Identification and Validation of Iron Metabolism-Related Biomarkers in Endometriosis: A Mendelian Randomization and Single-Cell Transcriptomics Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Identification and Enrichment Analyses of Differentially Expressed IM-RGs (DEIM-RGs)
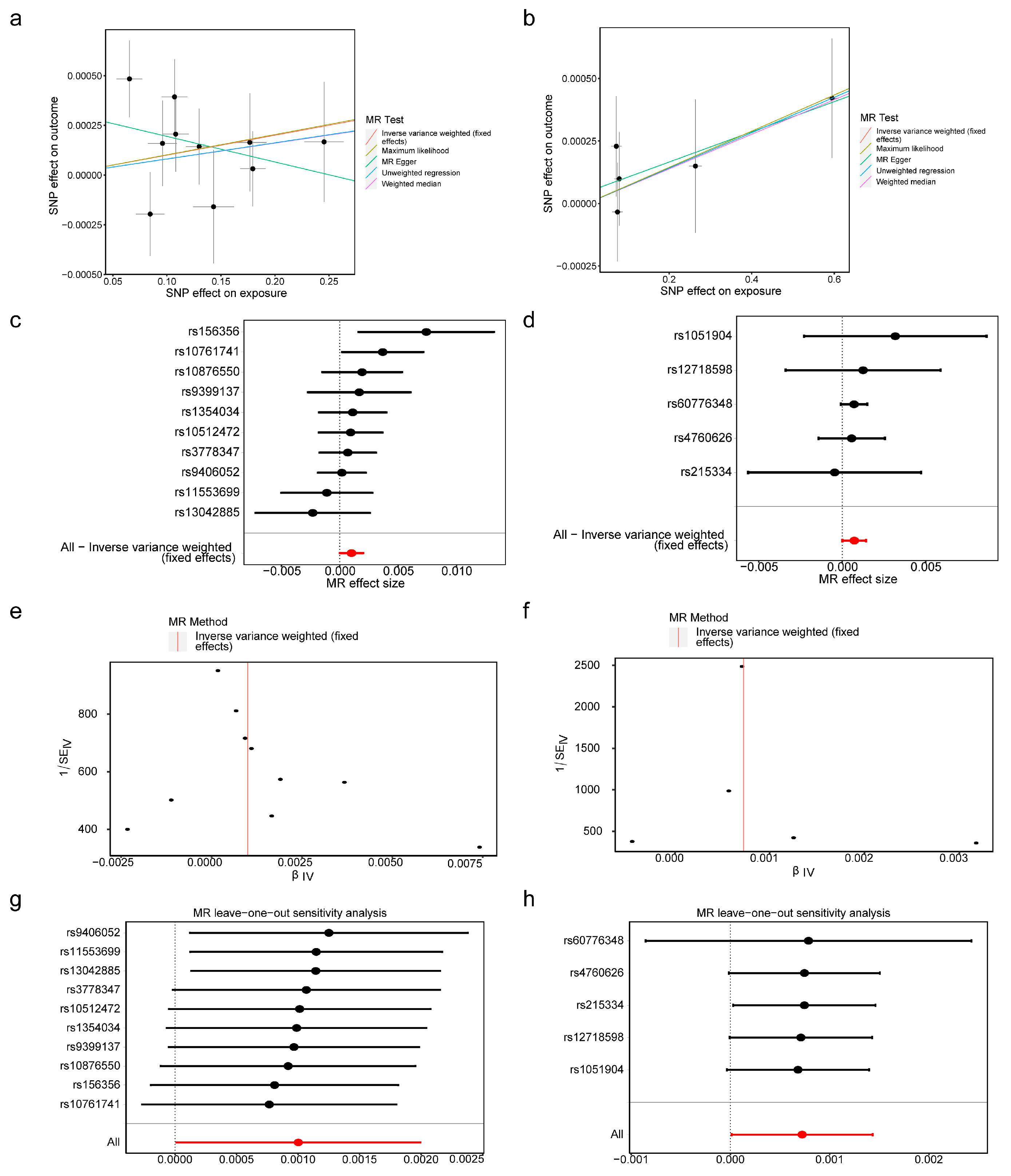
2.3. Screening of Instrumental Variables (IVs) in MR Analysis
2.4. Mendelian Randomization (MR) Analysis and Expression Verification
2.5. Functional Annotation of Biomarkers
2.6. Construction of Regulatory Network
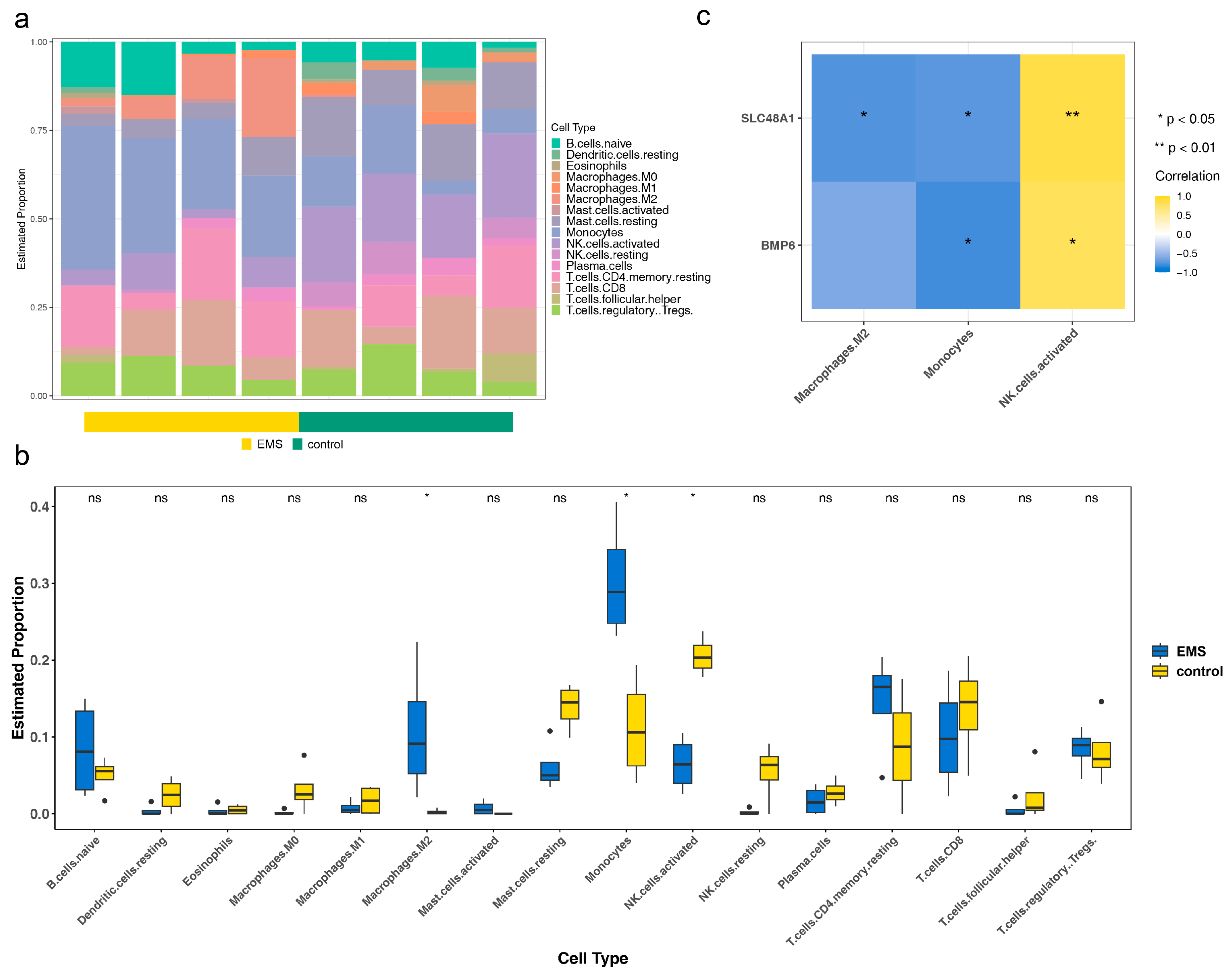
2.7. Immune Infiltration Analysis
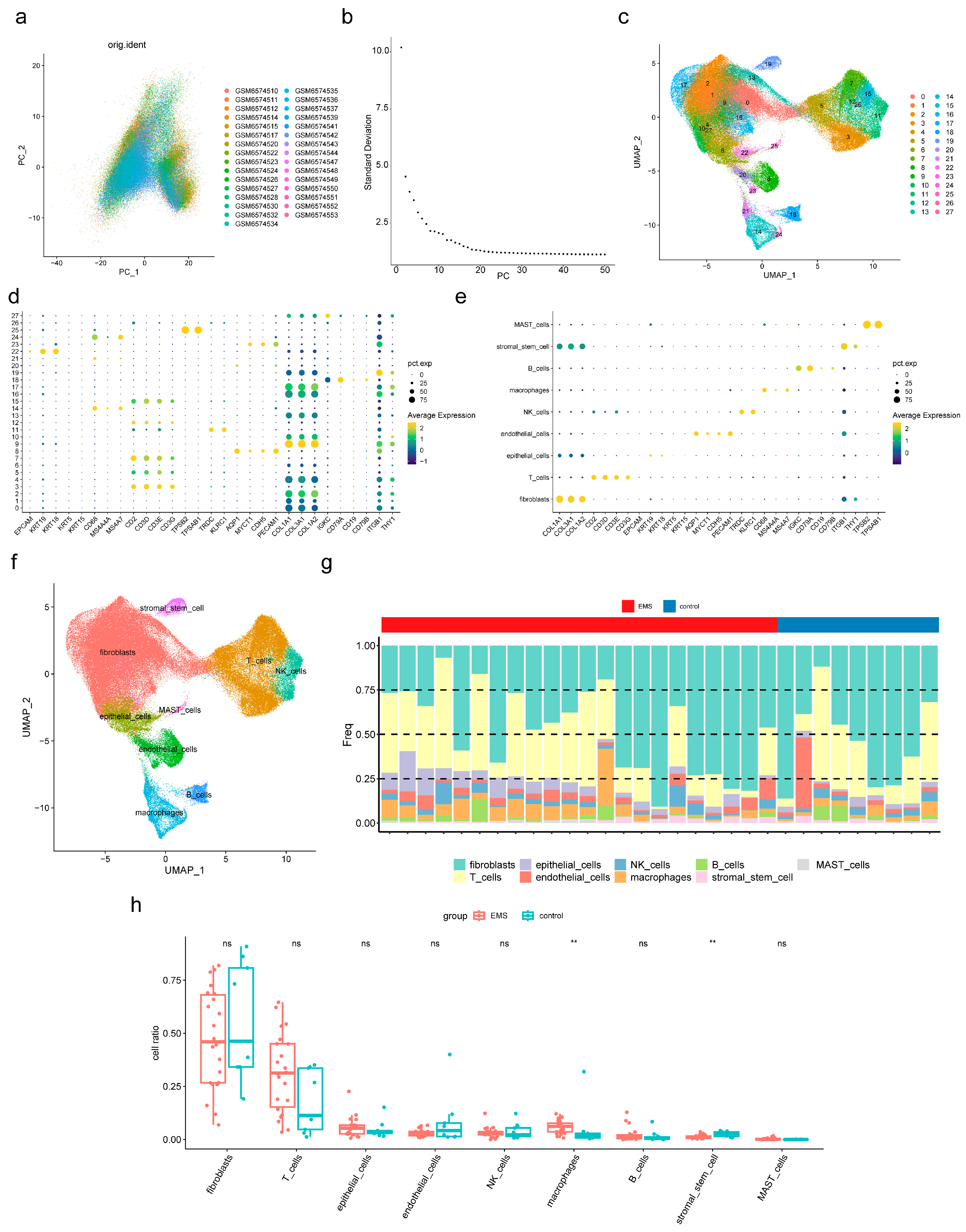
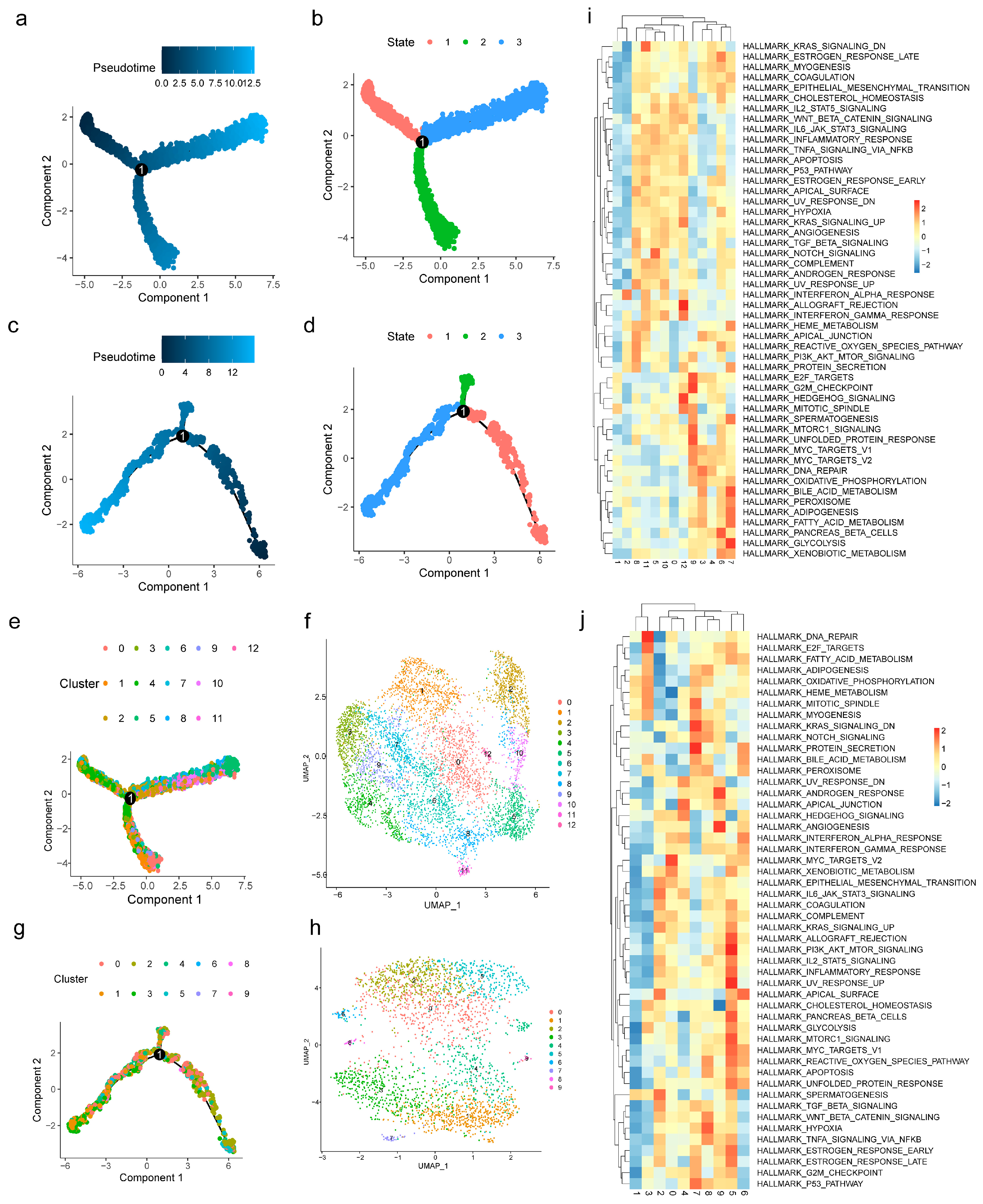
2.8. ScRNA-Seq Data Analysis
2.9. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
2.10. Statistical Analysis
3. Results
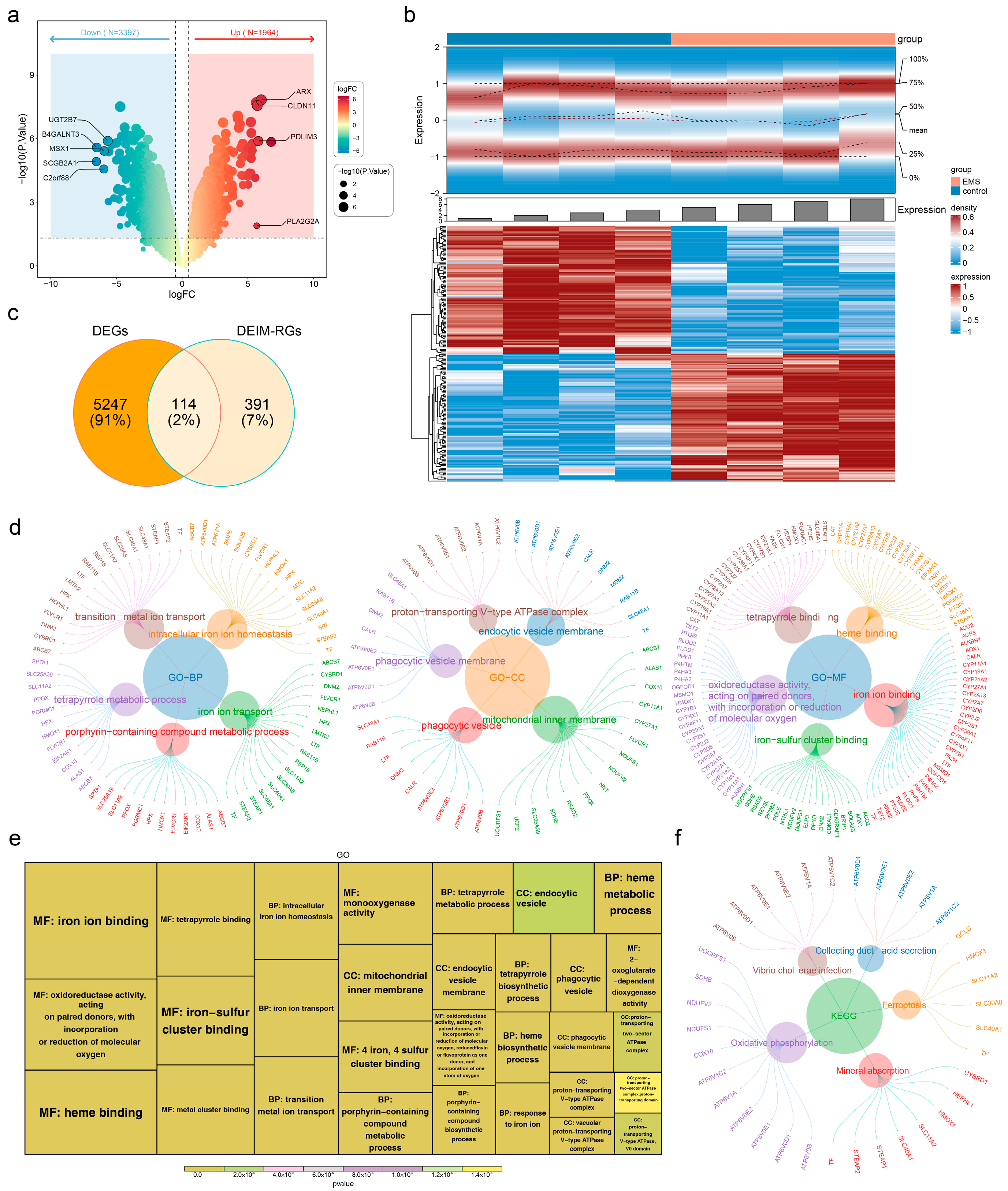
3.1. Screening and GO/KEGG Enrichment Analysis of DEIM-RGs
3.2. BMP6 and SLC48A1 Were Identified as Biomarkers
3.3. GeneMANIA and GSEA for BMP6 and SLC48A1
3.4. Network of lncRNA-miRNA-mRNA
3.5. The Roles of BMP6 and SLC48A1 in the Immune Microenvironment Might Be Consistent
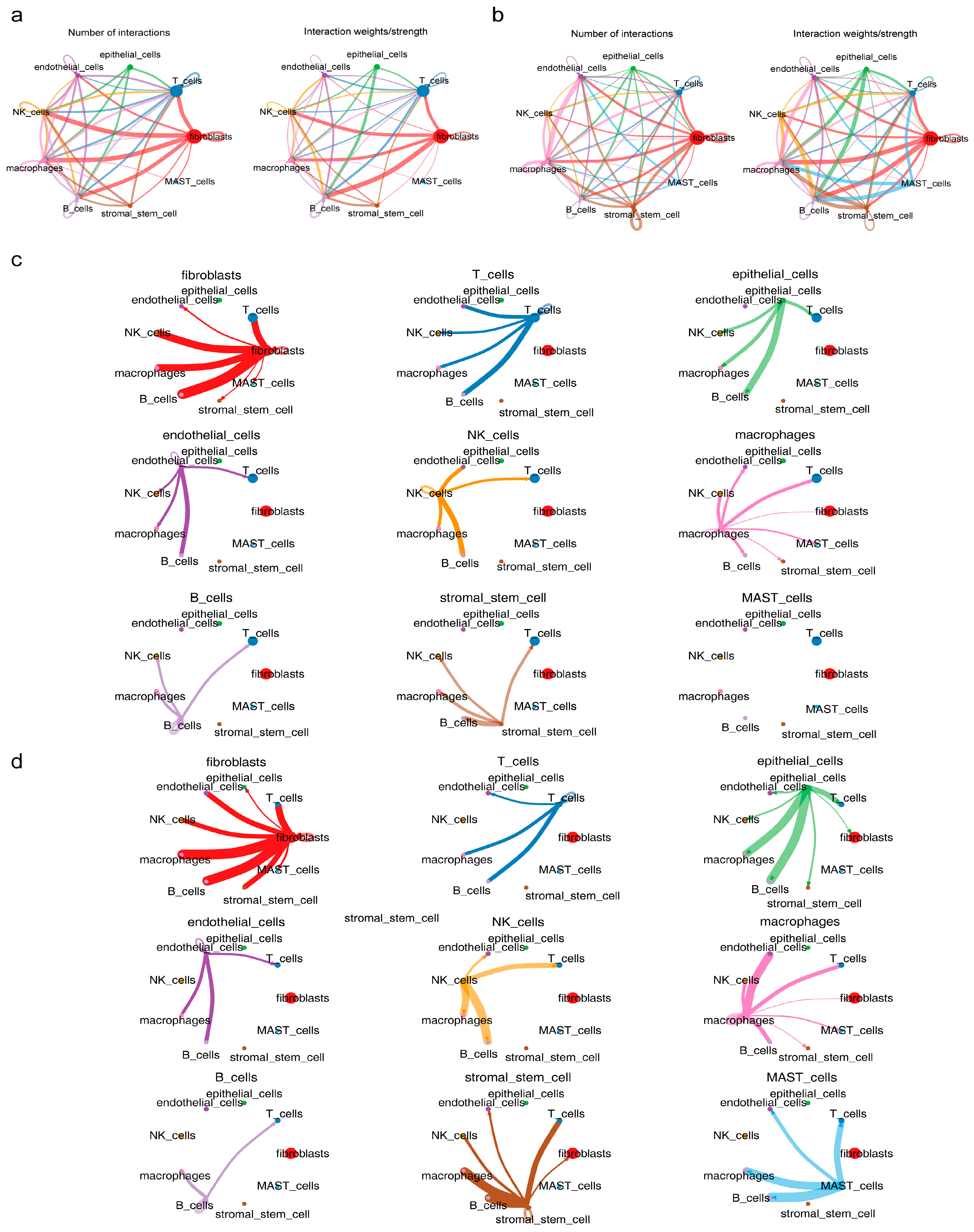
3.6. Macrophages and Stromal Stem Cells Were Designated as Key Cells
3.7. Functional Analysis of Key Cells
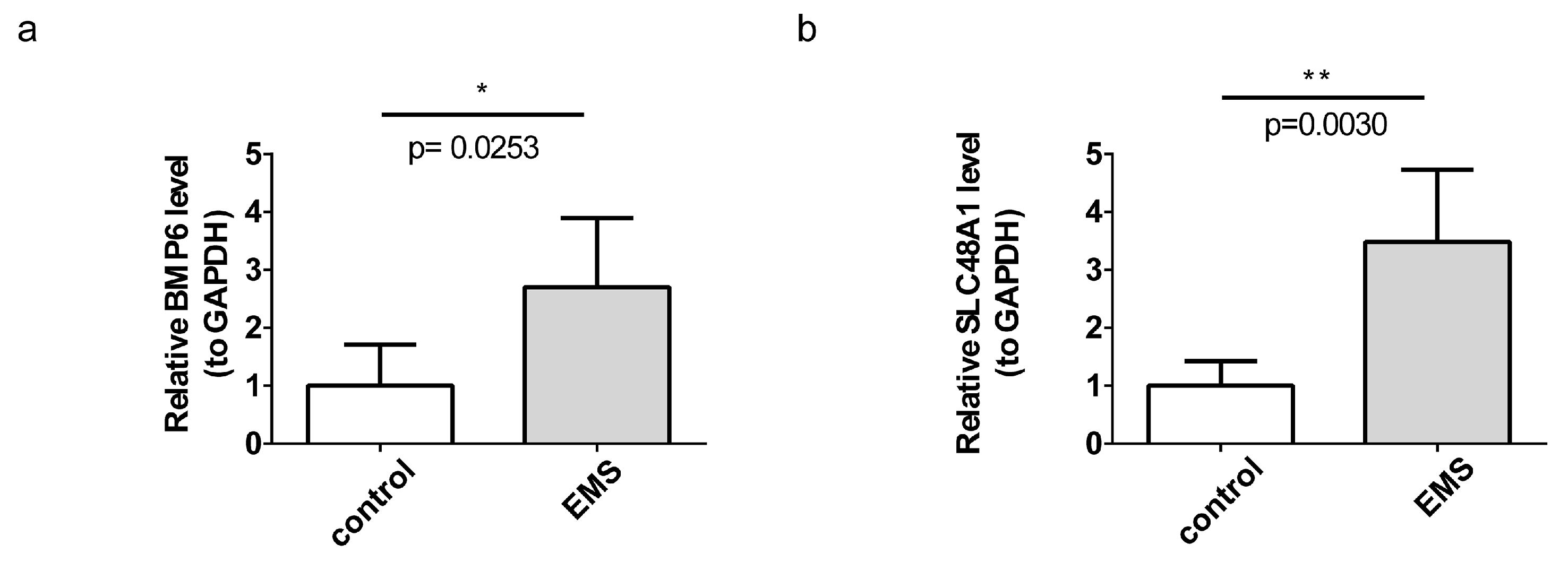
3.8. BMP6 and SLC48A1 Were Verified by RT-qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADAMTS13 | A Disintegrin and Metalloproteinase with Thrombospondin Motifs 13 |
| AHSG | Alpha-2-HS-Glycoprotein |
| ATF3/SLC7A11 | Activating Transcription Factor 3/Solute Carrier Family 7 Member 11 |
| BP | Biological process |
| BMP | Bone Morphogenetic Protein |
| BMP4 | Bone Morphogenetic Protein 4 |
| BMP6 | Bone Morphogenetic Protein 6 |
| BPs | Biological Processes |
| CCs | Cellular Components |
| CD16 | Cluster of Differentiation 16 |
| CD56 | Cluster of Differentiation 56 |
| cDNA | Complementary DNA |
| ceRNA | Competing endogenous RNA |
| CHRDL2 | Cysteine-Rich Dipeptidase-Like 2 |
| CI | Confidence Interval |
| CIBERSORT | Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts |
| DEGs | Differentially expressed genes |
| DEIM-RGs | Differentially Expressed IM-RGs |
| DNA | Deoxyribonucleic Acid |
| EC | Ectopic Endometrium |
| eQTL | Expression Quantitative Trait Loci |
| EU | Eutopic endometrium |
| FC | Fold change |
| FDR | False Discovery rate |
| GAPDH | Glyceraldehyde-3-Phosphate Dehydrogenase |
| GeneMANIA | Gene Microarray Analysis Network and Interaction Analysis |
| GEO | Gene expression omnibus |
| GGI | Gene-gene Interaction |
| GO | Gene ontology |
| GPL | Gene Expression Omnibus Platform |
| GSE | Gene Expression Series |
| GSEA | Gene set enrichment analysis |
| GSVA | Gene set variation analysis |
| GWAS | Genome-wide association study |
| HRG-1 | Heme Responsive Gene 1 |
| IEU | Integrative Epidemiology Unit |
| IL-33/ST2 | Interleukin-33/Suppressor of Tumorigenicity 2 |
| IM-RGs | Iron Metabolism-related Genes |
| IV | Instrumental Variable |
| IVs | Instrumental Variables |
| IVW | Inverse Variance Weighted |
| KEGG | Kyoto encyclopedia of genes and genomes |
| LOO | Leave-one-out |
| LD | Linkage Disequilibrium |
| MFs | Molecular Functions |
| MR | Mendelian Randomization |
| MSigDB | Molecular Signatures Database |
| MYC | Myelocytomatosis Oncogene |
| OR | Odd Ratio |
| NEAT1 | Nuclear Enriched Abundant Transcript 1 |
| NK | Natural Killer |
| PC | Principal Component |
| PCA | Principal Component Analysis |
| PINK1 | PTEN-Induced Kinase 1 |
| RNA | Ribonucleic Acid |
| ROC | Receiver operating characteristic |
| RT-qPCR | Reverse transcription-quantitative polymerase chain reaction |
| scRNA-seq | Single-cell RNA Sequencing |
| SLC | Solute Carrier Family |
| SLC48A1 | Solute Carrier Family 48 Member 1 |
| SNPs | Single Nucleotide Polymorphisms |
| TGFβ | Transforming Growth Factor Beta |
| UV | Ultraviolet radiation |
References
- Lamceva, J.; Uljanovs, R.; Strumfa, I. The Main Theories on the Pathogenesis of Endometriosis. Int. J. Mol. Sci. 2023, 24, 4254. [Google Scholar] [CrossRef] [PubMed]
- Zippl, A.L.; Reiser, E.; Seeber, B. Endometriosis and mental health disorders: Identification and treatment as part of a multimodal approach. Fertil. Steril. 2024, 121, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.M.; Bokor, A.; Heikinheimo, O.; Horne, A.; Jansen, F.; Kiesel, L.; King, K.; Kvaskoff, M.; Nap, A.; Petersen, K.; et al. ESHRE guideline: Endometriosis. Hum. Reprod. Open 2022, 2022, hoac009. [Google Scholar] [CrossRef]
- Crump, J.; Suker, A.; White, L. Endometriosis: A review of recent evidence and guidelines. Aust. J. Gen. Pract. 2024, 53, 11–18. [Google Scholar] [CrossRef]
- Kuznetsov, L.; Dworzynski, K.; Davies, M.; Overton, C. Diagnosis and management of endometriosis: Summary of NICE guidance. BMJ 2017, 358, j3935. [Google Scholar] [CrossRef]
- Xie, V. Effective biomarker measurement is key for biotherapeutic development. Bioanalysis 2022, 14, 451–453. [Google Scholar] [CrossRef]
- Raghunathan, R.; Turajane, K.; Wong, L.C. Biomarkers in Neurodegenerative Diseases: Proteomics Spotlight on ALS and Parkinson’s Disease. Int. J. Mol. Sci. 2022, 23, 9299. [Google Scholar] [CrossRef]
- Golestan, A.; Zareinejad, M.; Ramezani, A. Comprehensive biomarker profiles in hematological malignancies: Improving diagnosis, prognosis, and treatment. Biomark. Med. 2025, 19, 223–238. [Google Scholar] [CrossRef]
- Singh, S.S.; Allaire, C.; Al-Nourhji, O.; Bougie, O.; Bridge-Cook, P.; Duigenan, S.; Kroft, J.; Lemyre, M.; Leonardi, M.; Leyland, N.; et al. Guideline No. 449: Diagnosis and Impact of Endometriosis—A Canadian Guideline. J. Obstet. Gynaecol. Can. 2024, 46, 102450. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, Y.; Cheng, W.; Zhou, Z.; Ni, Z.; Yu, C. Double-edged roles of ferroptosis in endometriosis and endometriosis-related infertility. Cell Death Discov. 2023, 9, 306. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, Y.; Wang, B.; Yang, S. Ferroptosis: Iron-mediated cell death linked to disease pathogenesis. J. Biomed. Res. 2024, 38, 413–435. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.H.; Wang, H.L.; Liu, M.N.; Xu, T.M.; Zhang, K. Reflections on the complex mechanisms of endometriosis from the perspective of ferroptosis. Pathol. Res. Pract. 2024, 259, 155353. [Google Scholar] [CrossRef]
- Skarżyńska, E.; Wróbel, M.; Zborowska, H.; Kołek, M.F.; Mańka, G.; Kiecka, M.; Lipa, M.; Warzecha, D.; Spaczyński, R.; Piekarski, P.; et al. The Influence of Lactoferrin in Plasma and Peritoneal Fluid on Iron Metabolism in Women with Endometriosis. Int. J. Mol. Sci. 2023, 24, 1619. [Google Scholar] [CrossRef]
- Smith, G.D.; Ebrahim, S. Mendelian randomisation at 20 years: How can it avoid hubris, while achieving more? Lancet Diabetes Endocrinol. 2024, 12, 14–17. [Google Scholar] [CrossRef]
- Sanderson, E.; Glymour, M.M.; Holmes, M.V.; Kang, H.; Morrison, J.; Munafò, M.R.; Palmer, T.; Schooling, C.M.; Wallace, C.; Zhao, Q.; et al. Mendelian randomization. Nat. Rev. Methods Primers 2022, 2, 6. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, B.; Zu, Y. Identifying OGN as a Biomarker Covering Multiple Pathogenic Pathways for Diagnosing Heart Failure: From Machine Learning to Mechanism Interpretation. Biomolecules 2024, 14, 179. [Google Scholar] [CrossRef]
- Wang, L.; Li, X.; Wang, Y.; Li, G.; Dai, S.; Cao, M.; Meng, Z.; Ren, S. Endometriosis and epithelial ovarian cancer: A two-sample Mendelian randomization analysis. Sci. Rep. 2023, 13, 21992. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhou, L.; Liu, M.; Zhu, H.; Zhang, X.; Cai, E.; Xu, X.; Chen, T.; Cheng, H.; Liu, J.; et al. Single-cell analysis reveals insights into epithelial abnormalities in ovarian endometriosis. Cell Rep. 2024, 43, 113716. [Google Scholar] [CrossRef]
- Li, Y.; Liu, H.; Ye, S.; Zhang, B.; Li, X.; Yuan, J.; Du, Y.; Wang, J.; Yang, Y. The effects of coagulation factors on the risk of endometriosis: A Mendelian randomization study. BMC Med. 2023, 21, 195. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, X.; Wu, Y.; Zhang, B.; Wei, J.; Li, J.; Huang, Y.; Chen, L.; He, X. Bioinformatics identification and validation of biomarkers and infiltrating immune cells in endometriosis. Front. Immunol. 2022, 13, 944683. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fan, L.; Jin, L.; Lu, C.; Li, T.; Zhang, Z.; Xie, C.; Li, S.; Zhang, Y.; Ren, J.; et al. Integrated bioinformatic analysis of dysregulated microRNA-mRNA co-expression network in ovarian endometriosis. Acta Obstet. Gynecol. Scand. 2022, 101, 1074–1084. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.F.; Mishra, K.; Xie, Y.; Park, H.; Huang, K.Y.; Petretto, E.; Behmoaras, J. Systems level identification of a matrisome-associated macrophage polarisation state in multi-organ fibrosis. eLife 2023, 12, e85530. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Gustavsson, E.K.; Zhang, D.; Reynolds, R.H.; Garcia-Ruiz, S.; Ryten, M. ggtranscript: An R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics 2022, 38, 3844–3846. [Google Scholar] [CrossRef]
- Gao, C.H.; Yu, G.; Cai, P. ggVennDiagram: An Intuitive, Easy-to-Use, and Highly Customizable R Package to Generate Venn Diagram. Front. Genet. 2021, 12, 706907. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef]
- Li, Z.; Qi, W.; Zang, T.; Zhang, Z. The Causal Relationship Between Acne Vulgaris and BMI: A Mendelian Randomization Study. J. Cosmet. Dermatol. 2025, 24, e70092. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Zhang, Z.; Chen, Z.; Song, J.; Wang, B.; Jin, F. Association between periodontitis and breast cancer: Two-sample Mendelian randomization study. Clin. Oral Investig. 2023, 27, 2843–2849. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, H.; Yang, Z.; Lu, Y.; Zou, C. Causal associations between type 1 diabetes mellitus and cardiovascular diseases: A Mendelian randomization study. Cardiovasc. Diabetol. 2023, 22, 236. [Google Scholar] [CrossRef] [PubMed]
- Mao, R.; Yu, Q.; Li, J. The causal relationship between gut microbiota and inflammatory dermatoses: A Mendelian randomization study. Front. Immunol. 2023, 14, 1231848. [Google Scholar] [CrossRef]
- Xiang, M.; Wang, Y.; Gao, Z.; Wang, J.; Chen, Q.; Sun, Z.; Liang, J.; Xu, J. Exploring causal correlations between inflammatory cytokines and systemic lupus erythematosus: A Mendelian randomization. Front. Immunol. 2022, 13, 985729. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, S.; Tian, Y.; Si, H.; Zeng, Y.; Wu, Y.; Liu, Y.; Li, M.; Sun, K.; Wu, L.; et al. Genetic Causal Association between Iron Status and Osteoarthritis: A Two-Sample Mendelian Randomization. Nutrients 2022, 14, 3683. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Xu, S.; Gui, Y.; Chen, J.; Xu, J. Screening biomarkers for spinal cord injury using weighted gene co-expression network analysis and machine learning. Neural Regen. Res. 2024, 19, 2723–2734. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Wolock, S.L.; Lopez, R.; Klein, A.M. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 2019, 8, 281–291.e9. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhang, L.; Zhan, H.; Mo, Y.; Ren, Z.; Shao, A.; Lin, J. Single-cell transcriptomic analysis of endometriosis provides insights into fibroblast fates and immune cell heterogeneity. Cell Biosci. 2021, 11, 125. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Shen, H.H.; Zhang, T.; Yang, H.L.; Lai, Z.Z.; Zhou, W.J.; Mei, J.; Shi, J.W.; Zhu, R.; Xu, F.Y.; Li, D.J.; et al. Ovarian hormones-autophagy-immunity axis in menstruation and endometriosis. Theranostics 2021, 11, 3512–3526. [Google Scholar] [CrossRef]
- Li, Y.; Cai, L.; Guo, N.; Liu, C.; Wang, M.; Zhu, L.; Li, F.; Jin, L.; Sui, C. Oviductal extracellular vesicles from women with endometriosis impair embryo development. Front. Endocrinol. 2023, 14, 1171778. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Lu, X.; Lin, K.; Li, J.; Yuan, M.; Lin, K. m6A methylation of RNF43 inhibits the progression of endometriosis through regulating oxidative phosphorylation via NDUFS1. J. Cell Physiol. 2024, 239, e31367. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Fan, W.; Yu, X.; Liu, J.; Liu, P. Research into the mechanism of intervention of SanQi in endometriosis based on network pharmacology and molecular docking technology. Medicine 2022, 101, e30021. [Google Scholar] [CrossRef]
- Okoye, C.N.; Koren, S.A.; Wojtovich, A.P. Mitochondrial complex I ROS production and redox signaling in hypoxia. Redox Biol. 2023, 67, 102926. [Google Scholar] [CrossRef]
- Arroum, T.; Borowski, M.T.; Marx, N.; Schmelter, F.; Scholz, M.; Psathaki, O.E.; Hippler, M.; Enriquez, J.A.; Busch, K.B. Loss of respiratory complex I subunit NDUFB10 affects complex I assembly and supercomplex formation. Biol. Chem. 2023, 404, 399–415. [Google Scholar] [CrossRef] [PubMed]
- McGregor, L.; Acajjaoui, S.; Desfosses, A.; Saïdi, M.; Bacia-Verloop, M.; Schwarz, J.J.; Juyoux, P.; von Velsen, J.; Bowler, M.W.; McCarthy, A.A.; et al. The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination. Nat. Commun. 2023, 14, 8248. [Google Scholar] [CrossRef]
- Gibbs, E.T.; Lerner, C.A.; Watson, M.A.; Wong, H.S.; Gerencser, A.A.; Brand, M.D. Site IQ in mitochondrial complex I generates S1QEL-sensitive superoxide/hydrogen peroxide in both the reverse and forward reactions. Biochem. J. 2023, 480, 363–384. [Google Scholar] [CrossRef]
- Fang, X.; Deng, Q.; Yang, H.; Yan, Z.; Peng, Z.; Zhao, Y.; Liao, T.; Tu, Z.; Liu, J.; Liu, L.; et al. Causal association of immune cells and endometriosis: A Mendelian randomization study. Front. Endocrinol. 2024, 15, 1397670. [Google Scholar] [CrossRef]
- Yang, S.; Wang, H.; Li, D.; Li, M. An Estrogen-NK Cells Regulatory Axis in Endometriosis, Related Infertility, and Miscarriage. Int. J. Mol. Sci. 2024, 25, 3362. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Chung, Y.J.; Moon, S.W.; Choi, E.J.; Kim, M.R.; Chung, Y.J.; Lee, S.H. Single-cell profiling identifies distinct hormonal, immunologic, and inflammatory signatures of endometriosis-constituting cells. J. Pathol. 2023, 261, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Hosseinzadeh, R.; Moini, A.; Hosseini, R.; Fatehnejad, M.; Yekaninejad, M.S.; Javidan, M.; Changaei, M.; Feizisani, F.; Rajaei, S. A higher number of exhausted local PD1+, but not TIM3+, NK cells in advanced endometriosis. Heliyon 2024, 10, e23294. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Imanaka, S.; Yoshimoto, C.; Matsubara, S.; Shigetomi, H. Role of autophagy and ferroptosis in the development of endometriotic cysts (Review). Int. J. Mol. Med. 2024, 54, 78. [Google Scholar] [CrossRef]
- Liu, M.; Wu, K.; Wu, Y. The emerging role of ferroptosis in female reproductive disorders. Biomed. Pharmacother. 2023, 166, 115415. [Google Scholar] [CrossRef]
- Liu, M.N.; Chen, L.; Xu, T.M.; Zhang, K. Potential clinical implications of iron metabolism in ovarian endometriosis. J. Trace Elem. Med. Biol. 2022, 73, 127017. [Google Scholar] [CrossRef]
- Polak, G.; Barczyński, B.; Wertel, I.; Kwaśniewski, W.; Bednarek, W.; Derewianka-Polak, M.; Frąszczak, K.; Olajossy, M.; Kotarski, J. Disrupted iron metabolism in peritoneal fluid may induce oxidative stress in the peritoneal cavity of women with endometriosis. Ann. Agric. Environ. Med. 2018, 25, 587–592. [Google Scholar] [CrossRef]
- Shen, R.; Jia, R.; Liu, W.; Lin, Q.; Hai, Y.; He, Z. The Function and Regulation of BMP6 in Various Kinds of Stem Cells. Curr. Pharm. Des. 2015, 21, 3634–3643. [Google Scholar] [CrossRef]
- Zurawska, G.; Jończy, A.; Niklewicz, M.; Sas, Z.; Rumieńczyk, I.; Kulecka, M.; Piwocka, K.; Rygiel, T.P.; Mikula, M.; Mleczko-Sanecka, K. Iron-triggered signaling via ETS1 and the p38/JNK MAPK pathway regulates Bmp6 expression. Am. J. Hematol. 2024, 99, 543–554. [Google Scholar] [CrossRef]
- Latour, C.; Besson-Fournier, C.; Meynard, D.; Silvestri, L.; Gourbeyre, O.; Aguilar-Martinez, P.; Schmidt, P.J.; Fleming, M.D.; Roth, M.P.; Coppin, H. Differing impact of the deletion of hemochromatosis-associated molecules HFE and transferrin receptor-2 on the iron phenotype of mice lacking bone morphogenetic protein 6 or hemojuvelin. Hepatology 2016, 63, 126–137. [Google Scholar] [CrossRef]
- Galaris, D.; Barbouti, A.; Pantopoulos, K. Iron homeostasis and oxidative stress: An intimate relationship. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 118535. [Google Scholar] [CrossRef]
- Scutiero, G.; Iannone, P.; Bernardi, G.; Bonaccorsi, G.; Spadaro, S.; Volta, C.A.; Greco, P.; Nappi, L. Oxidative Stress and Endometriosis: A Systematic Review of the Literature. Oxid. Med. Cell Longev. 2017, 2017, 7265238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Chang, H.M.; Zhu, H.; Liu, R.Z.; Leung, P.C.K. BMP6 increases TGF-β1 production by up-regulating furin expression in human granulosa-lutein cells. Cell Signal 2019, 55, 109–118. [Google Scholar] [CrossRef]
- Akiyama, I.; Yoshino, O.; Osuga, Y.; Shi, J.; Takamura, M.; Harada, M.; Koga, K.; Hirota, Y.; Hirata, T.; Fujii, T.; et al. The role of bone morphogenetic protein 6 in accumulation and regulation of neutrophils in the human ovary. Reprod. Sci. 2014, 21, 772–777. [Google Scholar] [CrossRef]
- Simmons, W.R.; Wain, L.; Toker, J.; Jagadeesh, J.; Garrett, L.J.; Pek, R.H.; Hamza, I.; Bodine, D.M. Normal Iron Homeostasis Requires the Transporter SLC48A1 for Efficient Heme-Iron Recycling in Mammals. Front. Genome Ed. 2020, 2, 8. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Yoshimoto, C.; Matsubara, S.; Shigetomi, H.; Imanaka, S. Current Understanding of and Future Directions for Endometriosis-Related Infertility Research with a Focus on Ferroptosis. Diagnostics 2023, 13, 1926. [Google Scholar] [CrossRef]
- Dong, X.; Xu, L.; Wang, S.; Jiao, X.; Yan, S.; Huang, Y.; Yuan, M.; Wang, G. Endometrial stromal cell autophagy-dependent ferroptosis caused by iron overload in ovarian endometriosis is inhibited by the ATF4-xCT pathway. Mol. Hum. Reprod. 2023, 30, gaad046. [Google Scholar] [CrossRef]
- Grange, C.; Lux, F.; Brichart, T.; David, L.; Couturier, A.; Leaf, D.E.; Allaouchiche, B.; Tillement, O. Iron as an emerging therapeutic target in critically ill patients. Crit. Care 2023, 27, 475. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhao, X.; Zhang, L.; Xia, Q.; Peng, Y.; Zhang, H.; Yan, D.; Yang, Z.; Li, J. Iron overload inhibits cell proliferation and promotes autophagy via PARP1/SIRT1 signaling in endometriosis and adenomyosis. Toxicology 2022, 465, 153050. [Google Scholar] [CrossRef]
- Donnez, J.; Binda, M.M.; Donnez, O.; Dolmans, M.M. Oxidative stress in the pelvic cavity and its role in the pathogenesis of endometriosis. Fertil. Steril. 2016, 106, 1011–1017. [Google Scholar] [CrossRef]
- Abramiuk, M.; Grywalska, E.; Małkowska, P.; Sierawska, O.; Hrynkiewicz, R.; Niedźwiedzka-Rystwej, P. The Role of the Immune System in the Development of Endometriosis. Cells 2022, 11, 2028. [Google Scholar] [CrossRef]
- Ramírez-Pavez, T.N.; Martínez-Esparza, M.; Ruiz-Alcaraz, A.J.; Marín-Sánchez, P.; Machado-Linde, F.; García-Peñarrubia, P. The Role of Peritoneal Macrophages in Endometriosis. Int. J. Mol. Sci. 2021, 22, 792. [Google Scholar] [CrossRef]
- Kobayashi, H.; Imanaka, S. Understanding the molecular mechanisms of macrophage polarization and metabolic reprogramming in endometriosis: A narrative review. Reprod. Med. Biol. 2022, 21, e12488. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liang, Z.; Jiang, J.; Feng, X.; Liu, J.; Zhang, Z.; Wang, H.; Wang, N.; Gou, Y.; Li, Z.; et al. Macrophages originated IL-33/ST2 inhibits ferroptosis in endometriosis via the ATF3/SLC7A11 axis. Cell Death Dis. 2023, 14, 668. [Google Scholar] [CrossRef] [PubMed]
- Vallvé-Juanico, J.; George, A.F.; Sen, S.; Thomas, R.; Shin, M.G.; Kushnoor, D.; Vásquez, J.J.; Vo, K.C.; Irwin, J.C.; Roan, N.R.; et al. Deep immunophenotyping reveals endometriosis is marked by dysregulation of the mononuclear phagocytic system in endometrium and peripheral blood. BMC Med. 2022, 20, 158. [Google Scholar] [CrossRef]
- Fukui, A.; Mai, C.; Saeki, S.; Yamamoto, M.; Takeyama, R.; Kato, T.; Ukita, Y.; Wakimoto, Y.; Yamaya, A.; Shibahara, H. Pelvic endometriosis and natural killer cell immunity. Am. J. Reprod. Immunol. 2021, 85, e13342. [Google Scholar] [CrossRef] [PubMed]
- Koller, D.; Pathak, G.A.; Wendt, F.R.; Tylee, D.S.; Levey, D.F.; Overstreet, C.; Gelernter, J.; Taylor, H.S.; Polimanti, R. Epidemiologic and Genetic Associations of Endometriosis With Depression, Anxiety, and Eating Disorders. JAMA Netw. Open 2023, 6, e2251214. [Google Scholar] [CrossRef]
- Duarte-Silva, E.; Maes, M.; Alves Peixoto, C. Iron metabolism dysfunction in neuropsychiatric disorders: Implications for therapeutic intervention. Behav. Brain Res. 2025, 479, 115343. [Google Scholar] [CrossRef]
- Tortuyaux, R.; Avila-Gutierrez, K.; Oudart, M.; Mazaré, N.; Mailly, P.; Deschemin, J.C.; Vaulont, S.; Escartin, C.; Cohen-Salmon, M. Physiopathological changes of ferritin mRNA density and distribution in hippocampal astrocytes in the mouse brain. J. Neurochem. 2023, 164, 847–857. [Google Scholar] [CrossRef]

| Outcome | Exposure | Method | nSNP | p Value | or | or_lci95 | or_uci95 |
|---|---|---|---|---|---|---|---|
| ukb-b-10903 | eqtl-a-ENSG00000153162 (BMP6) | Inverse variance weighted | 10 | 0.04750698 | 1.001002221 | 1.000010995 | 1.001994429 |
| ukb-b-10903 | eqtl-a-ENSG00000211584 (SLC48A1) | Inverse variance weighted | 5 | 0.046119158 | 1.000723176 | 1.000012434 | 1.001434424 |
| Symbol | Exposure | Method | Q | Q_df | Q_p Value |
|---|---|---|---|---|---|
| SLC48A1 | eqtl-a-ENSG00000211584 | Inverse variance weighted | 1.048 | 4.000 | 0.902 |
| BMP6 | eqtl-a-ENSG00000153162 | Inverse variance weighted | 10.915 | 9.000 | 0.282 |
| Symbol | Exposure | Egger_Intercept | se | p Value |
|---|---|---|---|---|
| BMP6 | eqtl-a-ENSG00000153162 | 0.000323385 | 0.000198411 | 0.141775357 |
| SLC48A1 | eqtl-a-ENSG00000211584 | 4.52 × 10−5 | 0.000131705 | 0.753939952 |
| Exposure | Symbol | snp_r2.exposure | snp_r2.outcome | Correct_Causal_Direction | Steiger_p Value |
|---|---|---|---|---|---|
| eqtl-a-ENSG00000153162 | BMP6 | 0.031483363 | 3.22 × 10−5 | TRUE | 1.63 × 10−194 |
| eqtl-a-ENSG00000211584 | SLC48A1 | 0.042311398 | 1.09 × 10−5 | TRUE | 1.11 × 10−268 |
| Cell Clusters | Cell Subpopulations |
|---|---|
| 0, 1, 2, 4, 9, 10, 13, 16, 17, 27 | fibroblasts |
| 3, 5, 7, 12, 15 | T cells |
| 8, 20, 23 | endothelial cells |
| 6, 22 | epithelial cells |
| 11, 26 | NK cells |
| 14, 21, 24 | macrophages |
| 18 | B cells |
| 19 | stromal stem cell |
| 25 | MAST cells |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, J.; Lv, Z.; Luo, X. Identification and Validation of Iron Metabolism-Related Biomarkers in Endometriosis: A Mendelian Randomization and Single-Cell Transcriptomics Study. Curr. Issues Mol. Biol. 2025, 47, 831. https://doi.org/10.3390/cimb47100831
Du J, Lv Z, Luo X. Identification and Validation of Iron Metabolism-Related Biomarkers in Endometriosis: A Mendelian Randomization and Single-Cell Transcriptomics Study. Current Issues in Molecular Biology. 2025; 47(10):831. https://doi.org/10.3390/cimb47100831
Chicago/Turabian StyleDu, Juan, Zili Lv, and Xiaohong Luo. 2025. "Identification and Validation of Iron Metabolism-Related Biomarkers in Endometriosis: A Mendelian Randomization and Single-Cell Transcriptomics Study" Current Issues in Molecular Biology 47, no. 10: 831. https://doi.org/10.3390/cimb47100831
APA StyleDu, J., Lv, Z., & Luo, X. (2025). Identification and Validation of Iron Metabolism-Related Biomarkers in Endometriosis: A Mendelian Randomization and Single-Cell Transcriptomics Study. Current Issues in Molecular Biology, 47(10), 831. https://doi.org/10.3390/cimb47100831






