Identification and Validation of a Macrophage Phagocytosis-Related Gene Signature for Prognostic Prediction in Colorectal Cancer (CRC)
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Preparation
2.2. Macrophage Phagocytosis-Related Genes Identification and Enrichment Analysis
2.3. Construction and Validation of the Risk Model
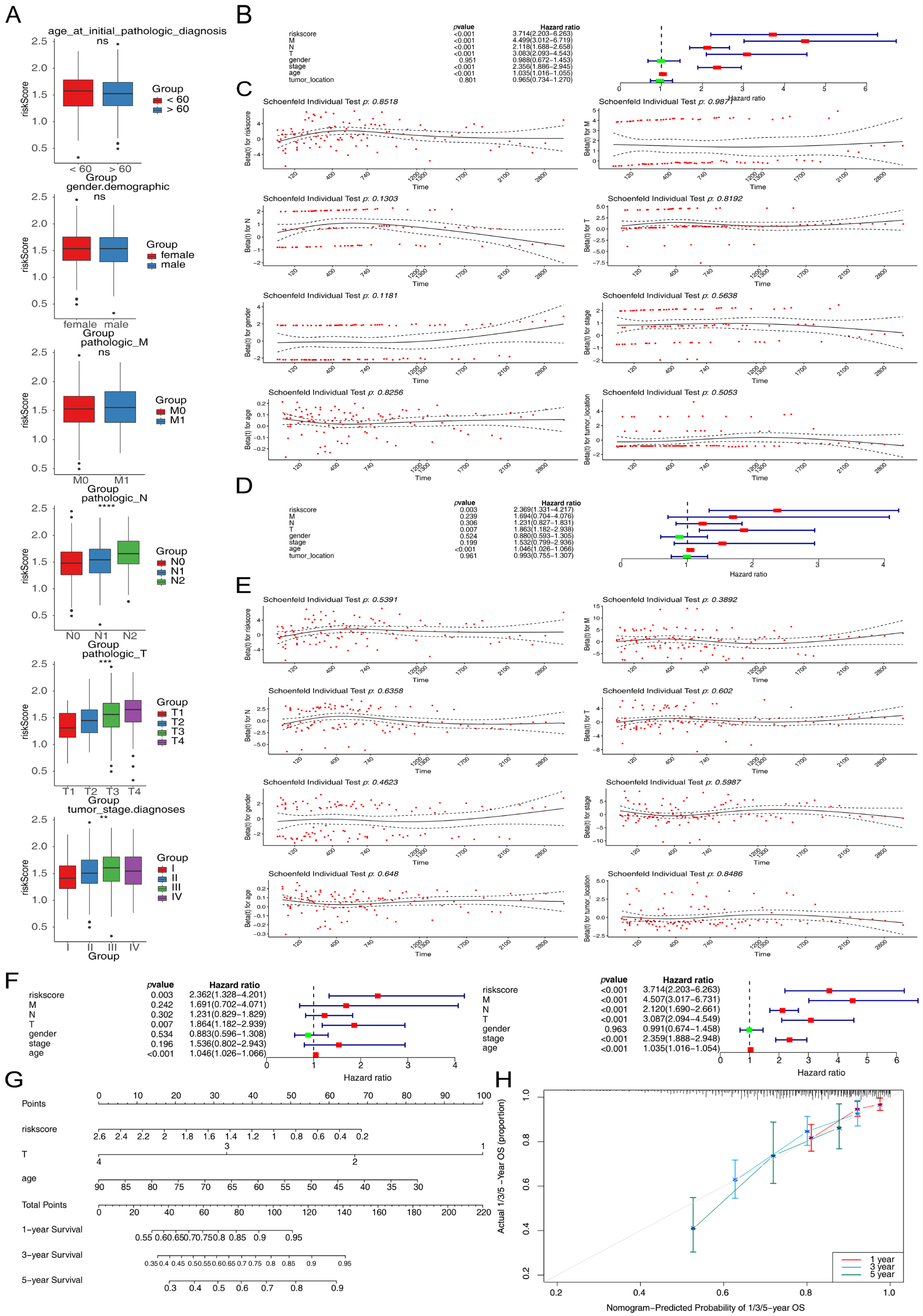
2.4. Clinical Correlation and Independent Prognostic Analysis
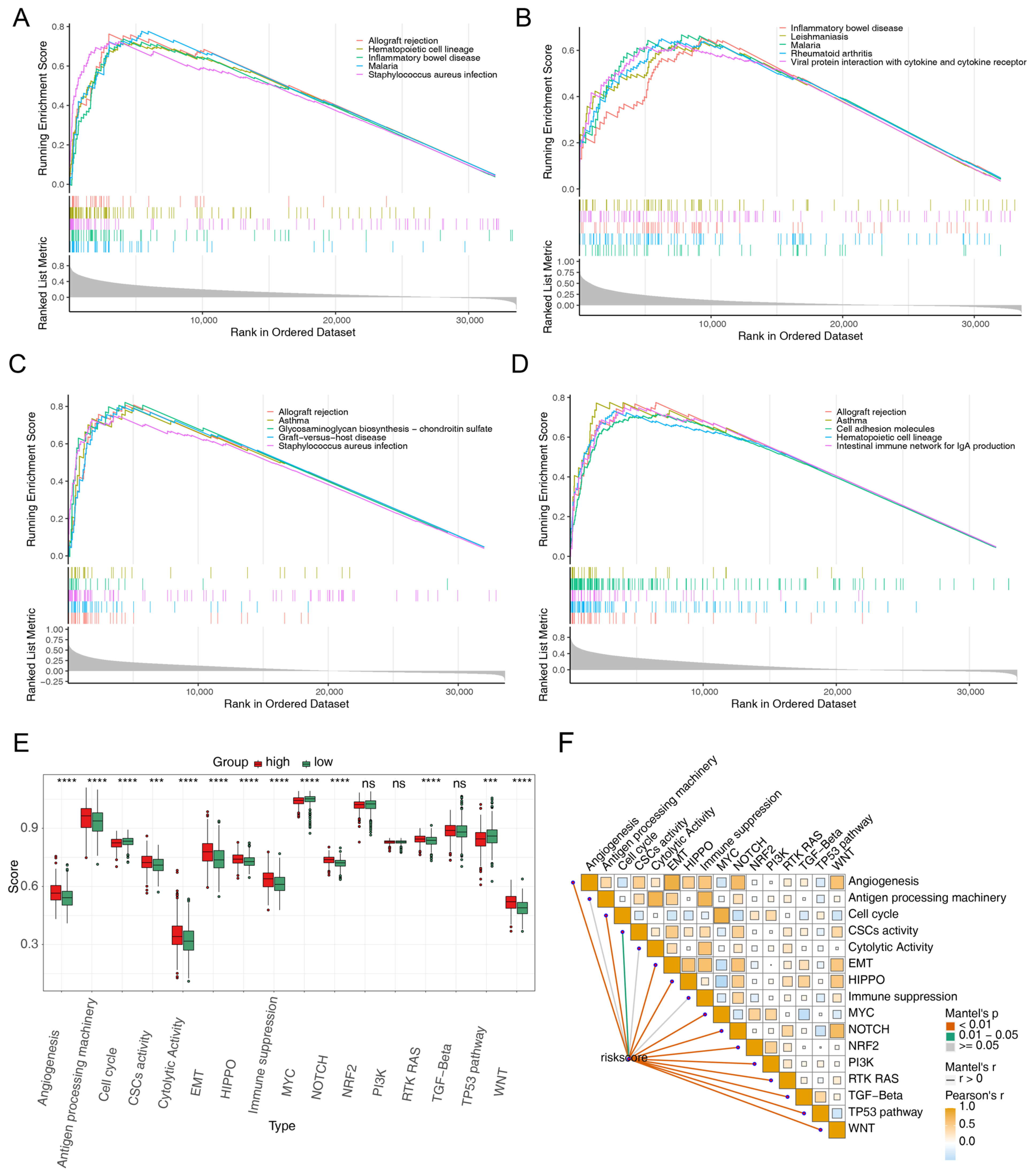
2.5. Single-Gene Gene Set Enrichment Analysis (GSEA)
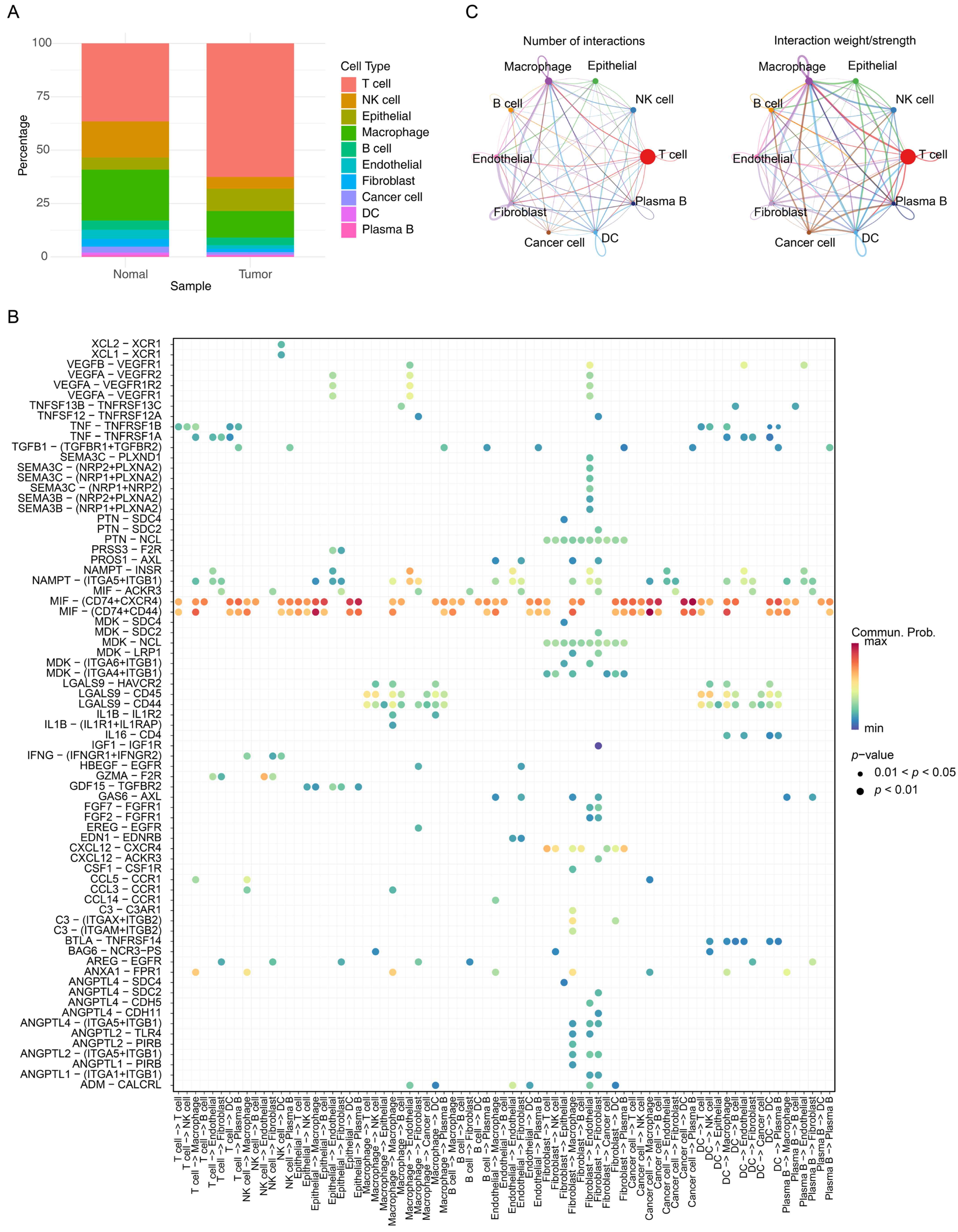
2.6. Tumor Microenvironment (TME) Analysis
2.7. Immunotherapy and Chemotherapy Response Analysis
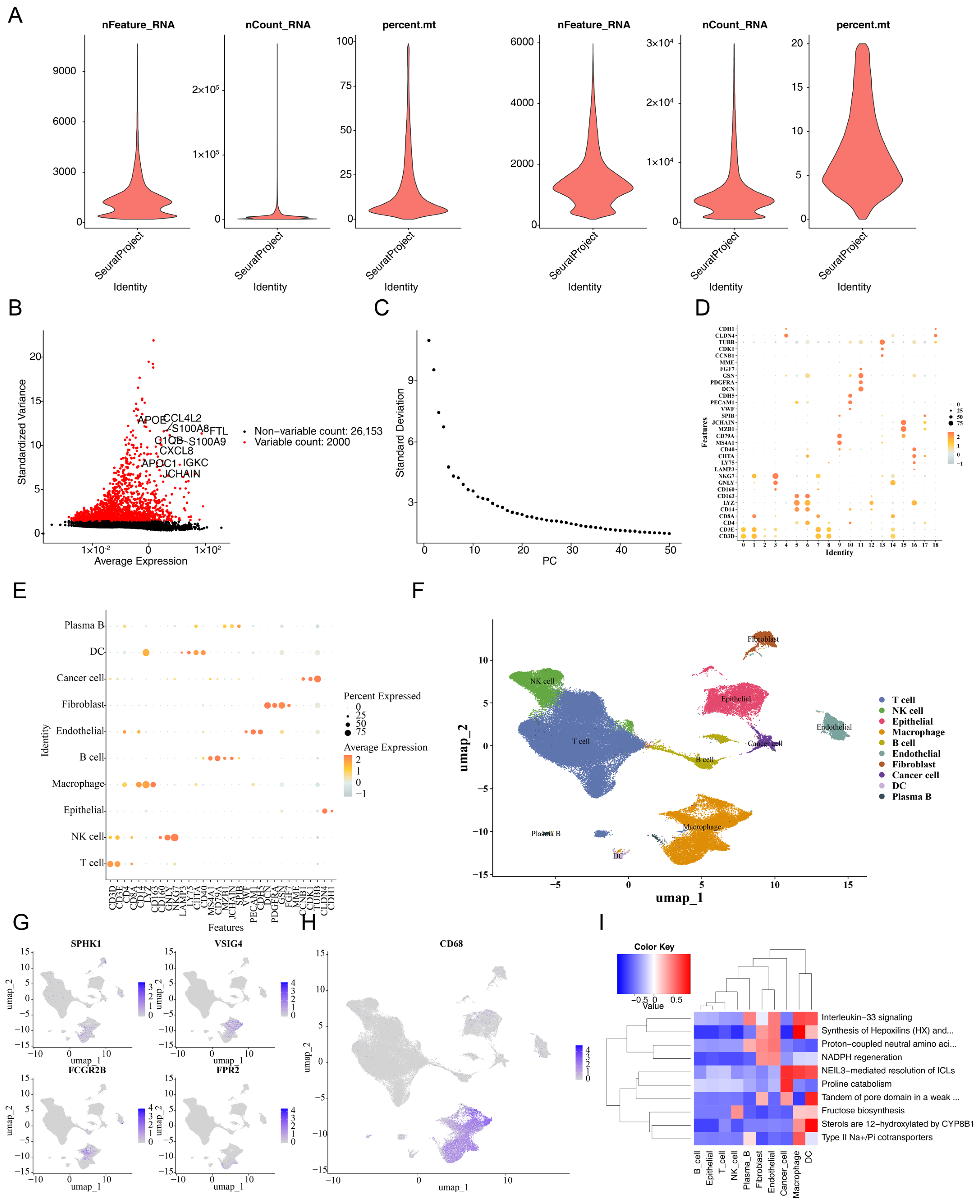
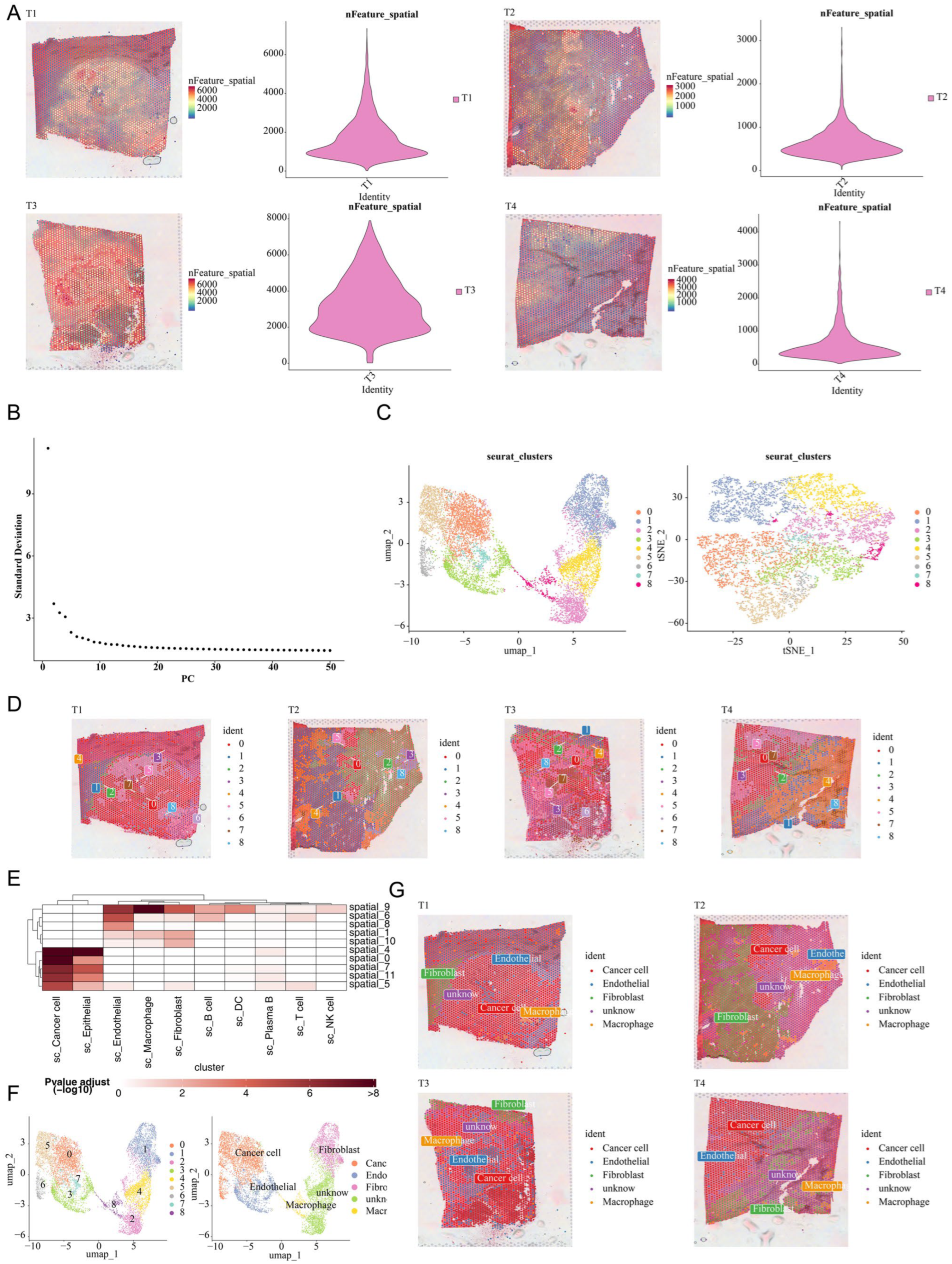
2.8. Single-Cell RNA Sequencing Analysis
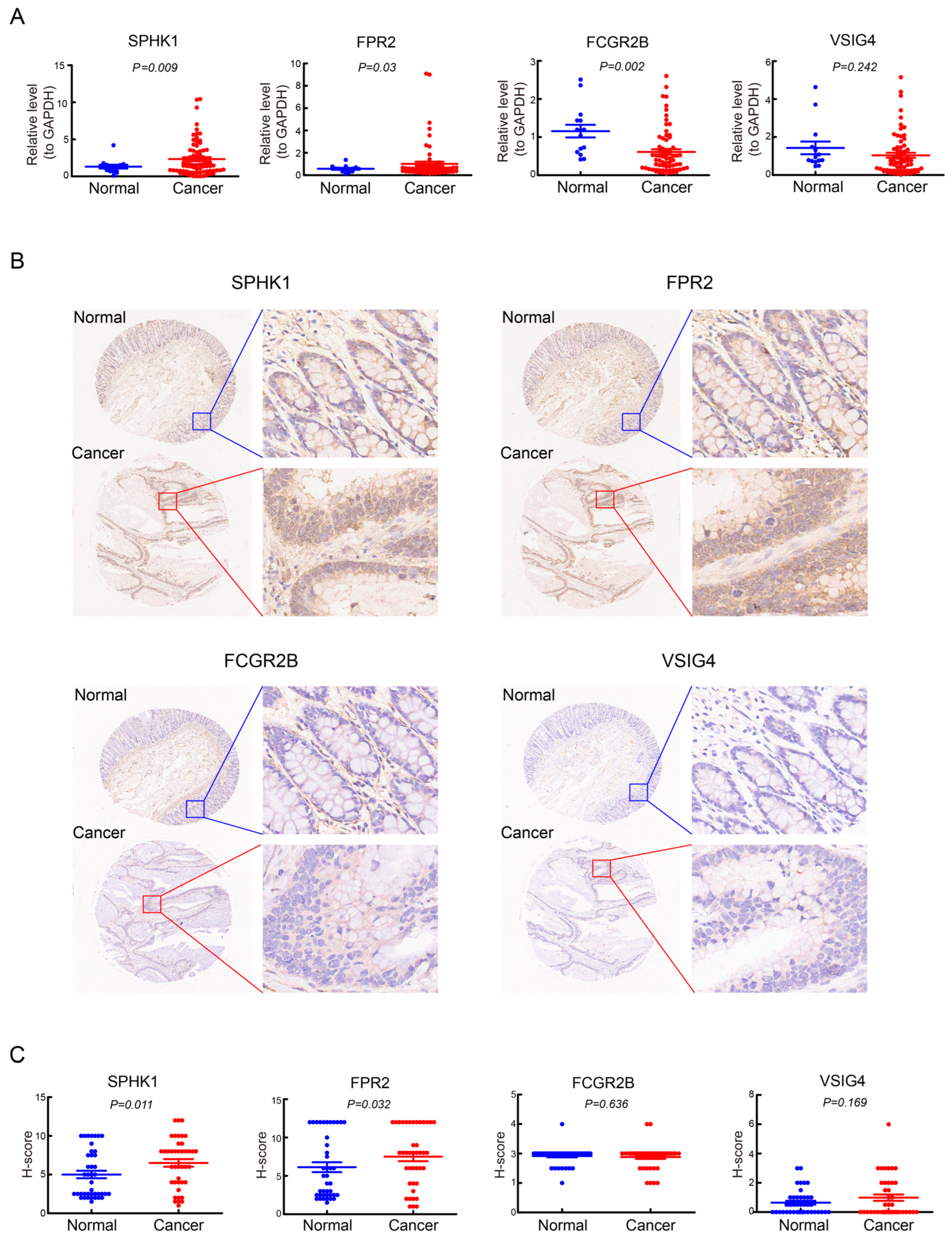
2.9. Quantitative Real-Time PCR (qRT-PCR)
2.10. Immunohistochemistry (IHC)
2.11. Spatial Transcriptomics Validation Analysis
2.12. Statistical Analysis
3. Results
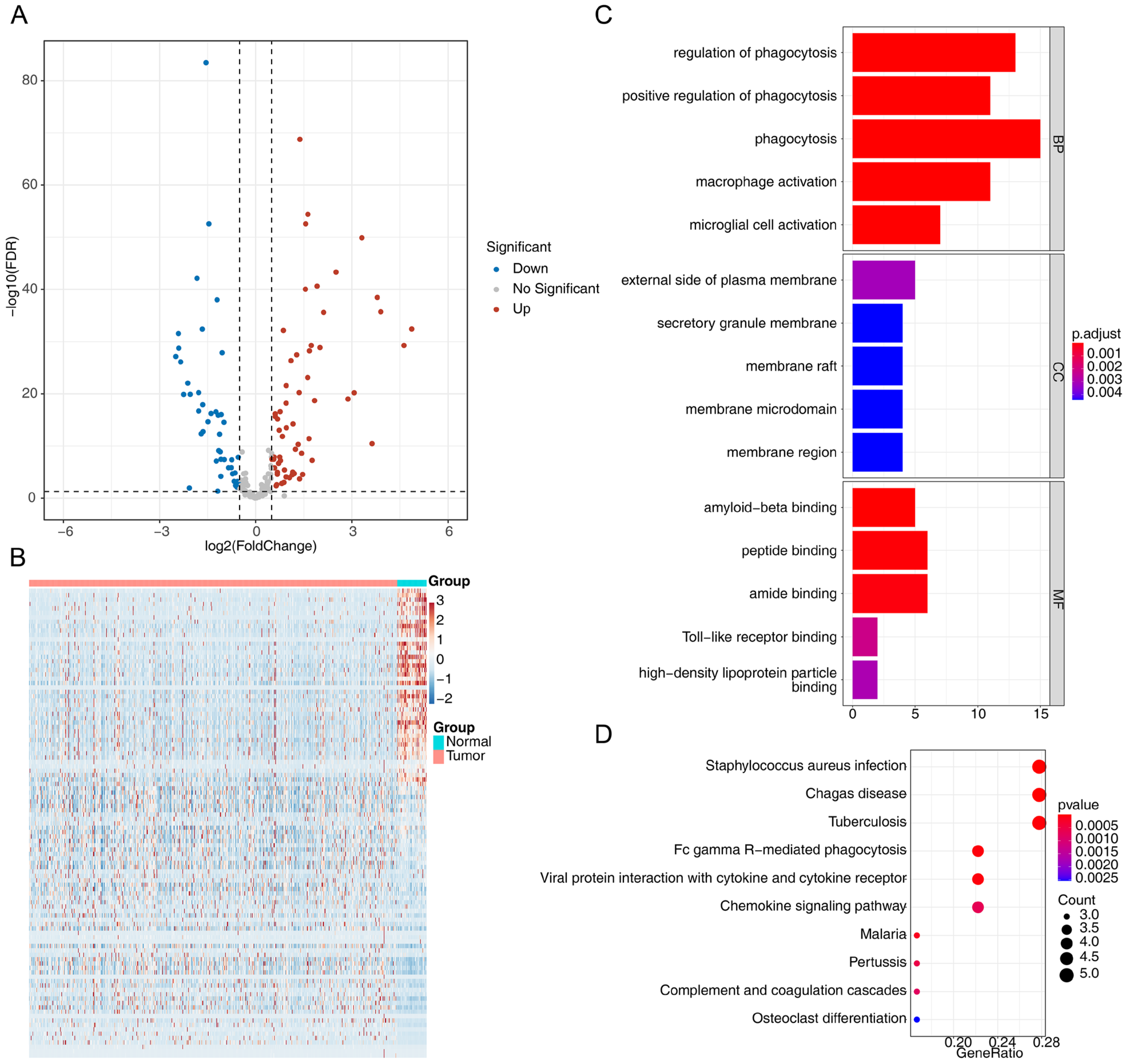
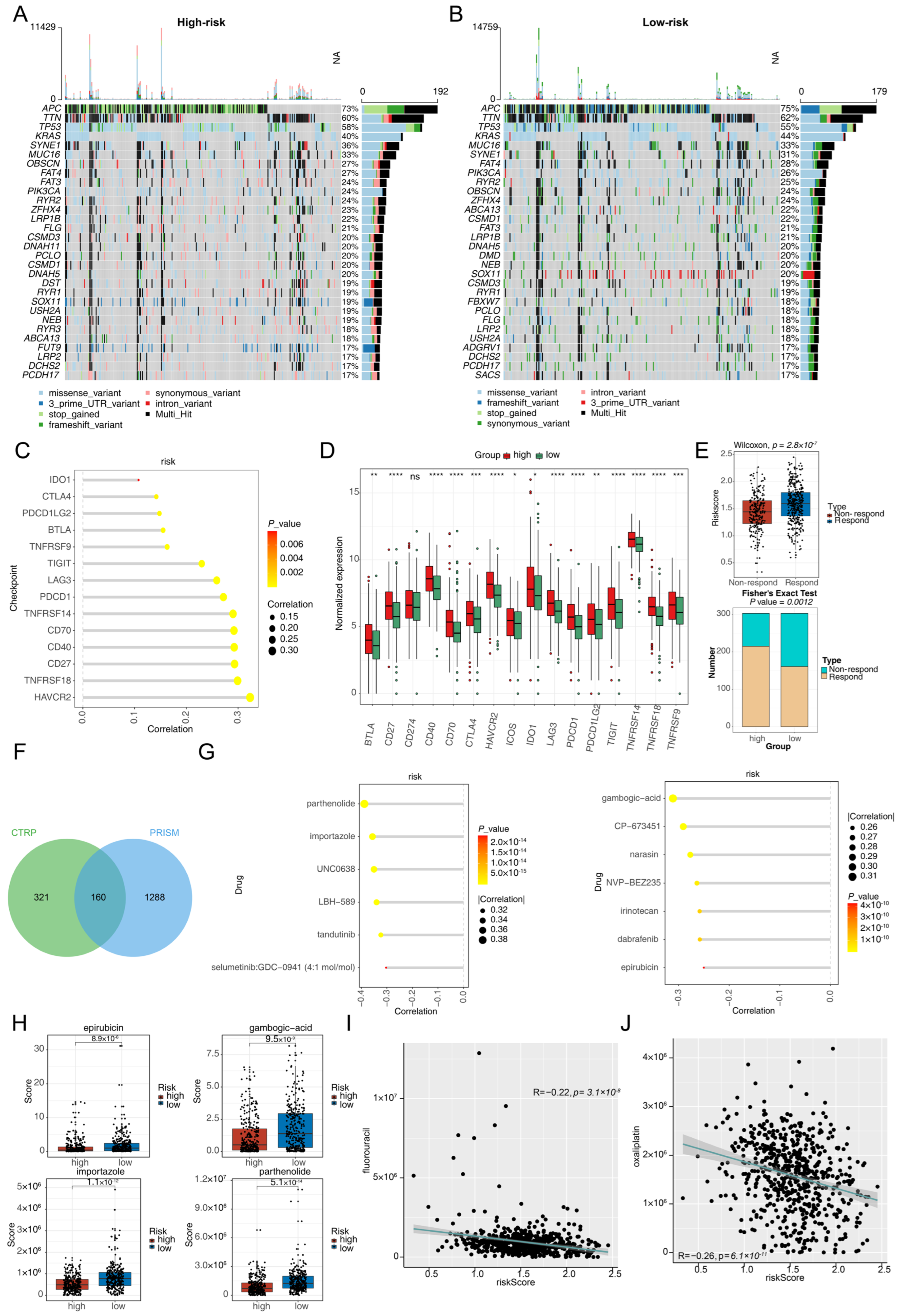
3.1. Identification of Differentially Expressed Macrophage Phagocytosis-Related Genes in CRC
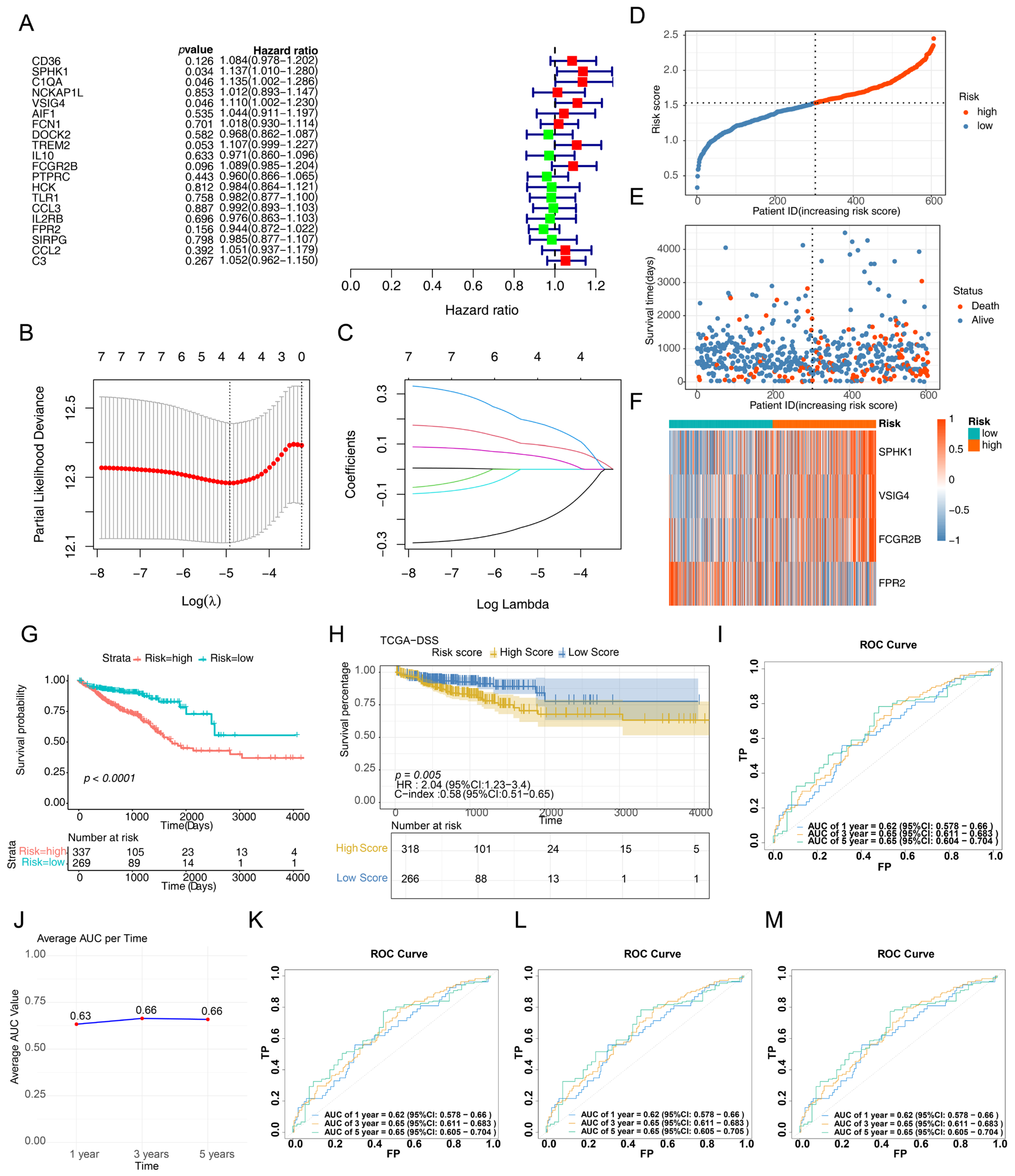
3.2. Development and Validation of a CRC-Related Risk Signature
3.3. Correlation Between Risk Scores and Clinicopathological Features
3.4. Functional Enrichment of the Prognostic Genes
3.5. Different Immune Infiltrates in the Two Risk Groups
3.6. Different Responses to Immunotherapy and Chemotherapy in the Two Risk Groups
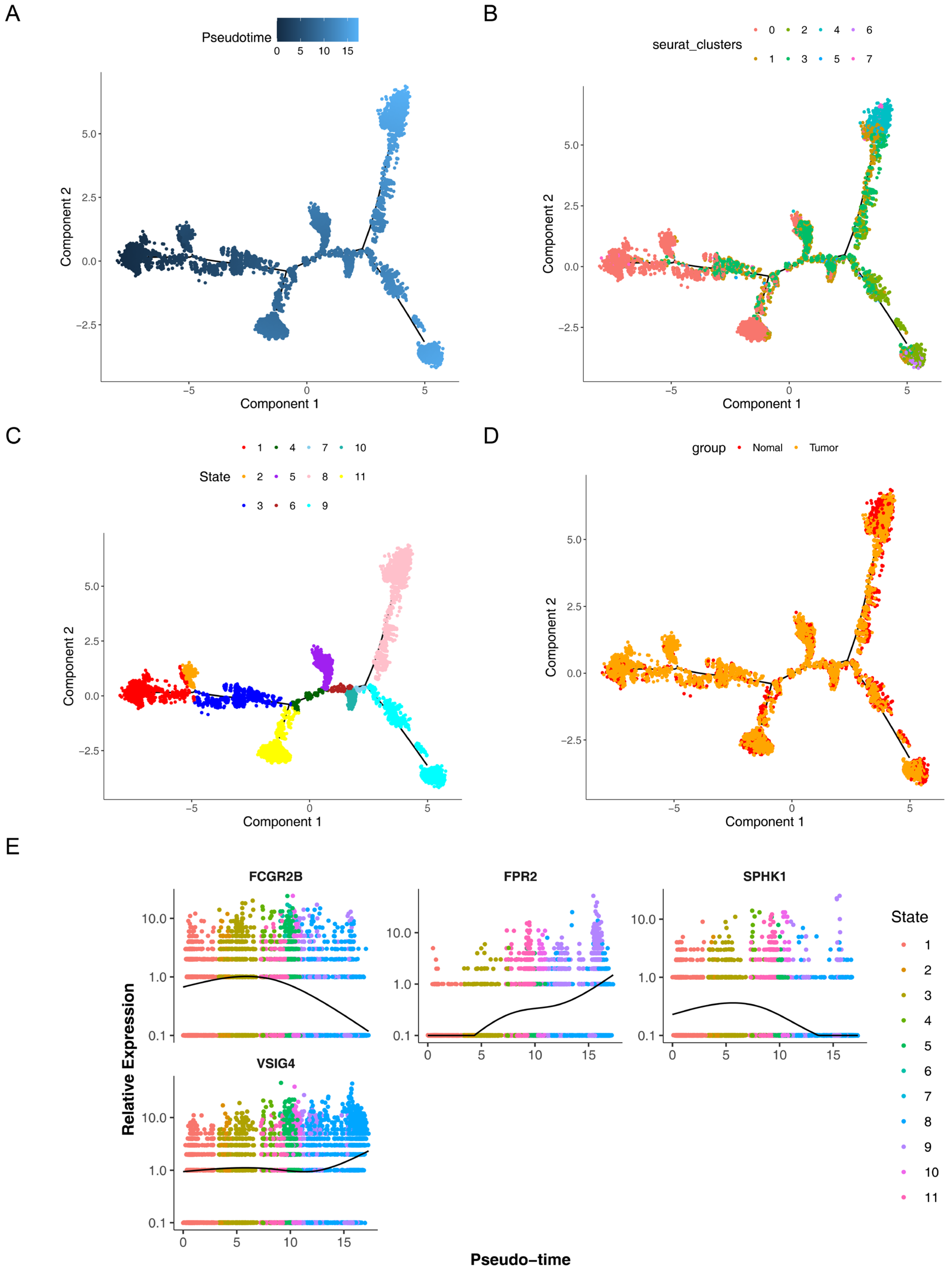
3.7. The Prognostic Genes Functioned Through Being Expressed in Macrophages
3.8. Spatial Validation of Macrophage Enrichment in CRC
3.9. Validation of the Expression of the Four Prognostic Genes in CRC
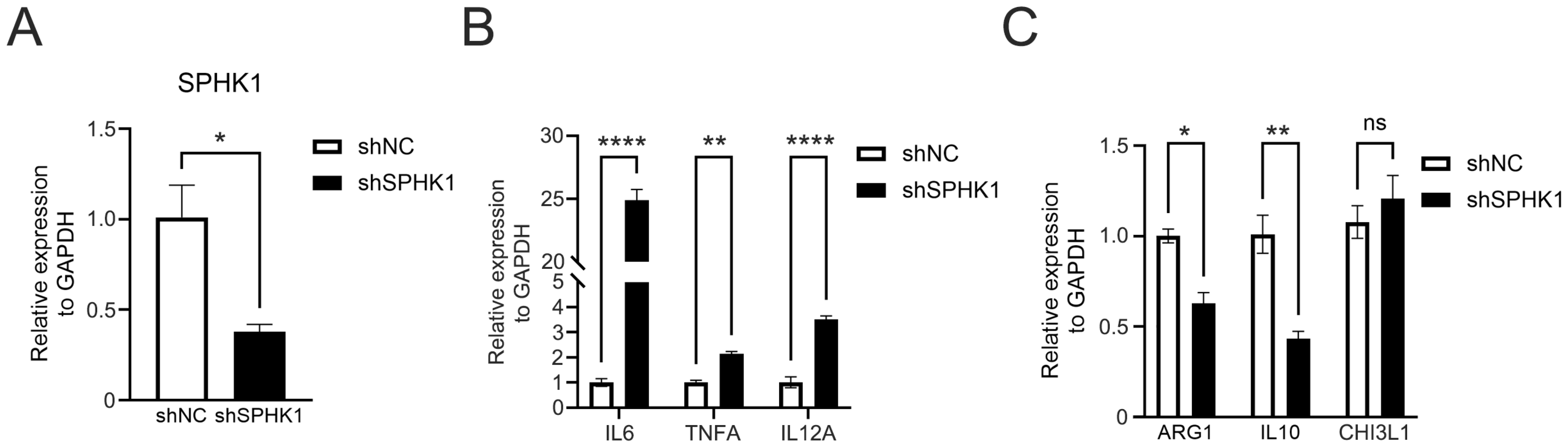
3.10. Knockdown of SPHK1 Promotes M1 Polarization of Macrophages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CRC | colorectal cancer |
| TCGA | The Cancer Genome Atlas |
| GEO | Gene Expression Omnibus |
| GSEA | gene set enrichment analysis |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| ssGSEA | single-sample GSEA |
| LASSO | least absolute shrinkage and selection operator |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| IHC | immunohistochemistry |
| TME | tumor microenvironment |
| TAMs | tumor-associated macrophages |
| MMPs | matrix metalloproteinases |
| PD-L1 | programmed cell death 1 ligand 1 |
| scRNA-seq | single-cell RNA sequencing |
| TMB | tumor mutation burden |
| TIDE | Tumor Immune Dysfunction and Exclusion |
| UMI | unique molecular identifier |
| VST | variance stabilizing transformation |
| UMAP | uniform manifold approximation and projection |
| AUC | area under the curve |
| MIF | macrophage migration inhibitory factor |
| SPHK1 | sphingosine kinase 1 |
| VSIG4 | v-set and immunoglobulin domain-containing 4 |
| FCGR2B | Fc gamma receptor IIb |
| FPR2 | formyl peptide receptor 2 |
| AT-SPM | aspirin-triggered specialized proresolving mediators |
References
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Rui, W.; Zhao, X.; Lin, X. Enhancing CAR-T cell efficacy in solid tumors by targeting the tumor microenvironment. Cell Mol. Immunol. 2021, 18, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Maacha, S.; Bhat, A.A.; Jimenez, L.; Raza, A.; Haris, M.; Uddin, S.; Grivel, J.-C. Extracellular vesicles-mediated intercellular communication: Roles in the tumor microenvironment and anti-cancer drug resistance. Mol. Cancer 2019, 18, 55. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef]
- Vitale, I.; Manic, G.; Coussens, L.M.; Kroemer, G.; Galluzzi, L. Macrophages and Metabolism in the Tumor Microenvironment. Cell Metab. 2019, 30, 36–50. [Google Scholar] [CrossRef]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef]
- DeNardo, D.G.; Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019, 19, 369–382. [Google Scholar] [CrossRef]
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef]
- Uribe-Querol, E.; Rosales, C. Phagocytosis: Our Current Understanding of a Universal Biological Process. Front. Immunol. 2020, 11, 1066. [Google Scholar] [CrossRef]
- Gordon, S. Phagocytosis: An Immunobiologic Process. Immunity 2016, 44, 463–475. [Google Scholar] [CrossRef]
- Georgoudaki, A.-M.; Prokopec, K.E.; Boura, V.F.; Hellqvist, E.; Sohn, S.; Östling, J.; Dahan, R.; Harris, R.A.; Rantalainen, M.; Klevebring, D.; et al. Reprogramming Tumor-Associated Macrophages by Antibody Targeting Inhibits Cancer Progression and Metastasis. Cell Rep. 2016, 15, 2000–2011. [Google Scholar] [CrossRef]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS−) vs. Alternatively Activated Macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef]
- Yamazaki, M.; Maruyama, S.; Abé, T.; Tsuneki, M.; Kato, H.; Izumi, K.; Tanuma, J.-I.; Cheng, J.; Saku, T. Rac1-dependent phagocytosis of apoptotic cells by oral squamous cell carcinoma cells: A possible driving force for tumor progression. Exp. Cell Res. 2020, 392, 112013. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Biller, L.H.; Schrag, D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA 2021, 325, 669–685. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, Y.; Hao, L.; Hou, A.; Chen, X.; Li, Y.; Wang, R.; Luo, P.; Ruan, Z.; Ou, J.; et al. Macrophages induce resistance to 5-fluorouracil chemotherapy in colorectal cancer through the release of putrescine. Cancer Lett. 2016, 381, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lim, S.Y.; Gordon-Weeks, A.N.; Tapmeier, T.T.; Im, J.H.; Cao, Y.; Beech, J.; Allen, D.; Smart, S.; Muschel, R.J. Recruitment of a myeloid cell subset (CD11b/Gr1mid) via CCL2/CCR2 promotes the development of colorectal cancer liver metastasis. Hepatology 2013, 57, 829–839. [Google Scholar] [CrossRef]
- Gordon, S.R.; Maute, R.L.; Dulken, B.W.; Hutter, G.; George, B.M.; McCracken, M.N.; Gupta, R.; Tsai, J.M.; Sinha, R.; Corey, D.; et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 2017, 545, 495–499. [Google Scholar] [CrossRef]
- Zhang, Q.W.; Liu, L.; Gong, C.-Y.; Shi, H.-S.; Zeng, Y.-H.; Wang, X.-Z.; Zhao, Y.-W.; Wei, Y.-Q. Prognostic significance of tumor-associated macrophages in solid tumor: A meta-analysis of the literature. PLoS ONE 2012, 7, e50946. [Google Scholar] [CrossRef]
- Feng, J.; Ren, J.; Li, X.; Zhang, X.; Yang, Q.; Wu, Z.; Cui, L.; Liao, L.; Gong, Y.; Cao, D. Phagocytosis-Regulators-Based Signature to Predict the Prognosis and Chemotherapy Resistance for Breast Cancer Patients. Int. J. Mol. Sci. 2022, 23, 10312. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef]
- Cao, P.W.; Liu, L.; Li, Z.H.; Cao, F.; Liu, F.B. Prognostic Value of Drug Targets Predicted Using Deep Bioinformatic Analysis of m6A-Associated lncRNA-Based Pancreatic Cancer Model Characteristics and Its Tumour Microenvironment. Front. Genet. 2022, 13, 853471. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000. [Google Scholar]
- Simon, N.; Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. J. Stat. Softw. 2011, 39, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liao, W.; Luo, Q.; Yang, D.; Pan, M. Histone Acetylation Regulator-Mediated Acetylation Patterns Define Tumor Malignant Pathways and Tumor Microenvironment in Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 761046. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Maeser, D.; Gruener, R.F.; Huang, R.S. oncoPredict: An R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief. Bioinform. 2021, 22, bbab260. [Google Scholar] [CrossRef]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef]
- Wang, M.; Xue, L.; Fei, Z.; Luo, L.; Zhang, K.; Gao, Y.; Liu, X.; Liu, C. Characterization of mitochondrial metabolism related molecular subtypes and immune infiltration in colorectal adenocarcinoma. Sci. Rep. 2024, 14, 24326. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.S.; Chen, X.M.; Zheng, H.X.; Shi, S.L.; Li, Y. Knockdown of Rab5a expression decreases cancer cell motility and invasion through integrin-mediated signaling pathway. J. Biomed. Sci. 2011, 18, 58. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, W.; Huo, M.; Wang, P.; Liu, X.; Wang, Y.; Li, Y.; Zhou, Z.; Xu, N.; Zhu, H. XBP1 regulates the protumoral function of tumor-associated macrophages in human colorectal cancer. Signal Transduct. Target Ther. 2021, 6, 357. [Google Scholar] [CrossRef]
- Wang, H.; Wen, C.; Chen, S.; Li, W.; Qin, Q.; He, L.; Wang, F.; Chen, J.; Ye, W.; Li, W.; et al. ROS/JNK/C-Jun Pathway is Involved in Chaetocin Induced Colorectal Cancer Cells Apoptosis and Macrophage Phagocytosis Enhancement. Front. Pharmacol. 2021, 12, 729367. [Google Scholar] [CrossRef]
- Wu, J.; Lin, L.; Luo, S.; Qiu, X.; Zhu, L.; Chen, D.; Wei, E.; Fu, Z.; Qin, M.; Liang, Z.; et al. SphK1-driven autophagy potentiates focal adhesion paxillin-mediated metastasis in colorectal cancer. Cancer Med. 2021, 10, 6010–6021. [Google Scholar] [CrossRef]
- Long, J.; Xie, Y.; Yin, J.; Lu, W.; Fang, S. SphK1 promotes tumor cell migration and invasion in colorectal cancer. Tumour Biol. 2016, 37, 6831–6836. [Google Scholar] [CrossRef]
- Bae, G.E.; Do, S.I.; Kim, K.; Park, J.H.; Cho, S.; Kim, H.S. Increased Sphingosine Kinase 1 Expression Predicts Distant Metastasis and Poor Outcome in Patients with Colorectal Cancer. Anticancer Res. 2019, 39, 663–670. [Google Scholar] [CrossRef]
- Pan, Z.; Chen, J.; Xu, T.; Cai, A.; Han, B.; Li, Y.; Fang, Z.; Yu, D.; Wang, S.; Zhou, J.; et al. VSIG4+ tumor-associated macrophages mediate neutrophil infiltration and impair antigen-specific immunity in aggressive cancers through epigenetic regulation of SPP1. J. Exp. Clin. Cancer Res. 2025, 44, 45. [Google Scholar] [CrossRef]
- Liu, B.; Cheng, L.; Gao, H.; Zhang, J.; Dong, Y.; Gao, W.; Yuan, S.; Gong, T.; Huang, W. The biology of VSIG4: Implications for the treatment of immune-mediated inflammatory diseases and cancer. Cancer Lett. 2023, 553, 215996. [Google Scholar] [CrossRef]
- Vogt, L.; Schmitz, N.; Kurrer, M.O.; Bauer, M.; Hinton, H.I.; Behnke, S.; Gatto, D.; Sebbel, P.; Beerli, R.R.; Sonderegger, I.; et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J. Clin. Investig. 2006, 116, 2817–2826. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, Y.; Wang, W.; Wei, C.; Liu, Z.; Li, Z.; Ye, Y.; Mao, Y.; Yuan, Y.; Huang, Z.; et al. Targeting VSIG4+ tissue-resident macrophages enhances T cell cytotoxicity and immunotherapy efficacy in cancer. Dev. Cell 2025. [Google Scholar] [CrossRef]
- Ma, D.; Wang, Y.; Shen, Q.; Liu, X.; Lu, W.; Li, S.; Yin, Q.; Xia, L.; Liu, G.; Chen, Y.; et al. VSIG4 as a tumor-associated macrophage marker predicting adverse prognosis in diffuse large B-cell lymphoma. Front. Immunol. 2025, 16, 1567035. [Google Scholar] [CrossRef] [PubMed]
- Menyhart, O.; Kakisaka, T.; Pongor, L.S.; Uetake, H.; Goel, A.; Gyorffy, B. Uncovering Potential Therapeutic Targets in Colorectal Cancer by Deciphering Mutational Status and Expression of Druggable Oncogenes. Cancers 2019, 11, 983. [Google Scholar] [CrossRef] [PubMed]
- Alessi, M.C.; Cenac, N.; Si-Tahar, M.; Riteau, B. FPR2: A Novel Promising Target for the Treatment of Influenza. Front. Microbiol. 2017, 8, 1719. [Google Scholar] [CrossRef] [PubMed]
- De Matteis, R.; Flak, M.B.; Gonzalez-Nunez, M.; Austin-Williams, S.; Palmas, F.; Colas, R.A.; Dalli, J. Aspirin activates resolution pathways to reprogram T cell and macrophage responses in colitis-associated colorectal cancer. Sci. Adv. 2022, 8, eabl5420. [Google Scholar] [CrossRef]
- Roghanian, A.; Stopforth, R.J.; Dahal, L.N.; Cragg, M.S. New revelations from an old receptor: Immunoregulatory functions of the inhibitory Fc gamma receptor, FcgammaRIIB (CD32B). J. Leukoc. Biol. 2018, 103, 1077–1088. [Google Scholar] [CrossRef]
- Sharp, P.E.; Martin-Ramirez, J.; Boross, P.; Mangsbo, S.M.; Reynolds, J.; Moss, J.; Pusey, C.D.; Cook, H.T.; Tarzi, R.M.; Verbeek, J.S. Increased incidence of anti-GBM disease in Fcgamma receptor 2b deficient mice, but not mice with conditional deletion of Fcgr2b on either B cells or myeloid cells alone. Mol. Immunol. 2012, 50, 49–56. [Google Scholar] [CrossRef]
- Kholod, O.; Basket, W.; Liu, D.; Mitchem, J.; Kaifi, J.; Dooley, L.; Shyu, C.-R. Identification of Immuno-Targeted Combination Therapies Using Explanatory Subgroup Discovery for Cancer Patients with EGFR Wild-Type Gene. Cancers 2022, 14, 4759. [Google Scholar] [CrossRef]
- Nowicka, M.; Hilton, L.K.; Ashton-Key, M.; Hargreaves, C.E.; Lee, C.; Foxall, R.; Carter, M.J.; Beers, S.A.; Potter, K.N.; Bolen, C.R.; et al. Prognostic significance of FCGR2B expression for the response of DLBCL patients to rituximab or obinutuzumab treatment. Blood Adv. 2021, 5, 2945–2957. [Google Scholar] [CrossRef] [PubMed]
- Nagelkerke, S.Q.; Dekkers, G.; Kustiawan, I.; van de Bovenkamp, F.S.; Geissler, J.; Plomp, R.; Wuhrer, M.; Vidarsson, G.; Rispens, T.; van den Berg, T.K.; et al. Inhibition of FcgammaR-mediated phagocytosis by IVIg is independent of IgG-Fc sialylation and FcgammaRIIb in human macrophages. Blood 2014, 124, 3709–3718. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Jia, H.; Bao, X.; Wu, Y.; Zhu, T.; Li, R.; Zhao, H. Tumor exosome promotes Th17 cell differentiation by transmitting the lncRNA CRNDE-h in colorectal cancer. Cell Death Dis. 2021, 12, 123. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yu, S. Diabetes mellitus is an independent risk factor for colorectal cancer. Dig. Dis. Sci. 2012, 57, 1586–1597. [Google Scholar] [CrossRef]
- Gong, Z.C.; Qian, J.; Zhang, Y.N.; Wang, W.; Kang, X.; Xu, W.; Wu, J.; Zheng, W. Colorectal cancer cells promote osteoclastogenesis and bone destruction through regulating EGF/ERK/CCL3 pathway. Biosci. Rep. 2020, 40, BSR20201175. [Google Scholar]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 2017, 152, 851–866.e824. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, W.; Chen, L.; Wang, X.; Mao, X.; Wu, Z.; Shi, H.; Qi, H.; Chen, L.; Huang, Y.; et al. VSIG4 Promotes Tumour-Associated Macrophage M2 Polarization and Immune Escape in Colorectal Cancer via Fatty Acid Oxidation Pathway. Clin. Transl. Med. 2025, 15, e70340. [Google Scholar] [CrossRef]
- He, T.; Hu, C.; Li, S.; Fan, Y.; Xie, F.; Sun, X.; Jiang, Q.; Chen, W.; Jia, Y.; Li, W. The role of CD8+ T-cells in colorectal cancer immunotherapy. Heliyon 2024, 10, e33144. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, D.; Qian, H.; Shi, Y.; Tao, Z. CD8+ T cell-based cancer immunotherapy. J. Transl. Med. 2024, 22, 394. [Google Scholar] [CrossRef]
- Al-Mterin, M.A.; Elkord, E. Myeloid-derived suppressor cells in colorectal cancer: Prognostic biomarkers and therapeutic targets. Explor. Target Antitumor. Ther. 2022, 3, 497–510. [Google Scholar] [CrossRef]
- Li, K.; Shi, H.; Zhang, B.; Ou, X.; Ma, Q.; Chen, Y.; Shu, P.; Li, D.; Wang, Y. Myeloid-derived suppressor cells as immunosuppressive regulators and therapeutic targets in cancer. Signal Transduct. Target Ther. 2021, 6, 362. [Google Scholar] [CrossRef]
- Fridlender, Z.G.; Sun, J.; Kim, S.; Kapoor, V.; Cheng, G.; Ling, L.; Worthen, G.S.; Albelda, S.M. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2” TAN. Cancer Cell 2009, 16, 183–194. [Google Scholar] [CrossRef]
- Xiao, L.; Cen, D.; Gan, H.; Sun, Y.; Huang, N.; Xiong, H.; Jin, Q.; Su, L.; Liu, X.; Wang, K.; et al. Adoptive Transfer of NKG2D CAR mRNA-Engineered Natural Killer Cells in Colorectal Cancer Patients. Mol. Ther. 2019, 27, 1114–1125. [Google Scholar] [CrossRef]
- Della Chiesa, M.; Setti, C.; Giordano, C.; Obino, V.; Greppi, M.; Pesce, S.; Marcenaro, E.; Rutigliani, M.; Provinciali, N.; Paleari, L.; et al. NK Cell-Based Immunotherapy in Colorectal Cancer. Vaccines 2022, 10, 1033. [Google Scholar] [CrossRef]
- Zahavi, D.; AlDeghaither, D.; O’Connell, A.; Weiner, L.M. Enhancing antibody-dependent cell-mediated cytotoxicity: A strategy for improving antibody-based immunotherapy. Antib. Ther. 2018, 1, 7–12. [Google Scholar] [CrossRef]
- Pohl, M.; Werner, N.; Munding, J.; Tannapfel, A.; Graeven, U.; Nickenig, G.; Schmiegel, W.; Reinacher-Schick, A. Biomarkers of anti-angiogenic therapy in metastatic colorectal cancer (mCRC): Original data and review of the literature. Z. Gastroenterol. 2011, 49, 1398–1406. [Google Scholar] [CrossRef]
- Pagès, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.-S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Tournigand, C.; André, T.; Achille, E.; Lledo, G.; Flesh, M.; Mery-Mignard, D.; Quinaux, E.; Couteau, C.; Buyse, M.; Ganem, G.; et al. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: A randomized GERCOR study. J. Clin. Oncol. 2004, 22, 229–237. [Google Scholar] [CrossRef]
- Kopetz, S.; Grothey, A.; Yaeger, R.; Van Cutsem, E.; Desai, J.; Yoshino, T.; Wasan, H.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N. Engl. J. Med. 2019, 381, 1632–1643. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Adachi, M.; Kawamura, R.; Sakamoto, H.; Hayashi, T.; Ishida, T.; Imai, K.; Shinomura, Y. Parthenolide-induced apoptosis in multiple myeloma cells involves reactive oxygen species generation and cell sensitivity depends on catalase activity. Apoptosis 2006, 11, 2225–2235. [Google Scholar] [CrossRef]
- Qi, Q.; You, Q.; Gu, H.; Zhao, L.; Liu, W.; Lu, N.; Guo, Q. Studies on the toxicity of gambogic acid in rats. J. Ethnopharmacol. 2008, 117, 433–438. [Google Scholar] [CrossRef]
- Tan, F.H.; Putoczki, T.L.; Lou, J.; Hinde, E.; Hollande, F.; Giraud, J.; Stylli, S.S.; Paradiso, L.; Zhu, H.-J.; Sieber, O.M.; et al. Ponatinib Inhibits Multiple Signaling Pathways Involved in STAT3 Signaling and Attenuates Colorectal Tumor Growth. Cancers 2018, 10, 526. [Google Scholar] [CrossRef]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Nagahashi, M.; Yamada, A.; Katsuta, E.; Aoyagi, T.; Huang, W.-C.; Terracina, K.P.; Hait, N.C.; Allegood, J.C.; Tsuchida, J.; Yuza, K.; et al. Targeting the SphK1/S1P/S1PR1 Axis That Links Obesity, Chronic Inflammation, and Breast Cancer Metastasis. Cancer Res. 2018, 78, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Pchejetski, D.; Doumerc, N.; Golzio, M.; Naymark, M.; Teissié, J.; Kohama, T.; Waxman, J.; Malavaud, B.; Cuvillier, O. Chemosensitizing effects of sphingosine kinase-1 inhibition in prostate cancer cell and animal models. Mol. Cancer Ther. 2008, 7, 1836–1845. [Google Scholar] [CrossRef]
- Chiang, N.; Dalli, J.; Colas, R.A.; Serhan, C.N. Identification of resolvin D2 receptor mediating resolution of infections and organ protection. J. Exp. Med. 2015, 212, 1203–1217. [Google Scholar] [CrossRef] [PubMed]
- Sazinsky, S.; Zafari, M.; Klebanov, B.; Ritter, J.; Nguyen, P.A.; Phennicie, R.T.; Wahle, J.; Kauffman, K.J.; Razlog, M.; Manfra, D.; et al. Antibodies Targeting Human or Mouse VSIG4 Repolarize Tumor-Associated Macrophages Providing the Potential of Potent and Specific Clinical Anti-Tumor Response Induced across Multiple Cancer Types. Int. J. Mol. Sci. 2024, 25, 6160. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, F.; Ravetch, J.V. Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 2008, 8, 34–47. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Tan, B.; Yang, J.; Liu, S. Identification and Validation of a Macrophage Phagocytosis-Related Gene Signature for Prognostic Prediction in Colorectal Cancer (CRC). Curr. Issues Mol. Biol. 2025, 47, 804. https://doi.org/10.3390/cimb47100804
Zhao X, Tan B, Yang J, Liu S. Identification and Validation of a Macrophage Phagocytosis-Related Gene Signature for Prognostic Prediction in Colorectal Cancer (CRC). Current Issues in Molecular Biology. 2025; 47(10):804. https://doi.org/10.3390/cimb47100804
Chicago/Turabian StyleZhao, Xibao, Binbin Tan, Jinxu Yang, and Shanshan Liu. 2025. "Identification and Validation of a Macrophage Phagocytosis-Related Gene Signature for Prognostic Prediction in Colorectal Cancer (CRC)" Current Issues in Molecular Biology 47, no. 10: 804. https://doi.org/10.3390/cimb47100804
APA StyleZhao, X., Tan, B., Yang, J., & Liu, S. (2025). Identification and Validation of a Macrophage Phagocytosis-Related Gene Signature for Prognostic Prediction in Colorectal Cancer (CRC). Current Issues in Molecular Biology, 47(10), 804. https://doi.org/10.3390/cimb47100804





