Abstract
Ferredoxins are proteins found in all biological kingdoms and are involved in essential biological processes including photosynthesis, lipid metabolism, and biogeochemical cycles. Ferredoxins are classified into different groups based on the iron-sulfur (Fe-S) clusters that they contain. A new subtype classification and nomenclature system, based on the spacing between amino acids in the Fe-S binding motif, has been proposed in order to better understand ferredoxins’ biological diversity and evolutionary linkage across different organisms. This new classification system has revealed an unparalleled diversity between ferredoxins and has helped identify evolutionarily linked ferredoxins between species. The current review provides the latest insights into ferredoxin functions and evolution, and the new subtype classification, outlining their potential biotechnological applications and the future challenges in streamlining the process.
1. Introduction
Ferredoxins are a large class of iron-sulfur (Fe-S) cluster containing proteins that are found across all the domains of life [1]. Ferredoxins play crucial roles in many fundamental biological processes including photosynthesis, lipid metabolism, cellular respiration, and the biogeochemical cycles of hydrogen, nitrogen, and sulfur [2,3,4,5], Fe-S cluster synthesis, steroidogenesis, bile acid production, and vitamin metabolism [6]. The involvement of ferredoxins in oxidation-reduction processes makes them essential proteins in organisms ranging from non-photosynthetic anaerobic bacteria to photosynthetic unicellular and multicellular life forms [3,6]. It is interesting to note that, apart from their primary role in electron transfer, ferredoxins are also found to have regulatory functions such as enhancing the expression of genes involved in photooxidative stress [7], remodeling of regulatory protein complexes [8], controlling cellular posttranslational lipoylation [9], and controlling copper-dependent cell death (cuproptosis) [10].
Ferredoxins were discovered in 1962 in the obligate anaerobic non-photosynthetic bacterium, Clostridium pasteurianum [11]. Due to their unique property of containing only iron and not heme or flavin cofactors, and for being able to transfer electrons to many protein redox partners, authors named them “ferredoxins” [11]. Based on the rudimentary function of electron transfer and internal sequence symmetry, ferredoxin proteins are thought to have evolved during abiogenesis in the burgeoning Earth [2,12,13,14,15,16]. Thus, these proteins are considered living protein fossils [2,12,13,14,15,16]. A well-known hypothesis is that ferredoxins evolved through tandem gene duplications encoding smaller proteins, which may have originated from duplicating even simpler ancestral peptides [11,12,13], and a recent study provided further evidence strengthening this hypothesis [1].
2. Ferredoxin Functions
Ferredoxins are involved in a range of biological processes, and any disruption of electron transfer from ferredoxins eventually disrupts that biological process. Important roles of ferredoxins in various biological processes include:
2.1. Pyruvate Synthesis/CO2 Fixation
The acetyl-CoA pathway is considered an ancient pathway for CO2 fixation that evolved >3.8 billion years ago in the autotrophs, where acetyl-CoA is converted to pyruvate by ferredoxin-dependent CO2 fixation [17] (Figure 1A). In this reaction, ferredoxins are directly reduced by hydrogen via native iron or flavin-based hydrogenases [17]. The role of ferredoxins in transferring electrons is key in pyruvate biosynthesis and its further bioconversion.
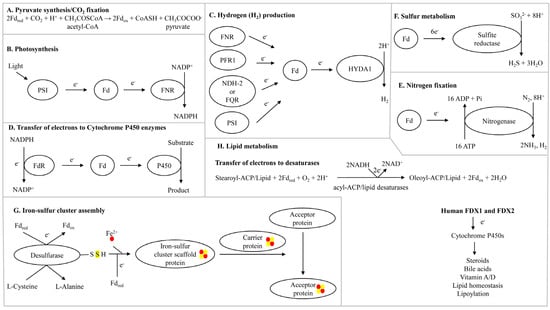
Figure 1.
Schematic diagram showing ferredoxin’s role in various biological processes. Abbreviations: PSI, photosystem I; FNR, ferredoxin-NADP+ reductase; FQR, ferredoxin-plastoquinone reductase; NDH-2, NAD(P)H-PQ oxidoreductase; PFR1, pyruvate:ferredoxin oxidoreductase; HYDA1, [FeFe]-hydrogenase; FdR, ferredoxin reductase; Fd, ferredoxin; e−, electrons; ATP, adenosine triphosphate; Pi, phosphate; SO32−, sulfite; H2S, hydrogen sulfide; CO2, carbon dioxide; H+, hydrogen ion; CoASH, coenzyme A; Fdox and Fered, ferredoxin-oxidized and ferredoxin-reduced; FDX1 and FDX1, human mitochondrial ferredoxins 1 and 2. Iron and sulfur atoms are presented with red and yellow dots.
2.2. Role in Photosynthesis
2.3. Role in the Production of Hydrogen
Hydrogen is a clean energy source, and producing this valuable gaseous molecule from organisms is an active area of research. Ferredoxins play a vital role in the production of this clean energy [18,19]. The production of hydrogen by the green alga Chlamydomonas reinhardtii has been well-studied, and the role of ferredoxin is well-defined [18]. Multiple pathways in C. reinhardtii that can transfer electrons to ferredoxins have been identified [18]. A specific ferredoxin (PetF) receives electrons from multiple donor partners under different conditions including PSI, FNR, NAD(P)H-PQ oxidoreductase (NDH-2), ferredoxin-plastoquinone reductase (FQR), or pyruvate:ferredoxin oxidoreductase (PFR1) (Figure 1C). The reduced ferredoxin subsequently transfers electrons to [FeFe]-hydrogenase HYDA1, which further catalyzes the reduction of protons into H2 (Figure 1C) [18].
2.4. Transfer of Electrons to Cytochrome P450 Monooxygenases
Cytochrome P450 monooxygenases (CYPs/P450s) perform various oxidative enzymatic reactions important in biology. P450s require electrons to perform their enzymatic activity, and ferredoxins transfer these electrons, particularly in bacterial P450 systems [20,21]. Ferredoxins not only transfer electrons to P450s but also modulate their functions; thus, the factors governing their molecular interactions are crucial for the successful outcome of the catalyzed reaction [20]. The P450 catalytic cycle starts with ferredoxin reductases (FdRs) extracting electrons from NADPH and reducing ferredoxins (Figure 1). The reduced ferredoxins then transfer the electrons to P450s in order to catalyze the molecular scission of atmospheric dioxygen (Figure 1D). Information on the transfer of electrons by ferredoxins to different mitochondrial human P450s that play a role in steroid metabolism (Figure 1D) has been reviewed elsewhere [5].
2.5. Role in Nitrogen Fixation
Specific microbes including cyanobacteria, rhizobia, green sulfur, and purple sulfur bacteria convert atmospheric nitrogen (N2) into bioavailable ammonia (NH3), which is essential for sustaining life [22]. Microbes use nitrogenase and ferredoxin enzymes to fix atmospheric nitrogen [23] (Figure 1E). Ferredoxins transfer electrons to nitrogenase, which converts N2 into NH3. During this reaction, 16 ATP molecules are utilized, and hydrogen is produced as a by-product [23] (Figure 1E).
2.6. Role in Sulfur Metabolism
Ferredoxins play a role in sulfur metabolism in archaea, bacteria, fungi, and plants. For example, they transfer electrons to sulfite reductase, which converts sulfite into hydrogen sulfide and water [24,25] (Figure 1F). This is an essential process for synthesizing sulfur-containing compounds in the cell.
2.7. Role in Iron-Sulfur Cluster Biosynthesis
The role of ferredoxins in transferring electrons for the biogenesis of iron-sulfur clusters have been elucidated [26,27] (Figure 1G). The mechanism of iron-sulfur cluster assembly has been studied in detail in cyanobacteria and humans [26,27]. The generally accepted pathway for iron-sulfur cluster biosynthesis can be divided into two main steps: (1) the assembly of the iron-sulfur cluster on a scaffold protein using iron and sulfur atoms and (2) the transfer of the iron-sulfur cluster to the acceptor protein [26,27] (Figure 1G). Two ferredoxins from human mitochondria (FDX1 and FDX2) have been shown to be involved in transferring electrons to iron-sulfur cluster assembly proteins, but FDX2 was found to transfer electrons at a faster rate than FDX1 [26] (Figure 1G). The reduced ferredoxins successfully transferred electrons to the iron-sulfur cluster assembly proteins, particularly cysteine desulfurases, whereby L-cysteine is converted to L-alanine with the simultaneous generation of sulfide [26] (Figure 1G). Reduced ferredoxins supported the iron-sulfur cluster assembly on the iron-sulfur cluster scaffold protein [26] (Figure 1G).
2.8. Role in Lipid Metabolism
Ferredoxins play essential roles in lipid metabolism [6]. For example, ferredoxins transfer electrons to acyl-ACP and acyl-lipid desaturases, leading to the generation of unsaturated fatty acids in plants and cyanobacteria [28,29] (Figure 1H).
Human FDX1 is involved in various processes associated with lipid metabolism, such as in the biogenesis of steroids and bile acids, vitamin A/D metabolism, and lipoylation of tricarboxylic acid (TCA) cycle enzymes [5,6] (Figure 1H). A thorough analysis of FDX1’s role has been previously reviewed [5] and so these processes are not elaborated in the present review. A study in mice revealed that FDX1 is essential for mammalian embryonic development and lipid homeostasis, and FDX1 deficiency led to the alteration of several classes of sterols and lipids, including cholesterol, triacylglycerides, acylcarnitines, ceramides, phospholipids, and lysophospholipids [6].
3. Classification of Ferredoxins-Types
Ferredoxins have been classified into different groups based upon the number of iron (Fe) atoms in their structure (Figure 2). These reported groups include 2Fe-2S, 3Fe-4S, 4Fe-4S, 7Fe-8S (3Fe-4S and 4Fe-4S), and 2[4Fe-4S] [30]. Each type of Fe-S cluster boasts a distinctive Fe-S sequence binding motif containing specific cysteine amino acids that coordinate with the Fe atom. The 2Fe-2S cluster type features four cysteines in its binding motif; the 3Fe-4S cluster type is three cysteines and a proline residue following the third cysteine; the 4Fe-4S cluster type consists of four cysteines and a proline following the fourth cysteine, and the 7Fe-8S ferredoxins encompass characteristics of both the 3Fe-4S and 4Fe-4S clusters (Figure 2). In the 2[4Fe-4S] ferredoxins the spacing between the cysteines binding to the Fe atom differs from that of the 4Fe-4S cluster type motif (Figure 2). The 2[4Fe-4S] proteins are bifurcated into two subfamilies: small proteins (approximately 55 amino acids) housing iso-potential Fe-S clusters and larger proteins named Alvin (Alv) ferredoxins, possessing Fe-S clusters with varying potentials [31]. Sequence analysis has uncovered an additional cysteine in 2[4Fe-4S]Alv ferredoxins, precisely three amino acids after the final cysteine of the second 4Fe-4S binding cluster [31].
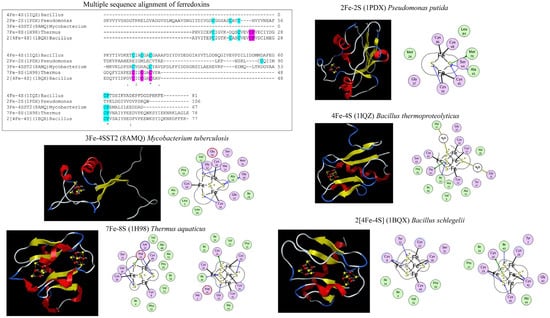
Figure 2.
Ferredoxin interactions with different iron-sulfur (Fe-S) clusters. Each ferredoxin type is presented with its type name, Protein Databank (PDB) identification number in parenthesis, and species name. The ferredoxin crystal structures include 2Fe-2s (1PDX) from Pseudomonas putida [32], 3Fe-4S (8AMQ–ferredoxin only) from Mycobacterium tuberculosis H37Rv [33], 4Fe-4S (1IQZ) from Bacillus thermoproteolyticus [34], 7Fe-8S (1H98) from Thermus aquaticus [35], and 2[4Fe-4S] (1BQX) from Bacillus schlegelii [36]. In multiple sequence alignment, the characteristics of residues interacting with iron atoms, especially cysteine residues, are highlighted along with the characteristic proline residue. Amino acid residues are highlighted with two colors to distinguish interactions with two Fe-S clusters. The individual ferredoxins are shown with their crystal structure and Fe-S cluster(s) interactions. The covalent interactions between the Fe atom and cysteine residues are shown with a solid line. In the crystal structure, Fe and S are colored red and yellow.
4. Evolution of Ferredoxins
Experiments mimicking the early conditions of Earth, such as the chemical evolution period, resulted in the formation of 4Fe-4S clusters, and thus it is now widely believed that 4Fe-4S clusters were the first to evolve among Fe-S cluster types [12,37,38]. Several lines of evidence also support these observations. 4Fe-4S clusters are more sensitive to oxygen than 2Fe-2S clusters [39,40,41]. This indicates that after the Great Oxidation Event species might have preferred 2Fe-2S clusters as they are inherently oxygen tolerant. The presence of abundant 4Fe-4S cluster-type proteins in anaerobes and 2Fe-2S cluster-type proteins in aerobes [42] strongly suggests that 4Fe-4S clusters were the first to evolve.
2[4Fe-4S] ferredoxins have a symmetrical arrangement of Fe-S binding motifs in their structure (Figure 2). Based on the symmetric arrangement of the Fe-S cluster binding motif of 2[4Fe-4S], it has been proposed that ferredoxins arose through the duplication of genes encoding even shorter and simpler ancestral peptides [16]. Many findings suggest that 2[4Fe-4S] ferredoxins have drifted from their symmetric roots via gene duplication followed by mutations ([28]). Additionally, studies report that gene duplication led to the growth and diversification of Fe-S cluster proteins [42].
Knowledge regarding the evolution of ferredoxins is scarce, but current data suggest that ferredoxins may have originated from a common ancestor and then undergone divergent evolution, leading to their diversity today [13,43,44,45,46].
5. Ferredoxin Subtype Classification and Nomenclature
Ferredoxins are classified into types based on their Fe-S clusters (see Section 3, Classification of Ferredoxins types). However, this classification will only help us to observe the abundance of cluster types in a species. This classification system does not help us to understand which ferredoxin types are conserved between and among the species of prokaryotes or eukaryotes, nor the diversity of ferredoxins within the cluster types. Therefore, ferredoxin subtype classification and nomenclature was proposed [1] (Figure 3).
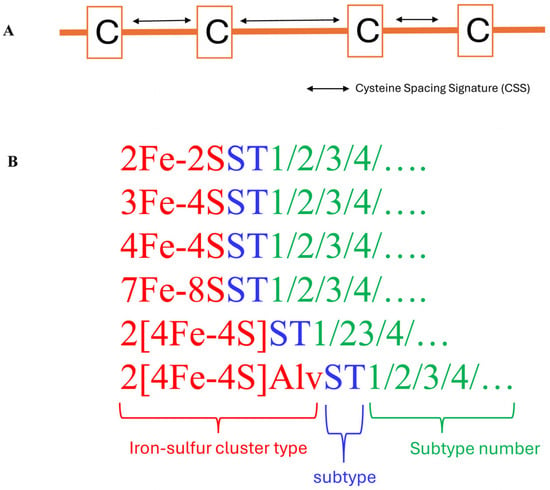
Figure 3.
Ferredoxins subtype classification criteria (A) and nomenclature (B). (A). The spacing pattern between the cysteine amino acids of the ferredoxin Fe-S cluster binding motif within the same cluster type is considered a cysteine spacing signature (CSS) and assigned to a particular subtype (Table 1). Ferredoxins with the same CSS are grouped under the same subtype. (B). To identify ferredoxins belonging to the same subtype, a nomenclature system was developed whereby each ferredoxin was represented by its type, followed by the abbreviation ST, indicating its subtype, and a number showing it belongs to that subtype number, such that ferredoxins that belong to a subtype have the same CSS.
Ferredoxin classification into different subtypes is based on the spacing pattern of amino acids between the conserved cysteine residues of the Fe-S cluster binding motif [1] (Figure 3). The number of amino acids between each of the cysteine residues of ferredoxin is considered a cysteine spacing signature (CSS), and is represented as the characteristic signature of a subtype [1] (Figure 3). This indicates that ferredoxins of a particular subtype have the same CSS (Table 1). To distinguish between ferredoxins belonging to the same subtype, a nomenclature system was proposed whereby a specific ferredoxin is identified by its Fe-S cluster type, followed by an ST abbreviation, indicating subtype, and a numerical number indicating its subtype [1] (Figure 3 and Table 1).

Table 1.
Cysteine spacing signature (CSS) sequences in ferredoxin subtypes. The CSS sequences (ferredoxin type, subtype, and CSS) were retrieved from published studies [1,47,48]. In the CSS sequences, C indicates a cysteine amino acid, P indicates proline, X indicates any amino acid, and the numerical number is an amino acid number.
6. Applications of Subtype Classification
Fe-S cluster type classification does not address the diversity of ferredoxins within cluster types. Also, it does not assist in identifying ferredoxins that are evolutionarily linked across species and belonging to different domains. However, ferredoxin subtype classification revealed unparalleled diversity within the ferredoxin cluster types and helped to identify evolutionarily linked ferredoxin in prokaryotes and eukaryotes and laterally/horizontally transferred ferredoxin genes between prokaryotes and eukaryotes.
6.1. Diversity Analysis/Enrichment/Evolutionary Linkage of Ferredoxins in Organisms
The subtype classification of ferredoxins enabled us to understand the diversity and enrichment (presence of a particular subtype of ferredoxin in organisms belonging to a specific group) of specific subtypes of ferredoxins and the evolutionary linkage of passing the same ferredoxins between species [1,48]. Analysis of ferredoxins in Alphaproteobacteria and Firmicutes revealed they share four Fe-S cluster-type ferredoxins, and two Fe-S cluster-type ferredoxins that are unique to Alphaproteobacteria [1] (Figure 4). Ferredoxin-subtype analysis revealed that these two groups of organisms have diverse subtypes within a particular Fe-S cluster type, and within the subtypes, a few subtypes are preferred/enriched, as these are present in many species [1]. The subtyping of ferredoxins also revealed that some ferredoxins have passed from Alphaproteobacteria to Firmicutes. Analysis of subtypes within a particular Fe-S cluster-type ferredoxin revealed that eight 2Fe-2S subtypes, three 2[4Fe-4S] subtypes, and a single 7Fe-8S subtype were shared between these two groups of organisms, indicating the common ancestral origin of these subtype of ferredoxins [1] (Figure 4).
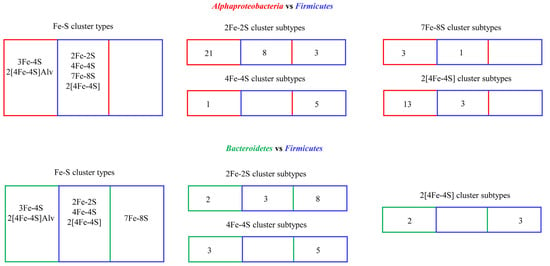
Figure 4.
Comparative analysis of ferredoxins in Alphaproteobacteria, Firmicutes, and Bacteroidetes. Each number indicates subtype quantity in a particular Fe-S cluster type, as reported in the literature [1,48].
In general, analysis of ferredoxins and their subtypes between Bacteroidetes and Firmicutes revealed that these phyla possess highly diverse Fe-S cluster-type ferredoxins, and within the common Fe-S cluster types, ferredoxins were found to be distinct as they belong to different subtypes [48] (Figure 4). Three Fe-S cluster types (2Fe-2S, 4Fe-4S, and 2[4Fe-4S]) are commonly present between both phyla (Figure 3) [48]. Two Fe-S cluster types, 3Fe-4S and 2[4Fe-4S]Alv, are unique to Bacteroidetes, and one Fe-S cluster type, 7Fe-8S, is unique to Firmicutes [48] (Figure 4). As observed for Alphaproteobacteria and Firmicutes, Bacteroidetes also contain a particular Fe-S cluster type and subtype species in this phyla enrich for these ferredoxins [48]. Subtyping analysis revealed that Bacteroidetes and Firmicutes only share three subtypes of 2Fe-2S cluster-type ferredoxins, indicating their common ancestral origin [48]. However, no common 4Fe-4S and 2[4Fe-4S] subtypes were found between these two bacterial groups (Figure 4), indicating that these ferredoxins are highly diverse in bacterial phyla [48].
6.2. Lateral/Horizontal Gene Transfer (LGT/HGT) of Ferredoxins
A handful of studies reported lateral/horizontal gene transfer (LGT/HGT) of ferredoxins [8,49,50]. These studies relied on the percentage similarity between ferredoxins. The ferredoxin domain of cyanobacterial origin was acquired by photosynthetic eukaryotes (Chlamydomonas reinhardtii) through HGT in chloroplast DnaJ-like proteins and further passed to Archaea (Nitrosopumilus maritimus and N. gargensis) [8]. Eukaryotic protists such as Giardia lamblia and Entamoeba histolytica possess ferredoxins that are suggested to have been acquired from anaerobic bacteria by LGT [49]. For example, the Archean Halobacterium salinarum is believed to have obtained a ferredoxin by LGT from a cyanobacterial species [50].
The subtype classification of ferredoxins helps to understand and provide insights into ferredoxin LGT across domains of life [1,47,48] (Figure 5). Since ferredoxin subtyping is not based on similarity but on CSS, ferredoxins belonging to the same subtype are expected to have a common evolutionary linkage [1]. Subtypes 3 and 9 in 2Fe-2S and subtypes 9 and 12 in 2[4Fe-4S] were found to be present across the Archaea, Bacteria, and Eukarya, indicating ferredoxin LGT from Archaea/Bacteria to Eukarya (Figure 5). LGT of subtype 24 in 2Fe-2S and subtype 9 in 4Fe-4S between Archaea and Eukarya and subtypes 1, 2, and 20 in 2Fe-2S and subtype 17 in 2[4Fe-4S] between Bacteria and Eukarya was observed (Figure 5) [1,47,48]. Many subtypes, such as 6,8,17,18, and 31 in 2Fe-2S; 3,8,10,15,18, and 20 in 2[4Fe-4S]; and 1,2, and 7 in 3Fe-4S, are commonly conserved between Archaea and Bacteria, indicating the common evolutionary origin of these ferredoxins [1,47,48] (Figure 5).
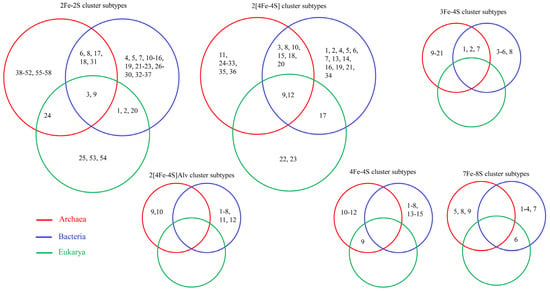
Figure 5.
Lateral/horizontal gene transfer (LGT/HGT) of ferredoxin in domains of life. Each number indicates the subtype number in a specific Fe-S cluster-type ferredoxin. Ferredoxin subtypes identified in different domains of life, such as Archaea, Bacteria, and Eukarya, were retrieved from published articles [1,47,48].
7. Challenges of Assigning Ferredoxins to Different Fe-S Cluster Types and Subtypes
Many databases, such as the National Center for Biotechnology (NCBI) (https://www.ncbi.nlm.nih.gov/ (accessed on 2 August 2024)), the Universal Protein Resource (UniProt) (https://www.uniprot.org/ (accessed on 2 August 2024)), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.genome.jp/kegg/ (accessed on 2 August 2024)), list known ferredoxins in species and ferredoxins can be identified by InterPro or Pfam ID and or by manually searching the name “ferredoxin”. However many ferredoxins are still not annotated, even in well-described genome databases [1,47,48]. Thus, manual searching and further grouping of ferredoxins still remains effective. Identification of Fe-S binding-motif characteristics remains a biological challenge concerning some ferredoxins such as 3Fe/4Fe-4S or 7Fe/8Fe-8S as they differ with only one Fe-atom, and thus manual identification is needed to classify ferredoxins belonging to 2[4Fe-4S]Alv. Another challenge concerns ferredoxins assigned to one Fe-S cluster type, such as 4Fe-4S or 2[4Fe-4S], which then show inter-conversion capability to 3Fe-4S and 7Fe-8S due to an oxidative or reductive environment [45,51,52,53,54]. Assigning these ferredoxins to their correct Fe-S cluster type is still challenging. In a recent study, based on the Fe-S binding-motif pattern, canonical motifs for each of the ferredoxin Fe-S cluster types have been proposed [1]. These canonical motifs are: CX3–5CX1–2CX22–82C, CX2–12CX30–44CX3C, and CX4–7CX29–35C for 2Fe-2S; CX5CX35–49CP for 3Fe-4S; CX2–5CX2–3CX30–45CP for 4Fe-4S; CX3–10CX3CPX17–40CX2CX2CX3CP for 7Fe-8S; CX2–7CX2–4CX2–3CX14–42CX1–2CX2–8CX3C for 2[4Fe-4S]; and CX2CX2CX3CX18–46CX2CX2–8CX3CX3C for 2[4Fe-4S]Alv [1]. Such motifs assist in sorting and assigning the putative ferredoxin proteins into different cluster types.
8. Conclusions and Future Perspectives
Undertaking ferredoxin classification and characterization has excellent applications in molecular biology studies, especially in identifying the evolutionarily linked ferredoxins between organisms and species and laterally/horizontally transferred ferredoxins between prokaryotes and eukaryotes. Ferredoxin subtyping also helps to understand the diversity within a ferredoxin cluster type; for example, establishing ferredoxins’ diverse roles in cellular metabolism, as observed between Bacteroides and Firmicutes [48]. Ferredoxin subtyping also shows that organisms may have ferredoxins belonging to the same cluster types, but they are diverse in function as they belong to different subtypes [1,48].
The current review described only a few subtype cysteine spacing signatures (CSSs) under each cluster type, as only a fraction of ferredoxins were annotated in this work concerning their subtyping. Further analysis is being carried out to identify and classify all known ferredoxins across the domains of life, especially in bacteria. Furthermore, ferredoxin databases are being developed where one can access ferredoxins’ information according to their types and subtypes. This database will also allow BLAST analysis options to be undertaken where researchers can identify the type and subtype to which a ferredoxin may belong. In the future, it would also be interesting to create super subtypes based on their percentage sequence identity, as the higher the sequence identity, the higher the chances that these ferredoxins may be involved in a similar biological process in cellular metabolic pathways with similar electron donor and receiver partners. Ferredoxin super-subtyping may help answer the challenging question: do particular ferredoxins have any unique specificity towards electron donors or receivers? This might be an interesting puzzle to solve and will assist in selecting a specific ferredoxin for the efficient transfer of electrons for biotechnologically valuable reactions.
Funding
This research received no external funding.
Acknowledgments
The author thanks the University of Zululand, South Africa, for the article-processing-fee payment.
Conflicts of Interest
The author declares no conflicts of interest.
References
- Nzuza, N.; Padayachee, T.; Chen, W.; Gront, D.; Nelson, D.R.; Syed, K. Diversification of ferredoxins across living organisms. Curr. Issues Mol. Biol. 2021, 43, 1374–1390. [Google Scholar] [CrossRef]
- Hall, D.; Cammack, R.; Rao, K. Role for ferredoxins in the origin of life and biological evolution. Nature 1971, 233, 136–138. [Google Scholar] [CrossRef]
- Mondal, J.; Bruce, B. Ferredoxin: The central hub connecting photosystem I to cellular metabolism. Photosynthetica 2018, 56, 279–293. [Google Scholar] [CrossRef]
- Braymer, J.J.; Freibert, S.A.; Rakwalska-Bange, M.; Lill, R. Mechanistic concepts of iron-sulfur protein biogenesis in Biology. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118863. [Google Scholar] [CrossRef] [PubMed]
- Ewen, K.M.; Ringle, M.; Bernhardt, R. Adrenodoxin—A versatile ferredoxin. IUBMB Life 2012, 64, 506–512. [Google Scholar] [CrossRef]
- Mohibi, S.; Zhang, Y.; Perng, V.; Chen, M.; Zhang, J.; Chen, X. Ferredoxin 1 is essential for embryonic development and lipid homeostasis. Elife 2024, 13, e91656. [Google Scholar] [CrossRef] [PubMed]
- Mustila, H.; Allahverdiyeva, Y.; Isojärvi, J.; Aro, E.; Eisenhut, M. The bacterial-type [4Fe–4S] ferredoxin 7 has a regulatory function under photooxidative stress conditions in the cyanobacterium Synechocystis sp. PCC 6803. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1837, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Petitjean, C.; Moreira, D.; López-García, P.; Brochier-Armanet, C. Horizontal gene transfer of a chloroplast DnaJ-Fer protein to Thaumarchaeota and the evolutionary history of the DnaK chaperone system in Archaea. BMC Evol. Biol. 2012, 12, 226. [Google Scholar] [CrossRef]
- Schulz, V.; Basu, S.; Freibert, S.-A.; Webert, H.; Boss, L.; Mühlenhoff, U.; Pierrel, F.; Essen, L.-O.; Warui, D.M.; Booker, S.J. Functional spectrum and specificity of mitochondrial ferredoxins FDX1 and FDX2. Nat. Chem. Biol. 2023, 19, 206–217. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef]
- Mortenson, L.; Valentine, R.; Carnahan, J. An electron transport factor from Clostridium pasteurianum. Biochem. Biophys. Res. Commun. 1962, 7, 448–452. [Google Scholar] [CrossRef]
- Venkateswara Rao, P.; Holm, R. Synthetic analogues of the active sites of iron—Sulfur proteins. Chem. Rev. 2004, 104, 527–560. [Google Scholar] [CrossRef] [PubMed]
- Cammack, R. Evolution and diversity in the iron-sulphur proteins. Chem. Scr. 1983, 21, 87–95. [Google Scholar]
- Hall, D.O.; Cammack, R.; Rao, K.K. The iron-sulphur proteins: Evolution of a ubiquitous protein from model systems to higher organisms. Orig. Life 1974, 5, 363–386. [Google Scholar] [CrossRef]
- Romero Romero, M.L.; Rabin, A.; Tawfik, D.S. Functional proteins from short peptides: Dayhoff’s hypothesis turns 50. Angew. Chem. Int. Ed. 2016, 55, 15966–15971. [Google Scholar] [CrossRef] [PubMed]
- Eck, R.V.; Dayhoff, M.O. Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science 1966, 152, 363–366. [Google Scholar] [CrossRef]
- Brabender, M.; Henriques Pereira, D.P.; Mrnjavac, N.; Schlikker, M.L.; Kimura, Z.I.; Sucharitakul, J.; Kleinermanns, K.; Tüysüz, H.; Buckel, W.; Preiner, M.; et al. Ferredoxin reduction by hydrogen with iron functions as an evolutionary precursor of flavin-based electron bifurcation. Proc. Natl. Acad. Sci. USA 2024, 121, e2318969121. [Google Scholar] [CrossRef] [PubMed]
- King, S.J.; Jerkovic, A.; Brown, L.J.; Petroll, K.; Willows, R.D. Synthetic biology for improved hydrogen production in Chlamydomonas reinhardtii. Microb. Biotechnol. 2022, 15, 1946–1965. [Google Scholar] [CrossRef]
- Koo, J.; Cha, Y. Investigation of the ferredoxin’s influence on the anaerobic and aerobic, enzymatic H2 production. Front. Bioeng. Biotechnol. 2021, 9, 641305. [Google Scholar] [CrossRef]
- Chiliza, Z.E.; Martínez-Oyanedel, J.; Syed, K. An overview of the factors playing a role in cytochrome P450 monooxygenase and ferredoxin interactions. Biophys. Rev. 2020, 12, 1217–1222. [Google Scholar] [CrossRef]
- He, J.; Liu, X.; Li, C. Engineering Electron Transfer Pathway of Cytochrome P450s. Molecules 2024, 29, 2480. [Google Scholar] [CrossRef] [PubMed]
- Sepp, S.-K.; Vasar, M.; Davison, J.; Oja, J.; Anslan, S.; Al-Quraishy, S.; Bahram, M.; Bueno, C.G.; Cantero, J.J.; Fabiano, E.C. Global diversity and distribution of nitrogen-fixing bacteria in the soil. Front. Plant Sci. 2023, 14, 1100235. [Google Scholar] [CrossRef] [PubMed]
- Addison, H.; Glatter, T.; KA Hochberg, G.; Rebelein, J.G. Two distinct ferredoxins are essential for nitrogen fixation by the iron nitrogenase in Rhodobacter capsulatus. mBio 2024, 15, e03314–e03323. [Google Scholar] [CrossRef]
- Takahashi, H.; Kopriva, S.; Giordano, M.; Saito, K.; Hell, R. Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 2011, 62, 157–184. [Google Scholar] [CrossRef]
- Kim, J.Y.; Nakayama, M.; Toyota, H.; Kurisu, G.; Hase, T. Structural and mutational studies of an electron transfer complex of maize sulfite reductase and ferredoxin. J. Biochem. 2016, 160, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Tonelli, M.; Frederick, R.O.; Markley, J.L. Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron–sulfur cluster biosynthesis. Biochemistry 2017, 56, 487–499. [Google Scholar] [CrossRef]
- Gao, F. Iron–sulfur cluster biogenesis and iron homeostasis in cyanobacteria. Front. Microbiol. 2020, 11, 165. [Google Scholar] [CrossRef]
- Mutter, A.C.; Tyryshkin, A.M.; Campbell, I.J.; Poudel, S.; Bennett, G.N.; Silberg, J.J.; Nanda, V.; Falkowski, P.G. De novo design of symmetric ferredoxins that shuttle electrons in vivo. Proc. Natl. Acad. Sci. USA 2019, 116, 14557–14562. [Google Scholar] [CrossRef] [PubMed]
- Cerone, M.; Smith, T.K. Desaturases: Structural and mechanistic insights into the biosynthesis of unsaturated fatty acids. Iubmb. Life 2022, 74, 1036–1051. [Google Scholar] [CrossRef]
- Hannemann, F.; Bichet, A.; Ewen, K.M.; Bernhardt, R. Cytochrome P450 systems--biological variations of electron transport chains. Biochim. Biophys. Acta 2007, 1770, 330–344. [Google Scholar] [CrossRef]
- Saridakis, E.; Giastas, P.; Efthymiou, G.; Thoma, V.; Moulis, J.-M.; Kyritsis, P.; Mavridis, I.M. Insight into the protein and solvent contributions to the reduction potentials of [4Fe–4S] 2+/+ clusters: Crystal structures of the Allochromatium vinosum ferredoxin variants C57A and V13G and the homologous Escherichia coli ferredoxin. JBIC J. Biol. Inorg. Chem. 2009, 14, 783–799. [Google Scholar] [CrossRef] [PubMed]
- Pochapsky, T.C.; Jain, N.U.; Kuti, M.; Lyons, T.A.; Heymont, J. A refined model for the solution structure of oxidized putidaredoxin. Biochemistry 1999, 38, 4681–4690. [Google Scholar] [CrossRef]
- Gilep, A.; Varaksa, T.; Bukhdruker, S.; Kavaleuski, A.; Ryzhykau, Y.; Smolskaya, S.; Sushko, T.; Tsumoto, K.; Grabovec, I.; Kapranov, I.; et al. Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450. Front. Mol. Biosci. 2022, 9, 1100032. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, K.; Okada, T.; Kakuta, Y.; Takahashi, Y. Atomic resolution structures of oxidized [4Fe-4S] ferredoxin from Bacillus thermoproteolyticus in two crystal forms: Systematic distortion of [4Fe-4S] cluster in the protein. J. Mol. Biol. 2002, 315, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Macedo-Ribeiro, S.; Martins, B.M.; Pereira, P.B.; Buse, G.; Huber, R.; Soulimane, T. New insights into the thermostability of bacterial ferredoxins: High-resolution crystal structure of the seven-iron ferredoxin from Thermus thermophilus. JBIC J. Biol. Inorg. Chem. 2001, 6, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Aono, S.; Bentrop, D.; Bertini, I.; Cosenza, G.; Luchinat, C. Solution structure of an artificial Fe8S8 ferredoxin: The D13C variant of Bacillus schlegelii Fe7S8 ferredoxin. Eur. J. Biochem. 1998, 258, 502–514. [Google Scholar] [CrossRef]
- Beinert, H.; Holm, R.H.; Münck, E. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science 1997, 277, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J. Iron–sulfur protein folds, iron–sulfur chemistry, and evolution. JBIC J. Biol. Inorg. Chem. 2008, 13, 157–170. [Google Scholar] [CrossRef]
- Imlay, J.A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006, 59, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, B.; Golbeck, J.H. Understanding of the binding interface between PsaC and the PsaA/PsaB heterodimer in photosystem I. Biochemistry 2009, 48, 5405–5416. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, K.-L.; Yu, L.-K.; Chen, Y.-H.; Cheng, Y.-H.; Hsieh, Y.-C.; Ke, S.-c.; Hung, K.-W.; Chen, C.-J.; Huang, T.-h. FeoC from Klebsiella pneumoniae contains a [4Fe-4S] cluster. J. Bacteriol. 2013, 195, 4726–4734. [Google Scholar] [CrossRef]
- Campbell, I.J.; Bennett, G.N.; Silberg, J.J. Evolutionary relationships between low potential ferredoxin and flavodoxin electron carriers. Front. Energy Res. 2019, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Yasunobu, K.T.; Tanaka, M. The evolution of iron-sulfur protein containing organisms. Syst. Zool. 1973, 22, 570–589. [Google Scholar] [CrossRef]
- Harayama, S.; Polissi, A.; Rekik, M. Divergent evolution of chloroplast-type ferredoxins. FEBS Lett. 1991, 285, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Bruschi, M.; Guerlesquin, F. Structure, function and evolution of bacterial ferredoxins. FEMS Microbiol. Lett. 1988, 54, 155–175. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J. The evolution of ferredoxins. Trends Ecol. Evol. 1988, 3, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Ngcobo, P.E.; Nkosi, B.V.Z.; Chen, W.; Nelson, D.R.; Syed, K. Evolution of cytochrome P450 enzymes and their redox partners in Archaea. Int. J. Mol. Sci. 2023, 24, 4161. [Google Scholar] [CrossRef] [PubMed]
- Nkosi, B.V.Z.; Padayachee, T.; Gront, D.; Nelson, D.R.; Syed, K. Contrasting health effects of Bacteroidetes and Firmicutes lies in their genomes: Analysis of P450s, ferredoxins, and secondary metabolite clusters. Int. J. Mol. Sci. 2022, 23, 5057. [Google Scholar] [CrossRef]
- Nixon, J.E.; Wang, A.; Field, J.; Morrison, H.G.; McArthur, A.G.; Sogin, M.L.; Loftus, B.J.; Samuelson, J. Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot. Cell 2002, 1, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, F.; Griffig, J.; Oesterhelt, D. The fdx gene encoding the [2Fe-2S] ferredoxin of Halobacterium salinarium (H. halobium). Mol. Gen. Genet. MGG 1993, 239, 66–71. [Google Scholar] [CrossRef]
- Ishizaka, M.; Chen, M.; Narai, S.; Tanaka, Y.; Ose, T.; Horitani, M.; Yao, M. Quick and Spontaneous Transformation between [3Fe-4S] and [4Fe-4S] Iron-Sulfur Clusters in the tRNA-Thiolation Enzyme TtuA. Int. J. Mol. Sci. 2023, 24, 833. [Google Scholar] [CrossRef] [PubMed]
- Ricagno, S.; de Rosa, M.; Aliverti, A.; Zanetti, G.; Bolognesi, M. The crystal structure of FdxA, a 7Fe ferredoxin from Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 2007, 360, 97–102. [Google Scholar] [CrossRef]
- Beinert, H.; Emptage, M.H.; Dreyer, J.-L.; Scott, R.A.; Hahn, J.E.; Hodgson, K.O.; Thomson, A.J. Iron-sulfur stoichiometry and structure of iron-sulfur clusters in three-iron proteins: Evidence for [3Fe-4S] clusters. Proc. Natl. Acad. Sci. USA 1983, 80, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Armengaud, J.; Gaillard, J.; Forest, E.; Jouanneau, Y. Characterization of a 2[4Fe-4S] ferredoxin obtained by chemical insertion of the Fe-S clusters into the apoferredoxin II from Rhodobacter capsulatus. Eur. J. Biochem. 1995, 231, 396–404. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).