp, and the IR regions varied from 25,193 to 26,242 bp in length. Moreover, A. tenerum encompassed 132 annotated genes, including 87 CDSs, 37 tRNA genes and eight rRNA genes (Table S2), which were highly similar to those of Momordica charantia and Lagenaria siceraria within the Cucurbitaceae, characterized by the absence of the infA gene and the retention of the ycf1 gene [39]. In this study, the chloroplast genome of A. tenerum exhibited a slightly larger size compared to other genera in the Cucurbitaceae, primarily due to a slight expansion of the LSC region. Taken together, the complete chloroplast genomes of Cucurbitaceae are highly conserved in terms of size, structure, gene order and content. These characteristics are valuable for the exploration of genome divergences and the identification of selection signals throughout evolutionary history.
4.2. Codon Usage Patterns and Their Drivers
Codon usage bias serves as a crucial indicator for studying the evolutionary relationships among plant chloroplast genomes [12,43] and is closely associated with the GC content of codons within chloroplast genomes [13,44,45,46]. Analysis of codon usage patterns identified significant variations in GC content among the first, second and third codons within the 51 selected CDSs, with GC3 exhibiting the lowest GC content (Table 1). No significant correlation was observed between GC12 and GC3 (Table 1). Additionally, the ENC values of these CDSs ranged from 35.34 to 56.22, with the majority exceeding 35 (Figure 2), indicating relatively weak codon bias in the chloroplast genome of A. tenerum. Moreover, most codons in the chloroplast genome of A. tenerum terminated with either A or U (Figure 3, Table S3). The similar weak codon usage bias and codon preferences were explored in other Cucurbitaceae species [42,47]. Previous studies revealed ENC values ranging from 55 to 56, with a preference for A and U at the third codon position on chloroplast genome genes in 11 Trichosanthes species [42]. Also, the genus Gynostemma demonstrated low codon usage bias and a similar preference for A and U at the third nucleotide position [47].
The formation of codon usage bias is a complex process influenced by natural selection, mutational pressures, genome size, and tRNA abundance [13,14,15]. A number of studies have indicated that natural selection and mutations are the primary drivers of CUB [13,44]. Analysis of A. tenerum’s CUB revealed a slight impact of mutational pressure, as indicated by the lack of significant correlation between GC3 and GC12 (Figure 4A). ENC-plot analysis of the 51 selected genes showed that the ENC ratio frequencies of 43 genes were distributed outside of the range −0.05 to 0.05 (Table 2), suggesting a significant influence of natural selection on the chloroplast genome’s CUB in A. tenerum. The PR2-plot illustrated uneven gene loci distribution in the chloroplast genome (Figure 4C), indicating the primary effect of natural selection on the third base during evolution. Overall, multivariate analysis identified natural selection as the primary influencing factor, followed by mutational pressure and other factors. These results align with studies in related angiosperms, such as the monocotyledons Zingiber [48] and the family Araceae [49,50], the basal angiosperm Manglietia [51], the important horticultural crop family Rutaceae [52] and Cucumis sativus within the Cucurbitaceae [53]. These results highlight the predominant role of natural selection in shaping codon usage bias, and are vital to explore the chloroplast genome evolution of angiosperms.
Additionally, eighteen optimal codons were identified based on their high frequency and expression levels. Notably, 17 of these optimal codons terminated with A or U. This pattern resembled the findings in other Cucurbitaceae genera. For instance, there were 12 optimal codons revealed by the Gynostemma, 11 of which ending with A or U [47]. Similarly, research on Cucumis sativus also showed a preference for A or U-ending optimal codons [53]. These consistent trends across different species underscore the universality and conservativeness of optimal codon usage.
5. Conclusions
Based on the assembly and annotation results, the chloroplast genome of A. tenerum was determined to be 160,579 bp in size, comprising the LSC, SSC, IRa, and IRb regions, exhibiting a quadripartite structure with an overall GC content of 36.5%. Additionally, the chloroplast genome of A. tenerum harbored 132 protein-coding genes. Notably, there were eighteen optimal codons, including GCA, AGA, CAA, GAA, GGA, GGU, CAU, AUA, AUU, UUA, UUG, AAA, UUU, UCU, ACA, UAU, GUA and GUU, all ending with A or U except for UUG. Analysis through Neutrality plot, ENC-plot and PR2-plot revealed that codon usage bias in the chloroplast genome of A. tenerum was mainly influenced by natural selection, with a preference for A and U bases. This study could offer insights into the evolution of A. tenerum and enhance our comprehension of codon patterns within angiosperm chloroplast genomes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cimb47100833/s1.
Author Contributions
Conceptualization, J.-S.Z.; formal analysis, J.-J.M. and J.-S.Z.; investigation, J.-J.M. and J.-S.Z.; data curation, J.-J.M. and J.-S.Z.; writing—original draft preparation, J.-J.M. and J.-S.Z.; writing—review and editing, J.-S.Z.; visualization, J.-J.M. and J.-S.Z.; supervision, J.-S.Z.; funding acquisition, J.-J.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Scientific Research Project of the Liaoning Education Department [LJKMZ20221808] and Anshan Normal University Project Funding-Doctoral Initiation Fund [23b02]. The APC was funded by the Scientific Research Project of the Liaoning Education Department [LJKMZ20221808].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The genome sequence of this study was deposited in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/ (accessed on 16 July 2025)) under accession no. PV938953.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Corriveau, J.L.; Coleman, A.W. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am. J. Bot. 1988, 75, 1443–1458. [Google Scholar] [CrossRef]
- Jansen, R.K.; Raubeson, L.A.; Boore, J.L.; dePamphilis, C.W.; Chumley, T.W.; Haberle, R.C.; Wyman, S.K.; Alverson, A.J.; Peery, R.; Herman, S.J.; et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Method Enzymol. 2005, 395, 348–384. [Google Scholar] [CrossRef]
- Cai, Z.-Q.; Guisinger, M.; Kim, H.-G.; Ruck, E.; Blazier, J.C.; McMurtry, V.; Kuehl, J.V.; Boore, J.; Jansen, R.K. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J. Mol. Evol. 2008, 67, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Jing, M.-L.; Bao, H.-J.; Ma, Y.-S.; Yang, X.-G. The complete chloroplast genome of Poa pratensis (Poaceae), a high-quality forage. Am. J. Plant Sci. 2021, 12, 1755–1760. [Google Scholar] [CrossRef]
- Chen, S.-L.; Yin, X.-M.; Han, J.-P.; Sun, W.; Yao, H.; Song, J.-Y.; Li, X.-W. DNA barcoding in herbal medicine: Retrospective and prospective. J. Pharm. Anal. 2023, 13, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-T.; Luo, Y.; Gan, L.; Ma, P.-F.; Gao, L.-M.; Yang, J.-B.; Cai, J.; Gitzendanner, M.A.; Fritsch, P.W.; Zhang, T.; et al. Plastid phylogenomic insights into relationships of all flowering plant families. BMC Biol. 2021, 19, 232. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, C.; Abdin, M.Z.; Kumar, S. Chloroplast genome transformation of medicinal plant Artemisia annua. Plant Biotechnol. 2020, 18, 2155–2157. [Google Scholar] [CrossRef]
- Dobrogojski, J.; Adamiec, M.; Luciński, R. The chloroplast genome: A review. Acta Physiol. Plant. 2020, 42, 98. [Google Scholar] [CrossRef]
- Cauz-Santos, L.A. Beyond conservation: The landscape of chloroplast genome rearrangements in angiosperms. New Phytol. 2025, 247, 2571–2580. [Google Scholar] [CrossRef]
- Li, N.; Li, Y.; Zheng, C.; Huang, J.; Zhang, S. Genome-wide comparative analysis of the codon usage patterns in plants. Genes Genom. 2016, 38, 723–731. [Google Scholar] [CrossRef]
- Iriarte, A.; Lamolle, G.; Musto, H. Codon usage bias: An endless tale. J. Mol. Evol. 2021, 89, 589–593. [Google Scholar] [CrossRef]
- Xu, C.; Cai, X.-N.; Chen, Q.-Z.; Zhou, H.-X.; Cai, Y.; Ben, A.-L. Factors affecting synonymous codon usage bias in chloroplast genome of Oncidium Gower Ramsey. Evol. Bioinform. 2011, 7, 271–278. [Google Scholar] [CrossRef]
- Romero, H.; Zavala, A.; Musto, H. Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces. Nucl. Acid. Res. 2000, 28, 2084–2090. [Google Scholar] [CrossRef] [PubMed]
- Duret, L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 2000, 16, 287–289. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.M.; Jeffrey, C. Actinostemma. In Flora of China; Wu, Z.-Y., Raven, P.H., Hong, D.-Y., Eds.; Science Press: Beijing, China, 2011; Volume 19, p. 18. [Google Scholar]
- Iwamoto, M.; Fujioka, T.; Okabe, H.; Mihashi, K.; Yamauchi, T. Studies on the constituents of Actinostemma lobatum Maxim. I. Structures of Actinostmmosides A, B, C and D, dammarane triterpene glycosides isolated from the herb. Chem. Pharm. Bull. 1987, 35, 553–561. [Google Scholar] [CrossRef][Green Version]
- Fujioka, T.; Iwase, Y.; Okabe, H.; Mihashi, K.; Yamauchi, T. Studies on the constituents of Actinostemma lobatum Maxim. II. Structures of actinostemmosides G and H, new dammarane triterpene glycosides isolated from the herb. Chem. Pharm. Bull. 1987, 35, 3870–3873. [Google Scholar] [CrossRef]
- Fujioka, T.; Iwamoto, M.; Iwase, Y.; Okabe, H.; Mihashi, K.; Yamauchi, T. Studies on the constituents of Actinostemma lobatum Maxim. III. Structures of actinostemmosides E and F, new baccharane-type triterpene glycosides isolated from the herb. Chem. Pharm. Bull. 1988, 36, 2772–2777. [Google Scholar] [CrossRef][Green Version]
- Fujioka, T.; Iwamoto, M.; Iwase, Y.; Hachiyama, S.; Okabe, H.; Yamauchi, T.; Mihashi, K. Studies on the constituents of Actinostemma lobatum Maxim. IV. Structures of lobatosides C, D and H, the dicrotalic acid esters of bayogenin bisdesmosides isolated from the herb. Chem. Pharm. Bull. 1989, 37, 1770–1775. [Google Scholar] [CrossRef][Green Version]
- Fujioka, T.; Iwamoto, M.; Iwase, Y.; Hachiyama, S.; Okabe, H.; Yamauchi, T.; Mihashi, K. Studies on the constituents of Actinostemma lobatum Maxim. V. Structures of lobatosides B, E, F and G, the dicrotalic acid esters of bayogenin bisdesmosides isolated from the herb. Chem. Pharm. Bull. 1989, 37, 2355–2360. [Google Scholar] [CrossRef]
- Fujioka, T.; Nagao, T.; Okabe, H.; Mihashi, K. Studies on the constituents of Actinostemma lobatum Maxim. VI. Structures of lobatosides I, J and K, oleanolic acid and gypsogenin glycosides isolated from the seed. Chem. Pharm. Bull. 1992, 40, 1105–1109. [Google Scholar] [CrossRef][Green Version]
- Fujioka, T.; Kashiwada, Y.; Okabe, H.; Mihashi, K.; Lee, K.H. Antitumor agents 171. Cytotoxicities of lobatosides B, C, D, and E, cyclic bisdesmosides isolated from Actinostemma lobatum maxim. Bioorg. Med. Chem. Lett. 1996, 6, 2807–2810. [Google Scholar] [CrossRef]
- Wu, Q.-N.; Wang, L.X.; Wang, Y.Z. Analyzing the inorganic elements and fat oil of the seeds in Actinostemma tenerum Griff. Nat. Prod. Res. Deve. 2001, 3, 33–35, (In Chinese with English abstract). [Google Scholar]
- Kim, D.K. Antioxidative constituents from the whole plant of Actinostemma lobatum maxim. J. Korean Soc. Appl. Biol. Chem. 2010, 53, 746–751. [Google Scholar] [CrossRef]
- Zhong, W.; Wang, Z.; Han, L.-N.; Gao, D.-Y.; Zhu, M.-M.; Jia, H.-T.; Xie, L.-L.; Xie, D.; Guo, H.-Y.; Zheng, L.-Y. Research progress on the chemical constituents and biological activities of Actinostemma lobatum Griff. Farm Prod. Proc. 2024, 11, 1–14, (In Chinese, with English abstract). [Google Scholar]
- Li, W.; Shi, S.-M.; Tang, Y.; Cao, J.-Q.; Yue, W.-W.; Zhao, Y.-Q. Chemical constituents from Actinostemma lobatum (I). Chin. Tradit. Herb. Drugs 2014, 45, 2143–2147, (In Chinese, with English abstract). [Google Scholar]
- Li, W.; Shi, S.-M.; Tang, Y.; Cao, J.-Q.; Zhao, Y.-Q. Chemical constituents from Actinostemma lobatum (II). Chin. Tradit. Herb. Drugs 2016, 47, 209–213, (In Chinese, with English abstract). [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; dePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq-versatile and accurate annotation of organelle genomes. Nucl. Acid. Res. 2017, 45, 6–11. [Google Scholar] [CrossRef]
- Chen, X.-H.; Zhao, Y.-D.; Xu, S.-H.; Zhou, Y.-Z.; Zhang, L.-J.; Qu, B.; Xu, Y.-F. Analysis of codon usage bias in the plastid genome of Diplandrorchis sinica (Orchidaceae). Curr. Issues Mol. Biol. 2024, 46, 9807–9820. [Google Scholar] [CrossRef] [PubMed]
- Vicario, S.; Moriyama, E.N.; Powell, J.R. Codon usage in twelve species of Drosophila. BMC Evol. Biol. 2007, 7, 226. [Google Scholar] [CrossRef]
- Tao, P.; Dai, L.; Luo, M.-C.; Tang, F.-Q.; Tien, P.; Pan, Z.-S. Analysis of synonymous codon usage in classical swine fever virus. Virus Genes 2009, 38, 104–112. [Google Scholar] [CrossRef]
- Yang, G.-F.; Su, K.-L.; Zhao, Y.-R.; Sun, J.; Song, Z.-B. Analysis of codon usage in the chloroplast genome of Medicago truncatula. Acta Prataculturae Sin. 2015, 24, 171–179. [Google Scholar]
- Sueoka, N. Translation-coupled violation of parity rule 2 in human genes is not the cause of heterogeneity of the DNA G+C content of third codon position. Gene 1999, 238, 53–58. [Google Scholar] [CrossRef]
- Sueoka, N. Near homogeneity of PR2-bias fingerprints in the human genome and their implications in phylogenetic analyses. J. Mol. Evol. 2001, 53, 469–476. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, T.; Yang, J.; Sun, J.-J.; Ju, M.-M.; Zhao, Y.-M.; Zhao, G.-F. Comparative analyses of chloroplast genomes of Cucurbitaceae species: Lights into selective pressures and phylogenetic relationships. Molecules 2018, 23, 2165. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.-W.; Yang, M.; Mo, C.-M.; Xie, W.-J.; Liu, C.; Wu, B.; Ma, X.-J. Complete chloroplast genomes of two Siraitia Merrill species: Comparative analysis, positive selection and novel molecular marker development. PLoS ONE 2019, 14, e0226865. [Google Scholar] [CrossRef] [PubMed]
- Bellot, S.; Mitchell, T.C.; Schaefer, H. Phylogenetic informativeness analyses to clarify past diversification processes in Cucurbitaceae. Sci. Rep. 2020, 10, 488. [Google Scholar] [CrossRef]
- Jiang, Z.-Z.; Hu, S.-B.; Yang, H.-D.; Guo, J. Comparative evolution of Trichosanthes based on chloroplast genomes. J. Anqing Norm. Univ. 2023, 29, 87–95, (In Chinese with English Abstract). [Google Scholar]
- Zhang, Y.; Shen, Z.-N.; Meng, X.-R.; Zhang, L.-M.; Liu, Z.-G.; Liu, M.-J.; Zhang, F.; Zhao, J. Codon usage patterns across seven Rosales species. BMC Plant Biol. 2022, 22, 65. [Google Scholar] [CrossRef]
- Liu, X.-Y.; Li, Y.; Ji, K.-K.; Zhu, J.; Ling, P.; Zhou, T.; Fan, L.-Y.; Xie, S.-Q. Genome-wide codon usage pattern analysis reveals the correlation between codon usage bias and gene expression in Cuscuta australis. Genomics 2020, 112, 2695–2702. [Google Scholar] [CrossRef]
- Sueoka, N. Intrastrand parity rules of DNA base composition and usage biases of synonymous codons. J. Mol. Evol. 1995, 40, 318–325. [Google Scholar] [CrossRef]
- Goetz, R.M.; Fuglsang, A. Correlation of codon bias measures with mRNA levels: Analysis of transcriptome data from Escherichia coli. Biochem. Biophys. Res. Commun. 2005, 327, 4–7. [Google Scholar] [CrossRef]
- Zhang, P.-P.; Xu, W.-B.; Lu, X.; Wang, L. Analysis of codon usage bias of chloroplast genomes in Gynostemma species. Physiol. Mol. Biol. Plants 2021, 27, 2727–2737. [Google Scholar] [CrossRef]
- Yang, Q.; Xin, C.; Xiao, Q.-S.; Lin, Y.-T.; Li, L.; Zhao, J.-L. Codon usage bias in chloroplast genes implicate adaptive evolution of four ginger species. Front. Plant Sci. 2023, 14, 1304264. [Google Scholar] [CrossRef]
- Jia, X.-B.; Wei, J.-Q.; Chen, Y.-W.; Zeng, C.-H.; Deng, C.; Zeng, P.-C.; Tang, Y.-F.; Zhou, Q.-H.; Huang, Y.-J.; Zhu, Q.-L. Codon usage patterns and genomic variation analysis of chloroplast genomes provides new insights into the evolution of Aroideae. Sci. Rep. 2025, 15, 4333. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, J.-S. Chloroplast genome evolution and codon usage in the medicinal plant Pothos chinensis (Araceae). Genes 2025, 16, 1017. [Google Scholar] [CrossRef]
- Luo, Y.; Luo, W.; Zhao, T.-X.; Yang, J.; Yuan, L.; Zhang, P.-Z.; Gong, Z.-X.; Li, H.-Z.; Sima, Y.; Xu, T. The complete chloroplast genomes of three Manglietia species and phylogenetic insight into the genus Manglietia Blume. Curr. Issues Mol. Biol. 2025, 47, 737. [Google Scholar] [CrossRef]
- Shen, L.-W.; Chen, S.-Q.; Liang, M.; Qu, S.; Feng, S.-J.; Wang, D.-W.; Wang, G. Comparative analysis of codon usage bias in chloroplast genomes of ten medicinal species of Rutaceae. BMC Plant Biol. 2024, 24, 424. [Google Scholar] [CrossRef]
- Niu, Y.; Luo, Y.-Y.; Wang, C.-L.; Liao, W.-B. Deciphering codon usage patterns in genome of Cucumis sativus in comparison with nine species of Cucurbitaceae. Agronomy 2021, 11, 2289. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).