Genetic Analysis of Vitamin C Content in Rapeseed Seedlings by the Major Gene Plus Polygene Mixed Effect Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Growth Conditions
2.3. Extraction and Determination of Vc Content
2.4. Statistical and Genetic Analysis
3. Results
3.1. Statistical Analysis of Vc Content in Six Generations from Two Crosses
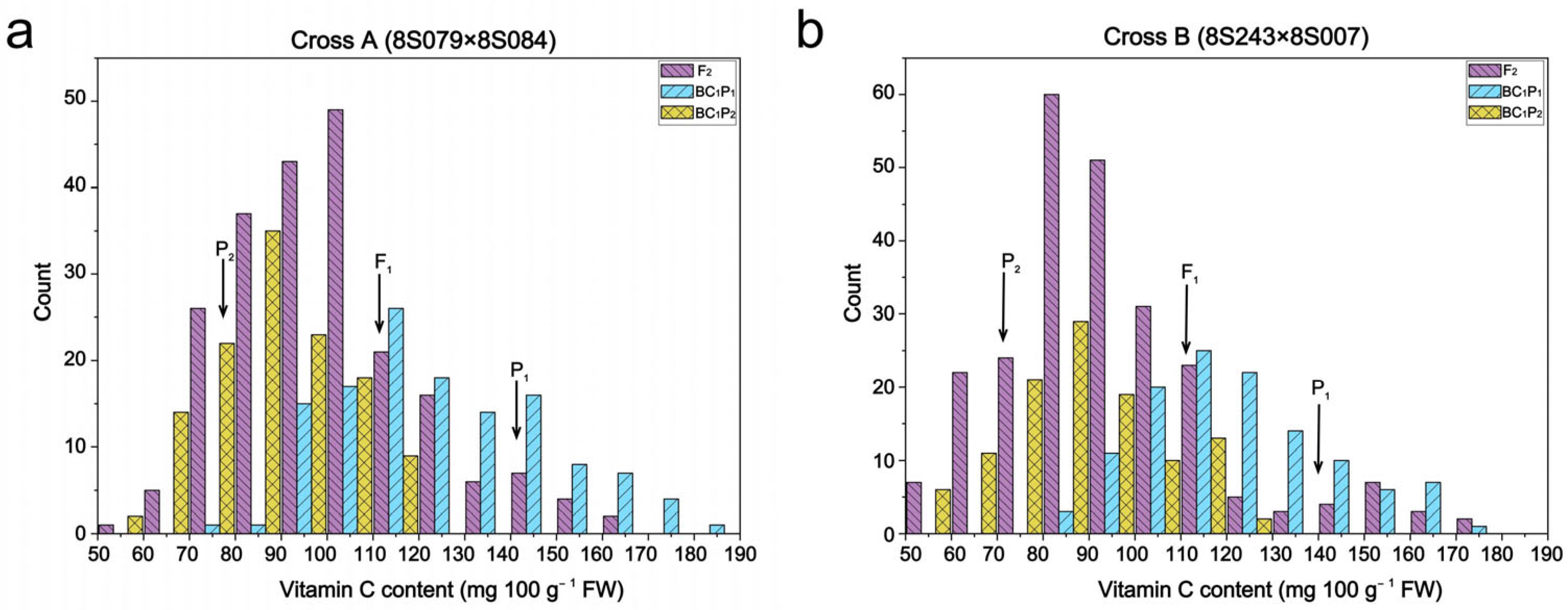
3.2. Distribution of Vc Content in Segregated Populations of Two Crosses
3.3. Selection and Testing for the Best Genetic Model of Vc Content
3.4. Estimation of Genetic Parameters for the Optimal Genetic Model of Vc Content
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ihien Katche, E.; Mason, A.S. Resynthesized rapeseed (Brassica napus): Breeding and genomics. Crit. Rev. Plant Sci. 2023, 42, 65–92. [Google Scholar] [CrossRef]
- Zheng, M.; Terzaghi, W.; Wang, H.; Hua, W. Integrated strategies for increasing rapeseed yield. Trends Plant Sci. 2022, 27, 742–745. [Google Scholar] [CrossRef]
- Xiao, Z.; Pan, Y.; Wang, C.; Li, X.; Lu, Y.; Tian, Z.; Kuang, L.; Wang, X.; Dun, X.; Wang, H. Multi-functional development and utilization of rapeseed: Comprehensive analysis of the nutritional value of rapeseed sprouts. Foods 2022, 11, 778. [Google Scholar] [CrossRef]
- Wu, X.; Chen, F.; Zhao, X.; Pang, C.; Shi, R.; Liu, C.; Sun, C.; Zhang, W.; Wang, X.; Zhang, J. QTL mapping and GWAS reveal the genetic mechanism controlling soluble solids content in Brassica napus shoots. Foods 2021, 10, 2400. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Pang, C.; Wu, X.; Zhao, X.; Chen, F.; Zhang, W.; Sun, C.; Fu, S.; Hu, M.; Zhang, J.; et al. Genetic dissection and germplasm selection of the low crude fiber component in Brassica napus L. shoots. Foods 2023, 12, 403. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N. Ascorbic acid metabolism and functions: A comparison of plants and mammals. Free Radic. Biol. Med. 2018, 122, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Zasowska-Nowak, A.; Nowak, P.J.; Ciałkowska-Rysz, A. High-dose vitamin C in advanced-stage cancer patients. Nutrients 2021, 13, 735. [Google Scholar] [CrossRef]
- Blaszczak, W.; Barczak, W.; Masternak, J.; Kopczyński, P.; Zhitkovich, A.; Rubiś, B. Vitamin C as a modulator of the response to cancer therapy. Molecules 2019, 24, 453. [Google Scholar] [CrossRef]
- Morelli, M.B.; Gambardella, J.; Castellanos, V.; Trimarco, V.; Santulli, G. Vitamin C and cardiovascular disease: An Update. Antioxidants 2020, 9, 1227. [Google Scholar] [CrossRef]
- Collins, B.J.; Mukherjee, M.S.; Miller, M.D.; Delaney, C.L. Effect of dietary or supplemental vitamin C intake on vitamin C levels in patients with and without cardiovascular disease: A systematic review. Nutrients 2021, 13, 2330. [Google Scholar] [CrossRef]
- Monacelli, F.; Acquarone, E.; Giannotti, C.; Borghi, R.; Nencioni, A. Vitamin C, aging and Alzheimer’s disease. Nutrients 2017, 9, 670. [Google Scholar] [CrossRef] [PubMed]
- Mumtaz, S.; Ali, S.; Tahir, H.M.; Kazmi, S.A.R.; Shakir, H.A.; Mughal, T.A.; Mumtaz, S.; Summer, M.; Farooq, M.A. Aging and its treatment with vitamin C: A comprehensive mechanistic review. Mol. Biol. Rep. 2021, 48, 8141–8153. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, A.E.; Szymczak-Tomczak, A.; Skrzypczak-Zielińska, M.; Rychter, A.M.; Zawada, A.; Dobrowolska, A.; Krela-Kaźmierczak, I. Vitamin C deficiency and the risk of osteoporosis in patients with an inflammatory bowel disease. Nutrients 2020, 12, 2263. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.; Fan, D.; He, Y.; Zhu, Y.; Zheng, Q. High-dose intravenous vitamin C attenuates hyperinflammation in severe coronavirus disease 2019. Nutrition 2021, 91–92, 111405. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M.; Amaya, I.; Valpuesta, V.; Botella, M.A. Vitamin C content in fruits: Biosynthesis and regulation. Front. Plant Sci. 2018, 9, 2006. [Google Scholar] [CrossRef] [PubMed]
- Bulley, S.; Laing, W. The regulation of ascorbate biosynthesis. Curr. Opin. Plant Biol. 2016, 33, 15–22. [Google Scholar] [CrossRef]
- Haroldsen, V.M.; Chi-Ham, C.L.; Kulkarni, S.; Lorence, A.; Bennett, A.B. Constitutively expressed DHAR and MDHAR influence fruit, but not foliar ascorbate levels in tomato. Plant Physiol. Biochem. 2011, 49, 1244–1249. [Google Scholar] [CrossRef]
- Zheng, X.; Gong, M.; Zhang, Q.; Tan, H.; Li, L.; Tang, Y.; Li, Z.; Peng, M.; Deng, W. Metabolism and regulation of ascorbic acid in fruits. Plants 2022, 11, 1602. [Google Scholar] [CrossRef] [PubMed]
- Ntagkas, N.; Woltering, E.; Nicole, C.; Labrie, C.; Marcelis, L.F.M. Light regulation of vitamin C in tomato fruit is mediated through photosynthesis. Environ. Exp. Bot. 2019, 158, 180–188. [Google Scholar] [CrossRef]
- Zhang, H.; Xiang, Y.; He, N.; Liu, X.; Liu, H.; Fang, L.; Zhang, F.; Sun, X.; Zhang, D.; Li, X.; et al. Enhanced vitamin C production mediated by an ABA-Induced PTP-like nucleotidase improves plant drought tolerance in Arabidopsis and Maize. Mol. Plant 2020, 13, 760–776. [Google Scholar] [CrossRef]
- Gallie, D.R. The role of l-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J. Exp. Bot. 2013, 64, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, S.; Khan, S.; Bhunia, R.K.; Kaur, K.; Tiwari, S. Metabolic engineering in food crops to enhance ascorbic acid production: Crop biofortification perspectives for human health. Physiol. Mol. Biol. Plants 2022, 28, 871–884. [Google Scholar] [CrossRef]
- Celi, G.E.A.; Gratão, P.L.; Lanza, M.; Reis, A.R.D. Physiological and biochemical roles of ascorbic acid on mitigation of abiotic stresses in plants. Plant Physiol. Biochem. 2023, 202, 107970. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Lorence, A.; Gruszewski, H.A.; Chevone, B.I.; Nessler, C.L. AMR1, an Arabidopsis gene that coordinately and negatively regulates the mannose/l-galactose ascorbic acid biosynthetic pathway. Plant Physiol. 2009, 150, 942–950. [Google Scholar] [CrossRef]
- Qin, C.; Qian, W.; Wang, W.; Wu, Y.; Yu, C.; Jiang, X.; Wang, D.; Wu, P. GDP-mannose pyrophosphorylase is a genetic determinant of ammonium sensitivity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2008, 105, 18308–18313. [Google Scholar] [CrossRef] [PubMed]
- Hoeberichts, F.A.; Vaeck, E.; Kiddle, G.; Coppens, E.; Van de Cotte, B.; Adamantidis, A.; Ormenese, S.; Foyer, C.H.; Zabeau, M.; Inzé, D.; et al. A temperature-sensitive mutation in the Arabidopsis thaliana phosphomannomutase gene disrupts protein glycosylation and triggers cell death. J. Biol. Chem. 2008, 283, 5708–5718. [Google Scholar] [CrossRef]
- Hu, T.; Ye, J.; Tao, P.; Li, H.; Zhang, J.; Zhang, Y.; Ye, Z. The tomato HD-Zip I transcription factor SlHZ24 modulates ascorbate accumulation through positive regulation of the D-mannose/L-galactose pathway. Plant J. 2016, 85, 16–29. [Google Scholar] [CrossRef]
- Ye, J.; Li, W.; Ai, G.; Li, C.; Liu, G.; Chen, W.; Wang, B.; Wang, W.; Lu, Y.; Zhang, J.; et al. Genome-wide association analysis identifies a natural variation in basic helix-loop-helix transcription factor regulating ascorbate biosynthesis via D-mannose/L-galactose pathway in tomato. PLoS Genet. 2019, 15, e1008149. [Google Scholar] [CrossRef]
- Chen, W.; Hu, T.; Ye, J.; Wang, B.; Liu, G.; Wang, Y.; Yuan, L.; Li, J.; Li, F.; Ye, Z.; et al. A CCAAT-binding factor, SlNFYA10, negatively regulates ascorbate accumulation by modulating the D-mannose/L-galactose pathway in tomato. Hortic. Res. 2020, 7, 200. [Google Scholar] [CrossRef]
- Liu, X.; Wu, R.; Bulley, S.M.; Zhong, C.; Li, D. Kiwifruit MYBS1-like and GBF3 transcription factors influence l-ascorbic acid biosynthesis by activating transcription of GDP-L-galactose phosphorylase 3. New Phytol. 2022, 234, 1782–1800. [Google Scholar] [CrossRef]
- Liu, X.; Bulley, S.M.; Varkonyi-Gasic, E.; Zhong, C.; Li, D. Kiwifruit bZIP transcription factor AcePosF21 elicits ascorbic acid biosynthesis during cold stress. Plant Physiol. 2023, 192, 982–999. [Google Scholar] [CrossRef]
- Chen, Y.; Shu, P.; Wang, R.; Du, X.; Xie, Y.; Du, K.; Deng, H.; Li, M.; Zhang, Y.; Grierson, D.; et al. Ethylene response factor AcERF91 affects ascorbate metabolism via regulation of GDP-galactose phosphorylase encoding gene (AcGGP3) in kiwifruit. Plant Sci. 2021, 313, 111063. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.W.; Kenis, K.; Keulemans, J. Genetic control of fruit vitamin C contents. Plant Physiol. 2006, 142, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Zorrilla-Fontanesi, Y.; Cabeza, A.; Domínguez, P.; Medina, J.J.; Valpuesta, V.; Denoyes-Rothan, B.; Sánchez-Sevilla, J.F.; Amaya, I. Quantitative trait loci and underlying candidate genes controlling agronomical and fruit quality traits in octoploid strawberry (Fragaria × ananassa). Theor. Appl. Genet. 2011, 123, 755–778. [Google Scholar] [CrossRef]
- Du, X.; Wang, H.; Liu, J.; Zhu, X.; Mu, J.; Feng, X.; Liu, H. Major gene with polygene inheritance analysis of shoot architecture traits in Viola cornuta. Sci. Hortic. 2022, 303, 111204. [Google Scholar] [CrossRef]
- Gai, J.; Wang, J.K. Identification and estimation of a QTL model and its effects. Theor. Appl. Genet. 1998, 97, 1162–1168. [Google Scholar] [CrossRef]
- Zheng, W.; Liu, Z.; Zhao, J.; Chen, W. Genetic Analysis of stripe disease resistance in rice restorer line C224 using major gene plus polygene mixed effect model. Rice Sci. 2012, 19, 202–206. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.; Wang, X.; Chen, G.; Zhang, J.; Zhou, R. Genetic analysis of cryotolerance in cotton during the overwintering period using mixed model of major gene and polygene. J. Integr. Agric. 2012, 11, 537–544. [Google Scholar] [CrossRef]
- Korir, P.C.; Wang, J.; Zhao, T.; Gai, J. Genetic analysis of tolerance to aluminum toxin at seedling stage in soybean based on major gene plus polygene mixed inheritance model. Front. Agric. China 2010, 4, 265–271. [Google Scholar] [CrossRef]
- Zhang, S.; Ma, C.; Zhu, J.; Wang, J.; Wen, Y.; Fu, T. Genetic analysis of oil content in Brassica napus L. using mixed model of major gene and polygene. Acta Genet. Sin. 2006, 33, 171–180. [Google Scholar] [CrossRef]
- Cao, X.; Cui, H.; Li, J.; Xiong, A.; Hou, X.; Li, Y. Heritability and gene effects for tiller number and leaf number in non-heading Chinese cabbage using joint segregation analysis. Sci. Hortiic. 2016, 203, 199–206. [Google Scholar] [CrossRef]
- Sun, X.; Liu, L.; Zhi, X.; Bai, J.; Cui, Y.; Shu, J.; Li, J. Genetic analysis of tomato internode length via mixed major gene plus polygene inheritance model. Sci. Hortic. 2019, 246, 759–764. [Google Scholar] [CrossRef]
- Dong, R.; Yu, B.; Yan, S.; Qiu, Z.; Lei, J.; Chen, C.; Li, Y.; Cao, B. Analysis of vitamin P content and inheritance models in eggplant. Hortic. Plant J. 2020, 6, 240–246. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, C.; Yang, X.; Dong, M.; Wang, Z.; Yan, F.; Wu, C.; Wang, J.; Liu, M.; Lin, M. Genetic analysis of mixed models of fruit sugar-acid fractions in a cross between jujube (Ziziphus jujuba Mill.) and wild jujube (Z. acido jujuba). Front. Plant Sci. 2023, 14, 1181903. [Google Scholar]
- Fan, Z.; Gao, Y.; Liu, R.; Wang, X.; Guo, Y.; Zhang, Q. The major gene and polygene effects of ornamental traits in bearded iris (Iris germanica) using joint segregation analysis. Scie Hortic. 2020, 260, 108882. [Google Scholar] [CrossRef]
- Ye, Y.; Wu, J.; Feng, L.; Ju, Y.; Cai, M.; Cheng, T.; Pan, H.; Zhang, Q. Heritability and gene effects for plant architecture traits of crape myrtle using major gene plus polygene inheritance analysis. Sci. Hortic. 2017, 225, 335–342. [Google Scholar] [CrossRef]
- Ren, X.; Li, H.; Yin, Y.; Duan, L.; Wang, Y.; Liang, X.; Wan, R.; Huang, T.; Zhang, B.; Xi, W.; et al. Genetic analysis of fruit traits in wolfberry (Lycium L.) by the major gene plus polygene model. Agronomy 2022, 12, 1403. [Google Scholar] [CrossRef]
- Qi, Z.; Li, J.; Raza, M.A.; Zou, X.; Cao, L.; Rao, L.; Chen, L. Inheritance of fruit cracking resistance of melon (Cucumis melo L.) fitting E-0 genetic model using major gene plus polygene inheritance analysis. Sci. Hortic. 2015, 189, 168–174. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Du, Y.; Ren, W.; Li, H.; Sun, W.; Ge, C.; Zhang, Y. SEA v2.0: An R software package for mixed major genes plus polygenes inheritance analysis of quantitative traits. Acta Agron. Sin. 2022, 48, 1416–1424. [Google Scholar] [CrossRef]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, J.; Liu, M.; Zhang, C.; Chen, Z.; Xiao, Z.; Jian, H.; et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1154. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Yoon, S.; Kwon, S.M.; Park, K.S.; Lee-Kim, Y.C. Kale juice improves coronary artery disease risk factors in hypercholesterolemic men. Biomed. Environ. Sci. 2008, 21, 91–97. [Google Scholar] [CrossRef]
- Kataya, H.A.; Hamza, A.A. Red cabbage (Brassica oleracea) ameliorates diabetic nephropathy in rats. Evid. Based Complement. Alternat. Med. 2008, 5, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Tiku, A.B.; Abraham, S.K.; Kale, R.K. Protective effect of the cruciferous vegetable mustard leaf (Brassica campestris) against in vivo chromosomal damage and oxidative stress induced by gamma-radiation and genotoxic chemicals. Environ. Mol. Mutagen. 2008, 49, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Akhlaghi, M.; Bandy, B. Dietary broccoli sprouts protect against myocardial oxidative damage and cell death during ischemia-reperfusion. Plant Foods Hum. Nutr. 2010, 65, 193–199. [Google Scholar] [CrossRef]
- Raiola, A.; Errico, A.; Petruk, G.; Monti, D.M.; Barone, A.; Rigano, M.M. Bioactive compounds in Brassicaceae vegetables with a role in the prevention of chronic diseases. Molecules 2017, 23, 15. [Google Scholar] [CrossRef]
- Heimler, D.; Vignolini, P.; Dini, M.G.; Vincieri, F.F.; Romani, A. Antiradical activity and polyphenol composition of local Brassicaceae edible varieties. Food Chem. 2006, 99, 464–469. [Google Scholar] [CrossRef]
- Fernandes, F.; Valentao, P.; Sousa, C.; Pereira, J.; Seabra, R.; Andrade, P. Chemical and antioxidative assessment of dietary turnip (Brassica rapa var. rapa L.). Food Chem. 2007, 105, 1003–1010. [Google Scholar] [CrossRef]
- Yang, J.; Zhu, Z.; Wang, Z.; Zhu, B. Effects of storage temperature on the contents of carotenoids and glucosinolates in pakchoi (Brassica rapa L. ssp. Chinensis Var. Communis). J. Food Biochem. 2010, 34, 1186–1204. [Google Scholar] [CrossRef]
- Pérez-Balibrea, S.; Moreno, D.A.; García-Viguera, C. Genotypic effects on the phytochemical quality of seeds and sprouts from commercial broccoli cultivars. Food Chem. 2011, 125, 348–354. [Google Scholar] [CrossRef]
- Domínguez-Perles, R.; Mena, P.; García-Viguera, C.; Moreno, D.A. Brassica foods as a dietary source of vitamin C: A review. Crit. Rev. Food Sci. Nutr. 2014, 54, 1076–1091. [Google Scholar] [CrossRef]
- Zhan, Z.; Hou, X.; Cao, S. Genetic analysis of vitamine C and soluble sugar contents in nonheading Chinese cabbage (Brassica campestris ssp. chinensis Makino). Acta Hortic. Sin. 1999, 26, 170–174. [Google Scholar]
- Li, H.; Han, Y.; Zhao, X.; Li, W. Analysis of genetic model on vitamin E composition contents in soybean seed in multiple locations. Oil Crop Sci. 2014, 36, 450–454. [Google Scholar]
- Li, H.; Xu, H.; Li, J.; Zhu, Q.; Chi, M.; Wang, J. Analysis of gene effect on chlorophyll content in maize. Crops 2019, 5, 46–51. [Google Scholar]
- Lin, T.; Wang, J.; Wang, L.; Chen, X.; Hou, X.; Li, Y. Major gene plus polygene inheritance of vitamin C content in non-heading Chinese cabbage. Acta Agron. Sin. 2014, 40, 1733–1739. [Google Scholar] [CrossRef]
- Wang, X.; Chen, L.; Hu, S.; Fu, L.; Xie, L.; Quan, H. Genetic analysis of vitamin C in pepper fruit. Chin. Agric. Sci. Bull. 2017, 33, 49–53. [Google Scholar]
- Song, Z.; Miao, H.; Zhang, S.; Wang, Y.; Zhang, S.; Gu, X. Genetic analysis and QTL mapping of fruit peduncle length in cucumber (Cucumis sativus L.). PLoS ONE 2016, 11, e0167845. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Si, L.; Min, Y.; Gao, P.; Meng, Q.; Li, K. Genetic analysis of vitamin C content of the south China type cucumber. Acta Agric. Boreali Sin. 2012, 27, 102–106. [Google Scholar]
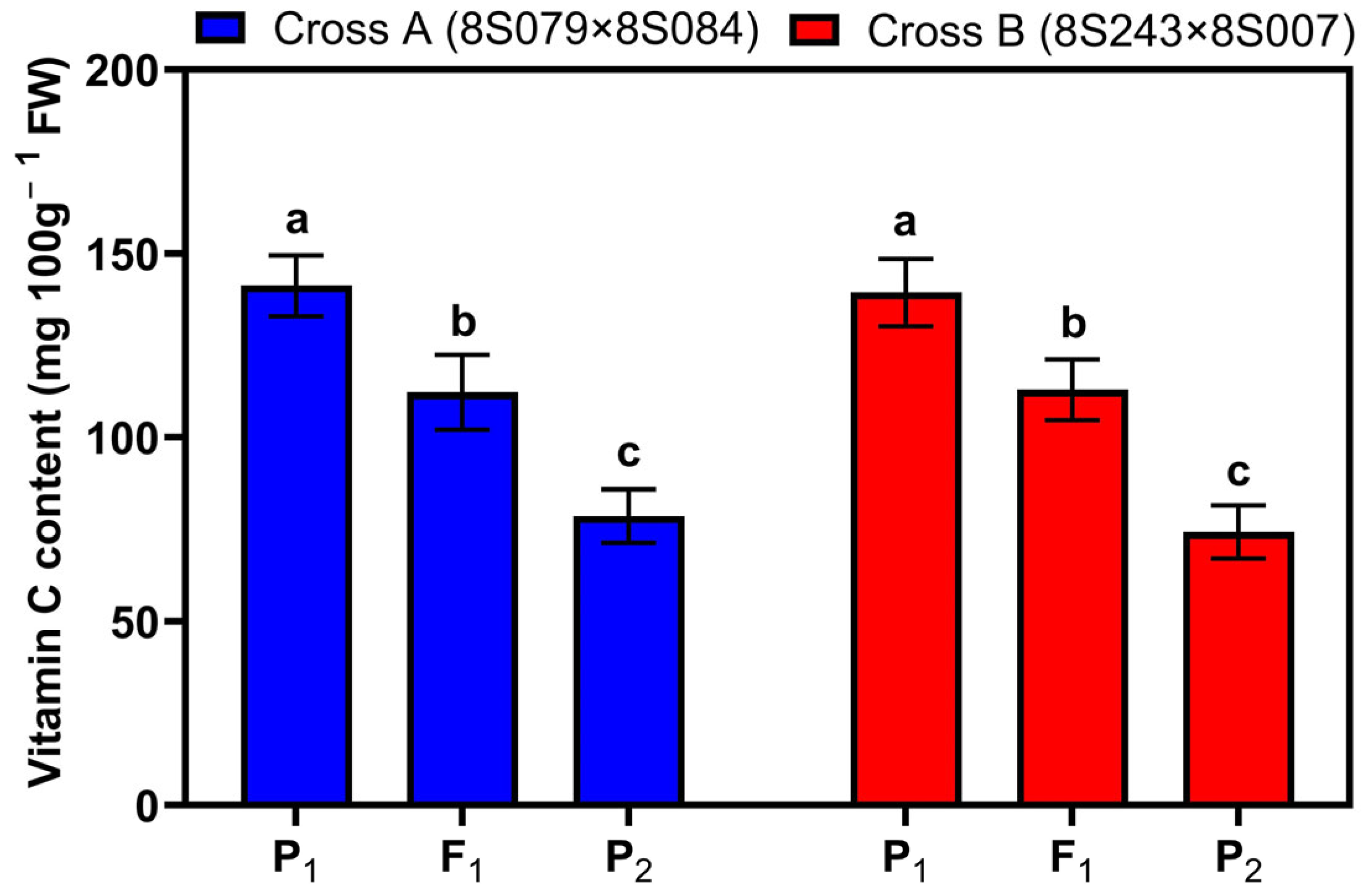
| Cross | Generation | No. of Plants | Minimum (mg 100 g−1) | Maximum (mg 100 g−1) | Mean (mg 100 g−1) | SD | Variance | CV (%) |
|---|---|---|---|---|---|---|---|---|
| Cross A 8S079 × 8S084 | P1 | 50 | 125.02 | 160.02 | 141.24 a | 8.27 | 68.37 | 5.85 |
| P2 | 50 | 62.76 | 90.27 | 78.58 f | 7.26 | 52.64 | 9.23 | |
| F1 | 46 | 96.64 | 135.68 | 112.22 c | 10.22 | 104.54 | 9.11 | |
| F2 | 217 | 57.70 | 163.86 | 101.00 d | 20.27 | 410.95 | 20.07 | |
| BC1P1 | 128 | 78.25 | 181.67 | 125.52 b | 23.17 | 536.94 | 18.46 | |
| BC1P2 | 123 | 55.30 | 118.36 | 87.40 e | 14.76 | 217.78 | 16.89 | |
| Cross B 8S243 × 8S007 | P1 | 50 | 122.79 | 154.89 | 139.34 a | 9.15 | 83.73 | 6.57 |
| P2 | 50 | 62.75 | 89.02 | 74.30 f | 7.30 | 53.24 | 9.82 | |
| F1 | 48 | 99.74 | 129.13 | 112.94 c | 8.26 | 68.20 | 7.31 | |
| F2 | 245 | 47.23 | 174.65 | 94.37 d | 23.64 | 558.84 | 25.05 | |
| BC1P1 | 119 | 85.32 | 170.50 | 122.81 b | 19.77 | 390.70 | 16.09 | |
| BC1P2 | 111 | 54.54 | 123.66 | 87.81 e | 16.96 | 287.71 | 19.32 |
| Cross | Generation | P1 | F1 | F1 | F2 | BC1P1 | BC1P2 |
|---|---|---|---|---|---|---|---|
| Cross A | Skewness | 0.21 | −0.18 | 0.25 | 0.76 | 0.36 | 0.08 |
| Kurtosis | −0.57 | −0.94 | −0.96 | 0.55 | −0.60 | −0.69 | |
| Cross B | Skewness | 0.09 | 0.35 | 0.12 | 0.95 | 0.50 | 0.18 |
| Kurtosis | −1.16 | −0.54 | −1.08 | 1.35 | −0.29 | −0.59 |
| Model | Maximum Likelihood | AIC Value | ||
|---|---|---|---|---|
| Cross A | Cross B | Cross A | Cross B | |
| 1MG-AD | −2587.82 | −2668.37 | 5183.64 | 5344.75 |
| 1MG-A | −2602.40 | −2671.64 | 5210.79 | 5349.28 |
| 1MG-EAD | −2735.58 | −2795.43 | 5477.15 | 5596.86 |
| 1MG-NCD | −2673.71 | −2749.70 | 5353.42 | 5505.40 |
| 2MG-ADI | −2574.38 | −2624.14 | 5168.76 | 5268.28 |
| 2MG-AD | −2579.41 | −2642.25 | 5170.81 | 5296.49 |
| 2MG-A | −2639.73 | −2664.71 | 5287.45 | 5337.41 |
| 2MG-EA | −2600.38 | −2665.74 | 5206.75 | 5337.48 |
| 2MG-CD | −2711.23 | −2768.11 | 5430.46 | 5544.22 |
| 2MG-EAD | −2711.23 | −2768.11 | 5428.46 | 5542.22 |
| PG-ADI | −2569.15 | −2637.90 | 5158.29 | 5295.79 |
| PG-AD | −2596.03 | −2681.62 | 5206.06 | 5377.24 |
| MX1-AD-ADI | −2555.84 | −2627.90 | 5135.68 | 5279.79 |
| MX1-AD-AD | −2562.51 | −2657.10 | 5143.03 | 5332.20 |
| MX1-A-AD | −2572.86 | −2649.33 | 5161.72 | 5314.66 |
| MX1-EAD-AD | −2595.26 | −2677.93 | 5206.52 | 5371.87 |
| MX1-NCD-AD | −2573.26 | −2657.18 | 5162.51 | 5330.35 |
| MX2-ADI-ADI | −2546.93 | −2603.42 | 5129.85 | 5242.84 |
| MX2-ADI-AD | −2548.58 | −2608.82 | 5127.17 | 5247.63 |
| MX2-AD-AD | −2561.92 | −2636.23 | 5145.85 | 5294.46 |
| MX2-A-AD | −2554.48 | −2609.53 | 5126.95 | 5237.06 |
| MX2-EA-AD | −2572.90 | −2635.76 | 5161.80 | 5287.52 |
| MX2-CD-AD | −2618.23 | −2689.42 | 5254.46 | 5396.84 |
| MX2-EAD-AD | −2595.26 | −2677.93 | 5206.51 | 5371.86 |
| Cross | Model | Generation | U12 | U22 | U32 | nW2 | Dn |
|---|---|---|---|---|---|---|---|
| Cross A | MX2-A-AD | P1 | 0.10 (0.75) | 0.20 (0.66) | 0.28 (0.60) | 0.10 (0.62) | 0.10 (0.60) |
| F1 | 0.01 (0.91) | 0.13 (0.72) | 1.00 (0.32) | 0.11 (0.57) | 0.11 (0.54) | ||
| P2 | 0.00 (0.96) | 0.04 (0.85) | 0.89 (0.34) | 0.08 (0.71) | 0.10 (0.64) | ||
| BC1P1 | 0.26 (0.61) | 0.06 (0.81) | 1.00 (0.32) | 0.14 (0.41) | 0.08 (0.33) | ||
| BC1P2 | 0.34 (0.56) | 0.45 (0.50) | 0.19 (0.67) | 0.06 (0.79) | 0.06 (0.77) | ||
| F2 | 0.24 (0.63) | 0.37 (0.54) | 0.30 (0.58) | 0.08 (0.71) | 0.61 (0.61) | ||
| MX2-AD-ADI | P1 | 0.15 (0.70) | 0.05 (0.83) | 0.37 (0.54) | 0.11 (0.54) | 0.12 (0.42) | |
| F1 | 0.40 (0.53) | 0.12 (0.73) | 1.18 (0.28) | 0.15 (0.40) | 0.15 (0.25) | ||
| P2 | 0.08 (0.78) | 0.00 (0.97) | 0.88 (0.35) | 0.08 (0.68) | 0.10 (0.64) | ||
| BC1P1 | 0.40 (0.84) | 0.01 (0.93) | 0.16 (0.69) | 0.03 (0.98) | 0.05 (0.92) | ||
| BC1P2 | 0.24 (0.62) | 0.28 (0.60) | 0.04 (0.84) | 0.05 (0.86) | 0.05 (0.91) | ||
| F2 | 0.02 (0.88) | 0.01 (0.92) | 0.00 (0.85) | 0.03 (0.98) | 0.04 (0.92) | ||
| Cross B | MX2-A-AD | P1 | 0.27 (0.60) | 0.60 (0.44) | 1.17 (0.28) | 0.12 (0.49) | 0.10 (0.66) |
| F1 | 0.13 (0.71) | 0.37 (0.54) | 1.05 (0.31) | 0.09 (0.65) | 0.11 (0.55) | ||
| P2 | 0.27 (0.60) | 0.19 (0.66) | 0.07 (0.79) | 0.07 (0.73) | 0.10 (0.63) | ||
| BC1P1 | 0.03 (0.86) | 0.16 (0.69) | 0.89 (0.34) | 0.08 (0.68) | 0.06 (0.85) | ||
| BC1P2 | 1.39 (0.24) | 1.27 (0.26) | 0.00 (0.96) | 0.15 (0.38) | 0.07 (0.63) | ||
| F2 | 0.45 (0.50) | 0.66 (0.42) | 0.43 (0.51) | 0.10 (0.58) | 0.55 (0.55) | ||
| MX2-ADI-ADI | P1 | 0.01 (0.93) | 0.04 (0.85) | 1.23 (0.27) | 0.11 (0.57) | 0.11 (0.48) | |
| F1 | 0.01 (0.94) | 0.03 (0.85) | 1.07 (0.30) | 0.08 (0.74) | 0.10 (0.65) | ||
| P2 | 0.06 (0.81) | 0.03 (0.86) | 0.05 (0.83) | 0.05 (0.88) | 0.09 (0.80) | ||
| BC1P1 | 0.15 (0.70) | 0.04 (0.84) | 0.46 (0.50) | 0.05 (0.85) | 0.05 (0.94) | ||
| BC1P2 | 0.00 (0.97) | 0.00 (0.99) | 0.04 (0.84) | 0.02 (1.00) | 0.04 (1.00) | ||
| F2 | 0.04 (0.85) | 0.09 (0.77) | 0.18 (0.67) | 0.06 (0.82) | 0.05 (0.63) |
| First-Order Genetic Parameter | Estimate | |
|---|---|---|
| Cross A | Cross B | |
| m | 107.11 | 103.92 |
| da | 22.43 | 30.72 |
| db | −6.34 | −9.61 |
| [d] | 19.65 | 15.95 |
| [h] | −0.52 | 3.20 |
| [h]/[d] | −0.03 | 0.20 |
| Second-Order Genetic Parameter | Estimate | |||||
|---|---|---|---|---|---|---|
| Cross A | Cross B | |||||
| BC1P1 | BC1P2 | F2 | BC1P1 | BC1P2 | F2 | |
| σ2p | 536.94 | 217.78 | 410.95 | 390.70 | 281.71 | 558.84 |
| σ2e | 71.93 | 71.93 | 71.93 | 67.07 | 67.07 | 67.07 |
| σ2mg | 225.76 | 119.60 | 339.02 | 289.72 | 147.33 | 491.77 |
| σ2pg | 239.25 | 26.25 | 0.00 | 33.91 | 73.31 | 0.00 |
| h2mg (%) | 42.05 | 54.92 | 82.50 | 74.15 | 51.21 | 88.00 |
| h2pg (%) | 44.56 | 12.05 | 0.00 | 8.68 | 25.48 | 0.00 |
| h2mg + pg (%) | 86.60 | 66.97 | 82.50 | 82.83 | 76.69 | 88.00 |
| 1 − h2mg + pg (%) | 13.40 | 33.03 | 17.50 | 17.17 | 23.31 | 12.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Wang, T.; Wang, X.; Wang, H.; Dun, X. Genetic Analysis of Vitamin C Content in Rapeseed Seedlings by the Major Gene Plus Polygene Mixed Effect Model. Curr. Issues Mol. Biol. 2024, 46, 9565-9575. https://doi.org/10.3390/cimb46090568
Wang C, Wang T, Wang X, Wang H, Dun X. Genetic Analysis of Vitamin C Content in Rapeseed Seedlings by the Major Gene Plus Polygene Mixed Effect Model. Current Issues in Molecular Biology. 2024; 46(9):9565-9575. https://doi.org/10.3390/cimb46090568
Chicago/Turabian StyleWang, Chao, Tao Wang, Xinfa Wang, Hanzhong Wang, and Xiaoling Dun. 2024. "Genetic Analysis of Vitamin C Content in Rapeseed Seedlings by the Major Gene Plus Polygene Mixed Effect Model" Current Issues in Molecular Biology 46, no. 9: 9565-9575. https://doi.org/10.3390/cimb46090568
APA StyleWang, C., Wang, T., Wang, X., Wang, H., & Dun, X. (2024). Genetic Analysis of Vitamin C Content in Rapeseed Seedlings by the Major Gene Plus Polygene Mixed Effect Model. Current Issues in Molecular Biology, 46(9), 9565-9575. https://doi.org/10.3390/cimb46090568







