Inflammation and Starvation Affect Housekeeping Gene Stability in Adipose Mesenchymal Stromal Cells
Abstract
1. Introduction
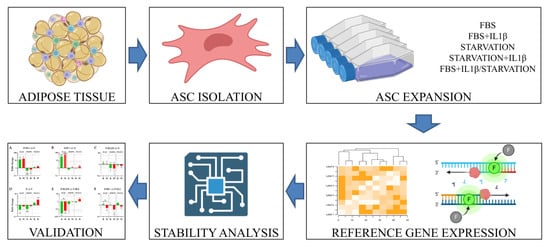
2. Materials and Methods
2.1. Ethics Statement
2.2. Adipose Mesenchymal Stromal Cells (ASCs) Isolation and Expansion
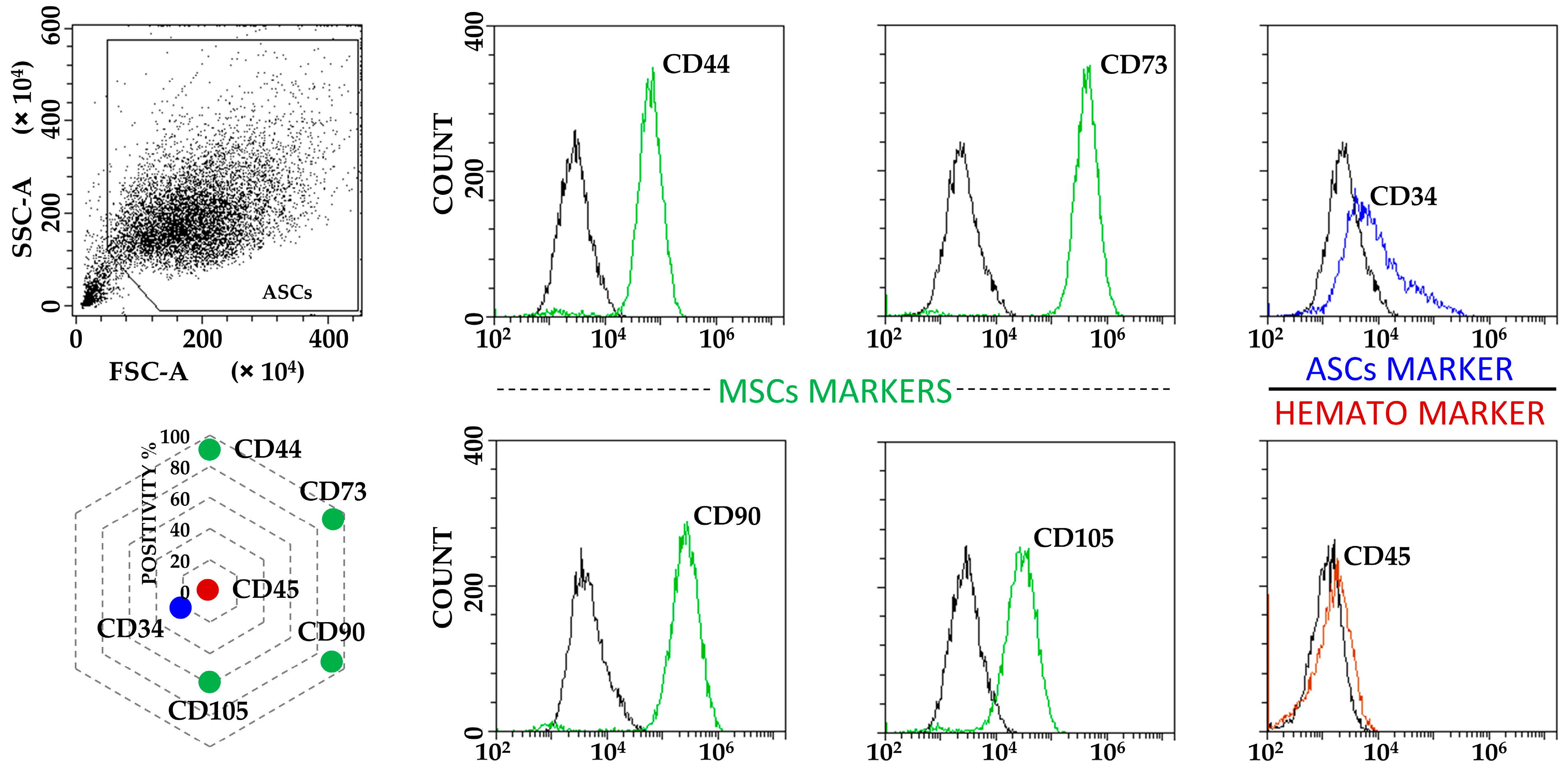
2.3. Flow Cytometry Analysis
2.4. RNA Extraction and mRNA Profiling
2.5. Data Analysis
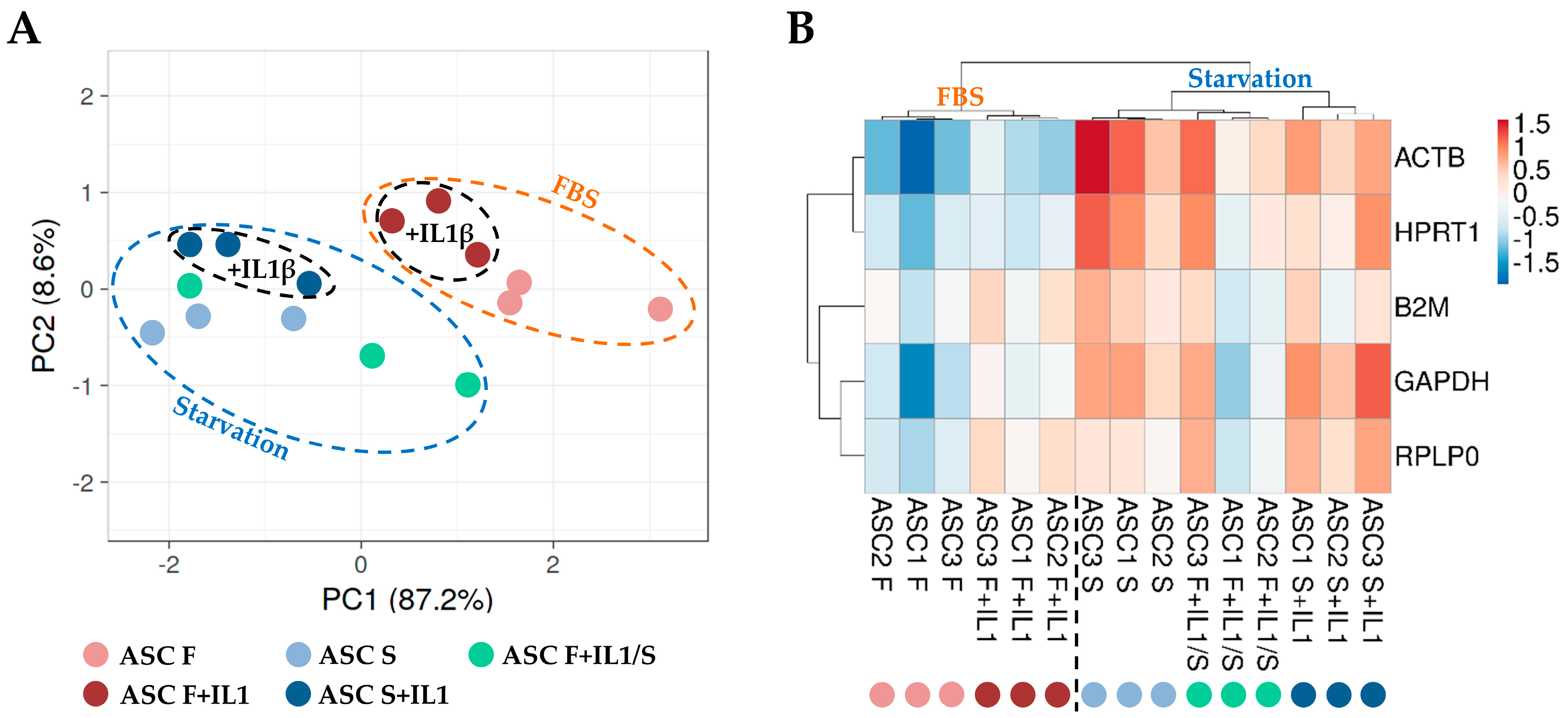
2.6. Principal Component Analysis and Hierarchical Clustering
2.7. Statistical Analyses
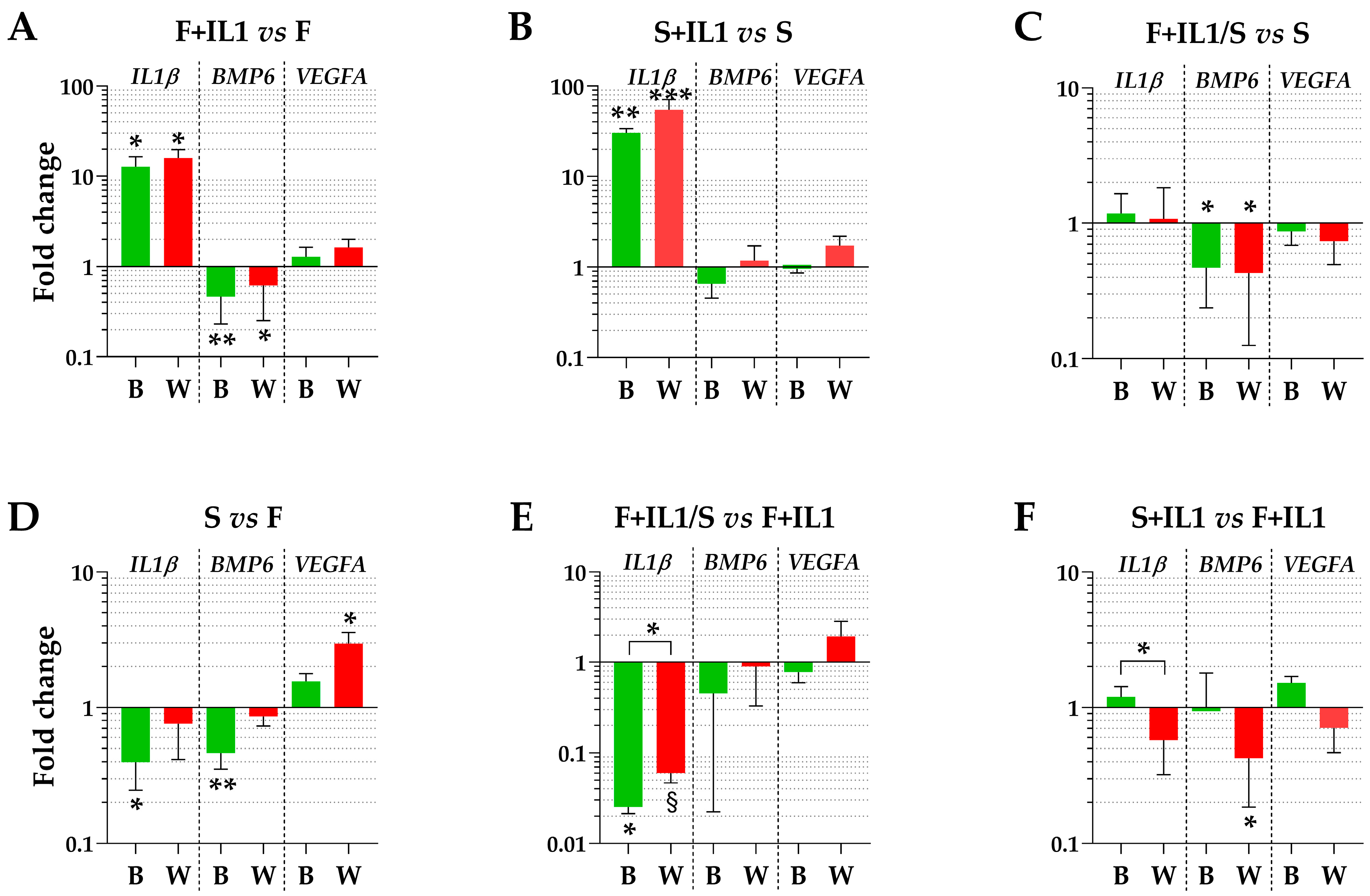
3. Results
3.1. ASCs Characterization
3.2. Expression of Candidate HKGs
3.3. HKGs Stability Analysis
3.4. Impact of HKG Choice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Loeser, R.F.; Goldring, S.R.; Scanzello, C.R.; Goldring, M.B. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheum. 2012, 64, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Huebner, K.; Frank, R.M.; Getgood, A. Ortho-Biologics for Osteoarthritis. Clin. Sports Med. 2019, 38, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Oberbauer, E.; Steffenhagen, C.; Wurzer, C.; Gabriel, C.; Redl, H.; Wolbank, S. Enzymatic and non-enzymatic isolation systems for adipose tissue-derived cells: Current state of the art. Cell Regen. 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, A.; Rubin, J.P.; Kokai, L.; Sowa, G.; Chen, J.; Onishi, K. Use of Adipose-Derived Orthobiologics for Musculoskeletal Injuries: A Narrative Review. PM&R 2020, 12, 805–816. [Google Scholar] [CrossRef]
- Czerwiec, K.; Zawrzykraj, M.; Deptuła, M.; Skoniecka, A.; Tymińska, A.; Zieliński, J.; Kosiński, A.; Pikuła, M. Adipose-Derived Mesenchymal Stromal Cells in Basic Research and Clinical Applications. Int. J. Mol. Sci. 2023, 24, 3888. [Google Scholar] [CrossRef] [PubMed]
- Cavallo, C.; Boffa, A.; de Girolamo, L.; Merli, G.; Kon, E.; Cattini, L.; Santo, E.; Grigolo, B.; Filardo, G. Bone marrow aspirate concentrate quality is affected by age and harvest site. Knee Surg. Sports Traumatol. Arthrosc. 2023, 31, 2140–2151. [Google Scholar] [CrossRef]
- Boffa, A.; Perucca Orfei, C.; Sourugeon, Y.; Laver, L.; Magalon, J.; Sánchez, M.; Tischer, T.; de Girolamo, L.; Filardo, G. Cell-based therapies have disease-modifying effects on osteoarthritis in animal models. A systematic review by the ESSKA Orthobiologic Initiative. Part 2: Bone marrow-derived cell-based injectable therapies. Knee Surg. Sports Traumatol. Arthrosc. 2023, 31, 3230–3242. [Google Scholar] [CrossRef] [PubMed]
- Strioga, M.; Viswanathan, S.; Darinskas, A.; Slaby, O.; Michalek, J. Same or not the same? Comparison of adipose tissue-derived versus bone marrow-derived mesenchymal stem and stromal cells. Stem Cells Dev. 2012, 21, 2724–2752. [Google Scholar] [CrossRef]
- Murphy, M.B.; Moncivais, K.; Caplan, A.I. Mesenchymal stem cells: Environmentally responsive therapeutics for regenerative medicine. Exp. Mol. Med. 2013, 45, e54. [Google Scholar] [CrossRef]
- Miceli, V.; Zito, G.; Bulati, M.; Gallo, A.; Busà, R.; Iannolo, G.; Conaldi, P.G. 2 Different priming strategies improve distinct therapeutic capabilities of mesenchymal stromal/stem cells: Potential implications for their clinical use. World J. Stem Cells 2023, 15, 400–420. [Google Scholar] [CrossRef]
- Cases-Perera, O.; Blanco-Elices, C.; Chato-Astrain, J.; Miranda-Fernández, C.; Campos, F.; Crespo, P.V.; Sánchez-Montesinos, I.; Alaminos, M.; Martín-Piedra, M.A.; Garzón, I. Development of secretome-based strategies to improve cell culture protocols in tissue engineering. Sci. Rep. 2022, 12, 10003. [Google Scholar] [CrossRef] [PubMed]
- Praveen Kumar, L.; Kandoi, S.; Misra, R.; Vijayalakshmi, S.; Rajagopal, K.; Verma, R.S. The mesenchymal stem cell secretome: A new paradigm towards cell-free therapeutic mode in regenerative medicine. Cytokine Growth Factor. Rev. 2019, 46, 1–9. [Google Scholar] [CrossRef]
- Krautgasser, C.; Mandl, M.; Hatzmann, F.M.; Waldegger, P.; Mattesich, M.; Zwerschke, W. Reliable reference genes for expression analysis of proliferating and adipogenically differentiating human adipose stromal cells. Cell Mol. Biol. Lett. 2019, 24, 14. [Google Scholar] [CrossRef]
- Fink, T.; Lund, P.; Pilgaard, L.; Rasmussen, J.G.; Duroux, M.; Zachar, V. Instability of standard PCR reference genes in adipose-derived stem cells during propagation, differentiation and hypoxic exposure. BMC Mol. Biol. 2008, 9, 98. [Google Scholar] [CrossRef] [PubMed]
- Dessels, C.; Pepper, M.S. Reference Gene Expression in Adipose-Derived Stromal Cells Undergoing Adipogenic Differentiation. Tissue Eng. Part. C Methods 2019, 25, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Ayanoğlu, F.B.; Elçin, A.E.; Elçin, Y.M. Evaluation of the stability of standard reference genes of adipose-derived mesenchymal stem cells during in vitro proliferation and differentiation. Mol. Biol. Rep. 2020, 47, 2109–2122. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Scherberich, A.; Di Maggio, N.; McNagny, K.M. A familiar stranger: CD34 expression and putative functions in SVF cells of adipose tissue. World J. Stem Cells 2013, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yi, G.; Sze, S.; Thon, M.R. Identifying clusters of functionally related genes in genomes. Bioinformatics 2007, 23, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Wu, W.; Qu, X. Mesenchymal stem cells in osteoarthritis therapy: A review. Am. J. Transl. Res. 2021, 13, 448–461. [Google Scholar] [PubMed]
- Goldring, M.B.; Otero, M. Inflammation in osteoarthritis. Curr. Opin. Rheumatol. 2011, 23, 471–478. [Google Scholar] [CrossRef]
- Jauković, A.; Kukolj, T.; Obradović, H.; Okić-Đorđević, I.; Mojsilović, S.; Bugarski, D. Inflammatory niche: Mesenchymal stromal cell priming by soluble mediators. World J. Stem Cells 2020, 12, 922–937. [Google Scholar] [CrossRef] [PubMed]
- Ragni, E.; Colombini, A.; Viganò, M.; Libonati, F.; Perucca Orfei, C.; Zagra, L.; de Girolamo, L. Cartilage Protective and Immunomodulatory Features of Osteoarthritis Synovial Fluid-Treated Adipose-Derived Mesenchymal Stem Cells Secreted Factors and Extracellular Vesicles-Embedded miRNAs. Cells 2021, 10, 1072. [Google Scholar] [CrossRef]
- Ragni, E.; Perucca Orfei, C.; Sinigaglia, F.; de Girolamo, L. Joint Tissue Protective and Immune-Modulating miRNA Landscape of Mesenchymal Stromal Cell-Derived Extracellular Vesicles under Different Osteoarthritis-Mimicking Conditions. Pharmaceutics 2022, 14, 1400. [Google Scholar] [CrossRef]
- Colombini, A.; Libonati, F.; Cangelosi, D.; Lopa, S.; De Luca, P.; Coviello, D.A.; Moretti, M.; de Girolamo, L. Inflammatory priming with IL-1β promotes the immunomodulatory behavior of adipose derived stem cells. Front. Bioeng. Biotechnol. 2022, 10, 1000879. [Google Scholar] [CrossRef]
- van Dalen, S.C.M.; Blom, A.B.; Walgreen, B.; Slöetjes, A.W.; Helsen, M.M.A.; Geven, E.J.W.; Huurne, M.T.; Vogl, T.; Roth, J.; van de Loo, F.A.J.; et al. IL-1β-Mediated Activation of Adipose-Derived Mesenchymal Stromal Cells Results in PMN Reallocation and Enhanced Phagocytosis: A Possible Mechanism for the Reduction of Osteoarthritis Pathology. Front. Immunol. 2019, 10, 1075. [Google Scholar] [CrossRef] [PubMed]
- Riveroll, A.L.; Skyba-Lewin, S.; Lynn, D.; Mubyeyi, G.; Abd-El-Aziz, A.; Kibenge, F.S.T.; Kibenge, M.J.T.; Cohen, A.M.; Esparza-Gonsalez, B.; McDuffee, L.; et al. Selection and Validation of Reference Genes for Gene Expression Studies in an Equine Adipose-Derived Mesenchymal Stem Cell Differentiation Model by Proteome Analysis and Reverse-Transcriptase Quantitative Real-Time PCR. Genes 2023, 14, 673. [Google Scholar] [CrossRef] [PubMed]
- Ragni, E.; Viganò, M.; Rebulla, P.; Giordano, R.; Lazzari, L. What is beyond a qRT-PCR study on mesenchymal stem cell differentiation properties: How to choose the most reliable housekeeping genes. J. Cell Mol. Med. 2013, 17, 168–180. [Google Scholar] [CrossRef] [PubMed]
- de Cássia Noronha, N.; Mizukami, A.; Caliári-Oliveira, C.; Gastaldi Cominal, J.; Rocha, J.L.M.; Tadeu Covas, D.; Swiech, K.; Malmegrim, K.C.R. Priming approaches to improve the efficacy of mesenchymal stromal cell-based therapies. Stem Cell Res. Ther. 2019, 10, 131. [Google Scholar] [CrossRef] [PubMed]
- Cimino, M.; Gonçalves, R.M.; Barrias, C.C.; Martins, M.C.L. Xeno-Free Strategies for Safe Human Mesenchymal Stem/Stromal Cell Expansion: Supplements and Coatings. Stem Cells Int. 2017, 2017, 6597815. [Google Scholar] [CrossRef] [PubMed]
- Bui, H.T.H.; Nguyen, L.T.; Than, U.T.T. Influences of Xeno-Free Media on Mesenchymal Stem Cell Expansion for Clinical Application. Tissue Eng. Regen. Med. 2021, 18, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Giannasi, C.; Niada, S.; Della Morte, E.; Casati, S.R.; De Palma, C.; Brini, A.T. Serum starvation affects mitochondrial metabolism of adipose-derived stem/stromal cells. Cytotherapy 2023, 25, 704–711. [Google Scholar] [CrossRef]
- Ferro, F.; Spelat, R.; Shaw, G.; Duffy, N.; Islam, M.N.; O’Shea, P.M.; O’Toole, D.; Howard, L.; Murphy, J.M. Survival/Adaptation of Bone Marrow-Derived Mesenchymal Stem Cells After Long-Term Starvation Through Selective Processes. Stem Cells 2019, 37, 813–827. [Google Scholar] [CrossRef]
- Wells, A.; Rodrigues, M.; Wells, A.W.; Nuschke, A. Starvation as an initiator of mesenchymal stem cell/multipotent stromal cell differentiation. J. Stem Cell Res. Ther. 2016, 1, 104–106. [Google Scholar] [CrossRef]
- Daheshia, M.; Yao, J.Q. The interleukin 1beta pathway in the pathogenesis of osteoarthritis. J. Rheumatol. 2008, 35, 2306–2312. [Google Scholar] [CrossRef]
- Sekiya, I.; Colter, D.C.; Prockop, D.J. BMP-6 enhances chondrogenesis in a subpopulation of human marrow stromal cells. Biochem. Biophys. Res. Commun. 2001, 284, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Nagao, M.; Hamilton, J.L.; Kc, R.; Berendsen, A.D.; Duan, X.; Cheong, C.W.; Li, X.; Im, H.; Olsen, B.R. Vascular Endothelial Growth Factor in Cartilage Development and Osteoarthritis. Sci. Rep. 2017, 7, 13027. [Google Scholar] [CrossRef] [PubMed]


| ACTB | B2M | GAPDH | HPRT1 | RPLP0 | |
|---|---|---|---|---|---|
| ASC1 F | 17.96 | 19.15 | 19.73 | 25.90 | 18.25 |
| ASC2 F | 18.71 | 19.78 | 20.71 | 26.44 | 18.54 |
| ASC3 F | 18.77 | 19.71 | 20.55 | 26.51 | 18.66 |
| ASC1 F+IL1 | 19.03 | 19.66 | 21.02 | 26.34 | 19.06 |
| ASC2 F+IL1 | 18.88 | 20.24 | 21.17 | 26.68 | 19.50 |
| ASC3 F+IL1 | 19.54 | 20.38 | 21.29 | 26.59 | 19.59 |
| ASC1 S | 21.09 | 20.42 | 22.20 | 28.01 | 19.36 |
| ASC2 S | 20.50 | 20.07 | 21.77 | 27.44 | 19.06 |
| ASC3 S | 21.50 | 20.68 | 22.18 | 28.30 | 19.38 |
| ASC1 S+IL1 | 20.78 | 20.43 | 22.33 | 27.39 | 19.86 |
| ASC2 S+IL1 | 20.34 | 19.64 | 21.96 | 27.12 | 19.48 |
| ASC3 S+IL1 | 20.69 | 20.18 | 22.59 | 28.02 | 19.98 |
| ASC1 F+IL1/S | 19.90 | 19.28 | 20.38 | 26.71 | 18.43 |
| ASC2 F+IL1/S | 20.30 | 19.58 | 21.04 | 27.20 | 18.90 |
| ASC3 F+IL1/S | 21.05 | 20.33 | 22.13 | 28.07 | 19.88 |
| Cond. | Geomean | DeltaCt | BestKeeper | NormFinder | geNorm | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| F | HPRT1 | 1.19 | HPRT1 | 0.14 | RPLP0 | 0.16 | HPRT1 | 0.04 | B2M|HPRT1 | 0.07 |
| B2M | 1.86 | B2M | 0.14 | B2M | 0.26 | B2M | 0.04 | |||
| ACTB | 3.22 | ACTB | 0.16 | HPRT1 | 0.26 | ACTB | 0.08 | ACTB | 0.11 | |
| RPLP0 | 3.34 | GAPDH | 0.22 | ACTB | 0.35 | GAPDH | 0.21 | GAPDH | 0.14 | |
| GAPDH | 4.23 | RPLP0 | 0.22 | GAPDH | 0.40 | RPLP0 | 0.22 | RPLP0 | 0.18 | |
| F+IL1 | GAPDH | 1.41 | RPLP0 | 0.18 | GAPDH | 0.09 | RPLP0 | 0.03 | GAPDH|HPRT1 | 0.12 |
| RPLP0 | 1.73 | GAPDH | 0.20 | HPRT1 | 0.13 | GAPDH | 0.06 | |||
| HPRT1 | 2.06 | HPRT1 | 0.22 | RPLP0 | 0.22 | HPRT1 | 0.15 | RPLP0 | 0.14 | |
| B2M | 4.23 | B2M | 0.24 | ACTB | 0.26 | B2M | 0.19 | B2M | 0.17 | |
| ACTB | 4.73 | ACTB | 0.34 | B2M | 0.29 | ACTB | 0.32 | ACTB | 0.24 | |
| S | B2M | 1.73 | B2M | 0.16 | RPLP0 | 0.14 | B2M | 0.07 | GAPDH|RPLP0 | 0.07 |
| RPLP0 | 2.00 | HPRT1 | 0.18 | GAPDH | 0.19 | HPRT1 | 0.11 | |||
| GAPDH | 2.06 | GAPDH | 0.19 | B2M | 0.21 | GAPDH | 0.14 | B2M | 0.12 | |
| HPRT1 | 2.83 | RPLP0 | 0.21 | HPRT1 | 0.32 | RPLP0 | 0.19 | HPRT1 | 0.17 | |
| ACTB | 5.00 | ACTB | 0.23 | ACTB | 0.35 | ACTB | 0.22 | ACTB | 0.19 | |
| S+IL1 | RPLP0 | 1.19 | RPLP0 | 0.17 | ACTB | 0.18 | RPLP0 | 0.04 | GAPDH|RPLP0 | 0.08 |
| GAPDH | 1.86 | GAPDH | 0.18 | RPLP0 | 0.20 | GAPDH | 0.04 | |||
| ACTB | 2.28 | ACTB | 0.21 | GAPDH | 0.22 | ACTB | 0.14 | ACTB | 0.12 | |
| B2M | 4.00 | B2M | 0.28 | B2M | 0.30 | B2M | 0.25 | B2M | 0.17 | |
| HPRT1 | 5.00 | HPRT1 | 0.32 | HPRT1 | 0.34 | HPRT1 | 0.31 | HPRT1 | 0.23 | |
| F+IL1/S | HPRT1 | 1.73 | HPRT1 | 0.13 | B2M | 0.40 | HPRT1 | 0.03 | ACTB|B2M | 0.06 |
| B2M | 2.00 | RPLP0 | 0.14 | ACTB | 0.42 | RPLP0 | 0.03 | |||
| ACTB | 2.06 | ACTB | 0.16 | HPRT1 | 0.50 | ACTB | 0.11 | HPRT1 | 0.11 | |
| RPLP0 | 2.83 | B2M | 0.19 | RPLP0 | 0.54 | B2M | 0.18 | RPLP0 | 0.12 | |
| GAPDH | 5.00 | GAPDH | 0.25 | GAPDH | 0.63 | GAPDH | 0.24 | GAPDH | 0.17 | |
| Cond. | Geomean | DeltaCt | BestKeeper | NormFinder | geNorm | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| F and F+IL1 | B2M | 1.57 | B2M | 0.24 | HPRT1 | 0.19 | B2M | 0.10 | ACTB|GAPDH | 0.21 |
| GAPDH | 2.00 | GAPDH | 0.26 | B2M | 0.33 | GAPDH | 0.17 | |||
| ACTB | 2.28 | ACTB | 0.28 | ACTB | 0.34 | ACTB | 0.19 | B2M | 0.24 | |
| HPRT1 | 3.34 | RPLP0 | 0.29 | GAPDH | 0.42 | RPLP0 | 0.21 | RPLP0 | 0.25 | |
| RPLP0 | 4.23 | HPRT1 | 0.32 | RPLP0 | 0.45 | HPRT1 | 0.27 | HPRT1 | 0.28 | |
| S and S+IL1 | B2M | 1.32 | B2M | 0.33 | GAPDH | 0.20 | B2M | 0.18 | ACTB|B2M | 0.18 |
| GAPDH | 2.00 | GAPDH | 0.34 | B2M | 0.27 | GAPDH | 0.20 | |||
| ACTB | 2.45 | ACTB | 0.35 | RPLP0 | 0.27 | ACTB | 0.26 | HPRT1 | 0.24 | |
| HPRT1 | 3.94 | HPRT1 | 0.37 | ACTB | 0.32 | HPRT1 | 0.28 | GAPDH | 0.32 | |
| RPLP0 | 3.98 | RPLP0 | 0.44 | HPRT1 | 0.40 | RPLP0 | 0.41 | RPLP0 | 0.37 | |
| S and F+IL1/S | HPRT1 | 1.41 | HPRT1 | 0.19 | RPLP0 | 0.37 | HPRT1 | 0.04 | ACTB|HPRT1 | 0.08 |
| B2M | 2.21 | B2M | 0.21 | B2M | 0.42 | B2M | 0.09 | |||
| ACTB | 2.28 | ACTB | 0.21 | ACTB | 0.49 | ACTB | 0.13 | B2M | 0.12 | |
| RPLP0 | 3.34 | GAPDH | 0.30 | HPRT1 | 0.51 | GAPDH | 0.25 | GAPDH | 0.19 | |
| GAPDH | 4.23 | RPLP0 | 0.32 | GAPDH | 0.60 | RPLP0 | 0.28 | RPLP0 | 0.25 | |
| F+IL1 and S+IL1 | GAPDH | 1.41 | GAPDH | 0.38 | RPLP0 | 0.23 | GAPDH | 0.16 | GAPDH|HPRT1 | 0.17 |
| HPRT1 | 1.86 | HPRT1 | 0.40 | B2M | 0.29 | HPRT1 | 0.17 | |||
| RPLP0 | 2.45 | RPLP0 | 0.42 | HPRT1 | 0.49 | RPLP0 | 0.20 | ACTB | 0.29 | |
| ACTB | 3.94 | ACTB | 0.54 | GAPDH | 0.57 | ACTB | 0.49 | RPLP0 | 0.38 | |
| B2M | 3.98 | B2M | 0.60 | ACTB | 0.73 | B2M | 0.57 | B2M | 0.47 | |
| F and S | HPRT1 | 1.73 | HPRT1 | 0.42 | RPLP0 | 0.39 | HPRT1 | 0.10 | B2M|RPLP0 | 0.15 |
| RPLP0 | 2.00 | GAPDH | 0.44 | B2M | 0.42 | GAPDH | 0.10 | |||
| B2M | 2.06 | B2M | 0.51 | HPRT1 | 0.82 | B2M | 0.41 | HPRT1 | 0.37 | |
| GAPDH | 2.83 | RPLP0 | 0.55 | GAPDH | 0.86 | RPLP0 | 0.49 | GAPDH | 0.39 | |
| ACTB | 5.00 | ACTB | 0.74 | ACTB | 1.28 | ACTB | 0.72 | ACTB | 0.53 | |
| F+IL1 and F+IL1/S | GAPDH | 1.32 | GAPDH | 0.44 | GAPDH | 0.36 | GAPDH | 0.08 | B2M|RPLP0 | 0.14 |
| RPLP0 | 2.06 | RPLP0 | 0.47 | B2M | 0.40 | HPRT1 | 0.29 | |||
| B2M | 2.21 | B2M | 0.52 | RPLP0 | 0.43 | RPLP0 | 0.35 | GAPDH | 0.24 | |
| HPRT1 | 3.36 | HPRT1 | 0.52 | HPRT1 | 0.47 | B2M | 0.43 | HPRT1 | 0.41 | |
| ACTB | 5.00 | ACTB | 0.73 | ACTB | 0.63 | ACTB | 0.69 | ACTB | 0.53 | |
| Cond. | Geomean | DeltaCt | BestKeeper | NormFinder | geNorm | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| ALL | HPRT1 | 1.86 | HPRT1 | 0.48 | B2M | 0.40 | HPRT1 | 0.20 | B2M|RPLP0 | 0.34 |
| B2M | 2.00 | GAPDH | 0.48 | RPLP0 | 0.46 | GAPDH | 0.22 | |||
| RPLP0 | 2.06 | RPLP0 | 0.54 | HPRT1 | 0.62 | RPLP0 | 0.40 | GAPDH | 0.44 | |
| GAPDH | 2.63 | B2M | 0.57 | GAPDH | 0.71 | B2M | 0.45 | HPRT1 | 0.47 | |
| ACTB | 5.00 | ACTB | 0.66 | ACTB | 0.90 | ACTB | 0.60 | ACTB | 0.54 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragni, E.; Piccolo, S.; Taiana, M.; Visconte, C.; Grieco, G.; de Girolamo, L. Inflammation and Starvation Affect Housekeeping Gene Stability in Adipose Mesenchymal Stromal Cells. Curr. Issues Mol. Biol. 2024, 46, 842-855. https://doi.org/10.3390/cimb46010054
Ragni E, Piccolo S, Taiana M, Visconte C, Grieco G, de Girolamo L. Inflammation and Starvation Affect Housekeeping Gene Stability in Adipose Mesenchymal Stromal Cells. Current Issues in Molecular Biology. 2024; 46(1):842-855. https://doi.org/10.3390/cimb46010054
Chicago/Turabian StyleRagni, Enrico, Simona Piccolo, Michela Taiana, Caterina Visconte, Giulio Grieco, and Laura de Girolamo. 2024. "Inflammation and Starvation Affect Housekeeping Gene Stability in Adipose Mesenchymal Stromal Cells" Current Issues in Molecular Biology 46, no. 1: 842-855. https://doi.org/10.3390/cimb46010054
APA StyleRagni, E., Piccolo, S., Taiana, M., Visconte, C., Grieco, G., & de Girolamo, L. (2024). Inflammation and Starvation Affect Housekeeping Gene Stability in Adipose Mesenchymal Stromal Cells. Current Issues in Molecular Biology, 46(1), 842-855. https://doi.org/10.3390/cimb46010054