Rapid Visual Detection of Elite Erect Panicle Dense and Erect Panicle 1 Allele for Marker-Assisted Improvement in Rice (Oryza sativa L.) Using the Loop-Mediated Isothermal Amplification Method
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Genomic DNA Isolation
2.2. The InDel and LAMP Marker Design
2.3. PCR Amplification and Detection
2.4. LAMP Amplification and Colorimetric LAMP Assay
2.4.1. LAMP Amplification and Agarose Gel Electrophoresis Detection
2.4.2. Colorimetric LAMP Assay
2.5. Application of LAMP Colorimetric Assay in the Improvement of the Heavy-Panicle Elite Hybrid Rice Restorer Line SHUHUI498
3. Results
3.1. InDel and LAMP Markers
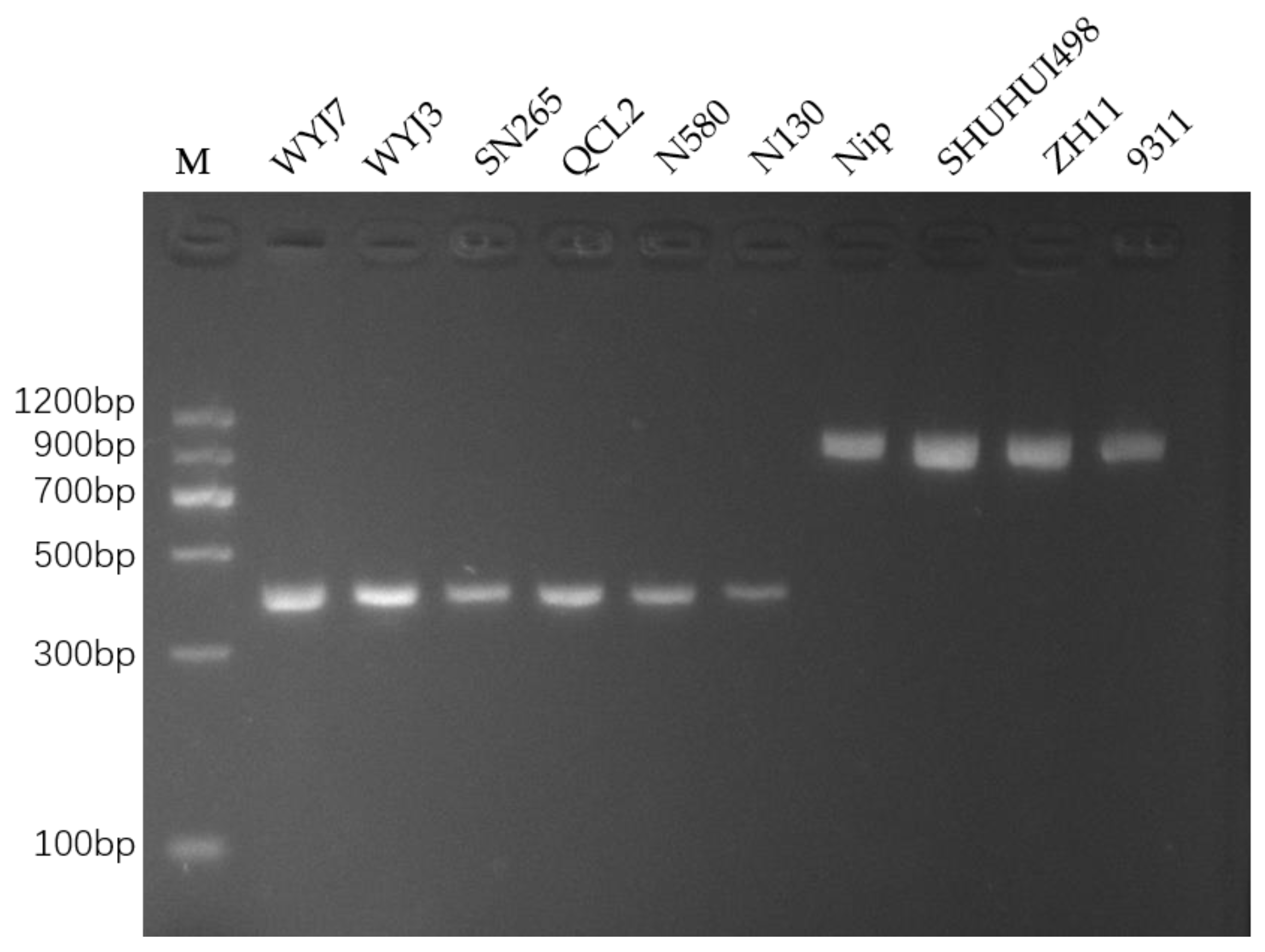
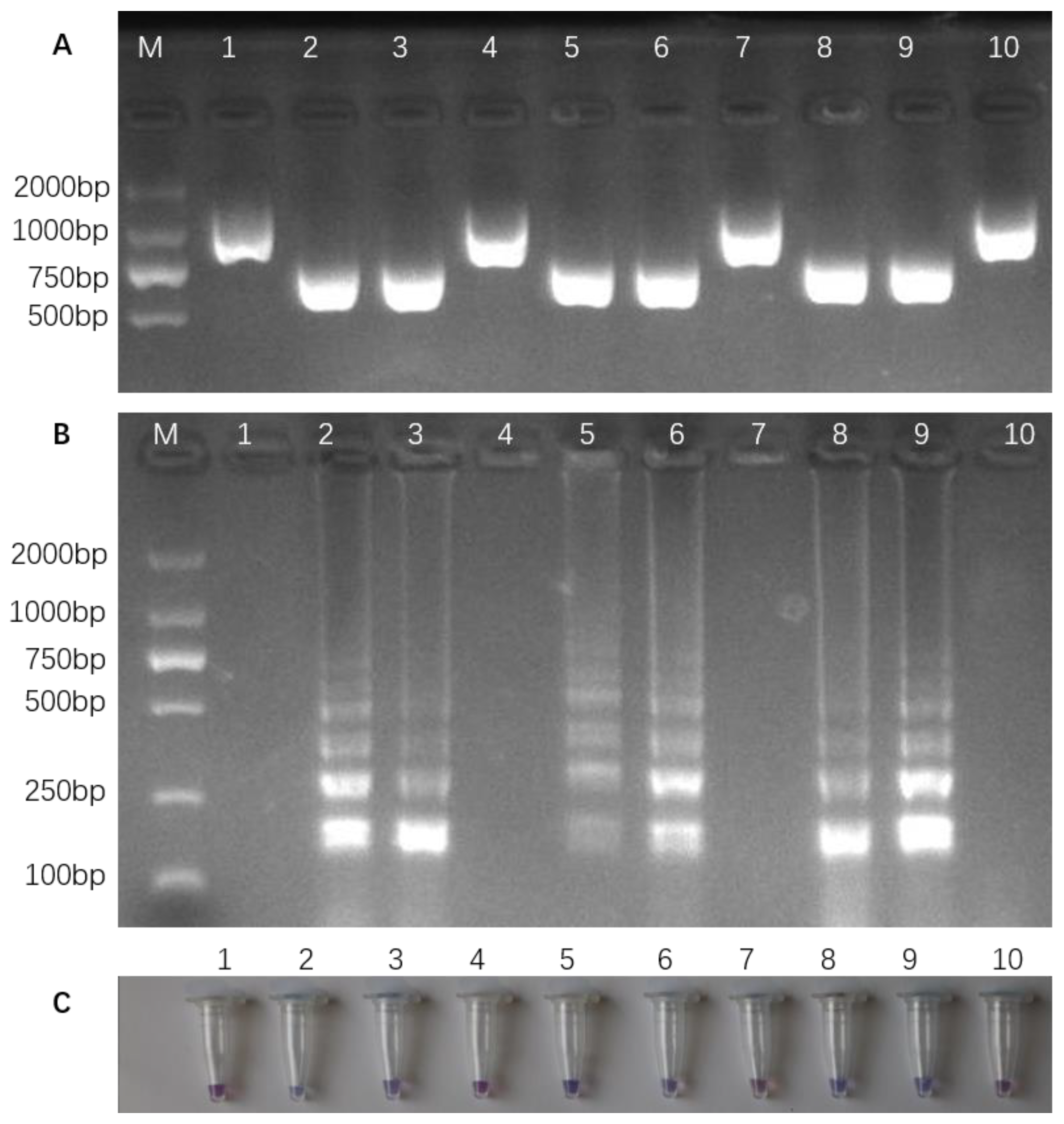
3.2. Optimizing PCR and Detecting InDel Markers in Different Rice Varieties
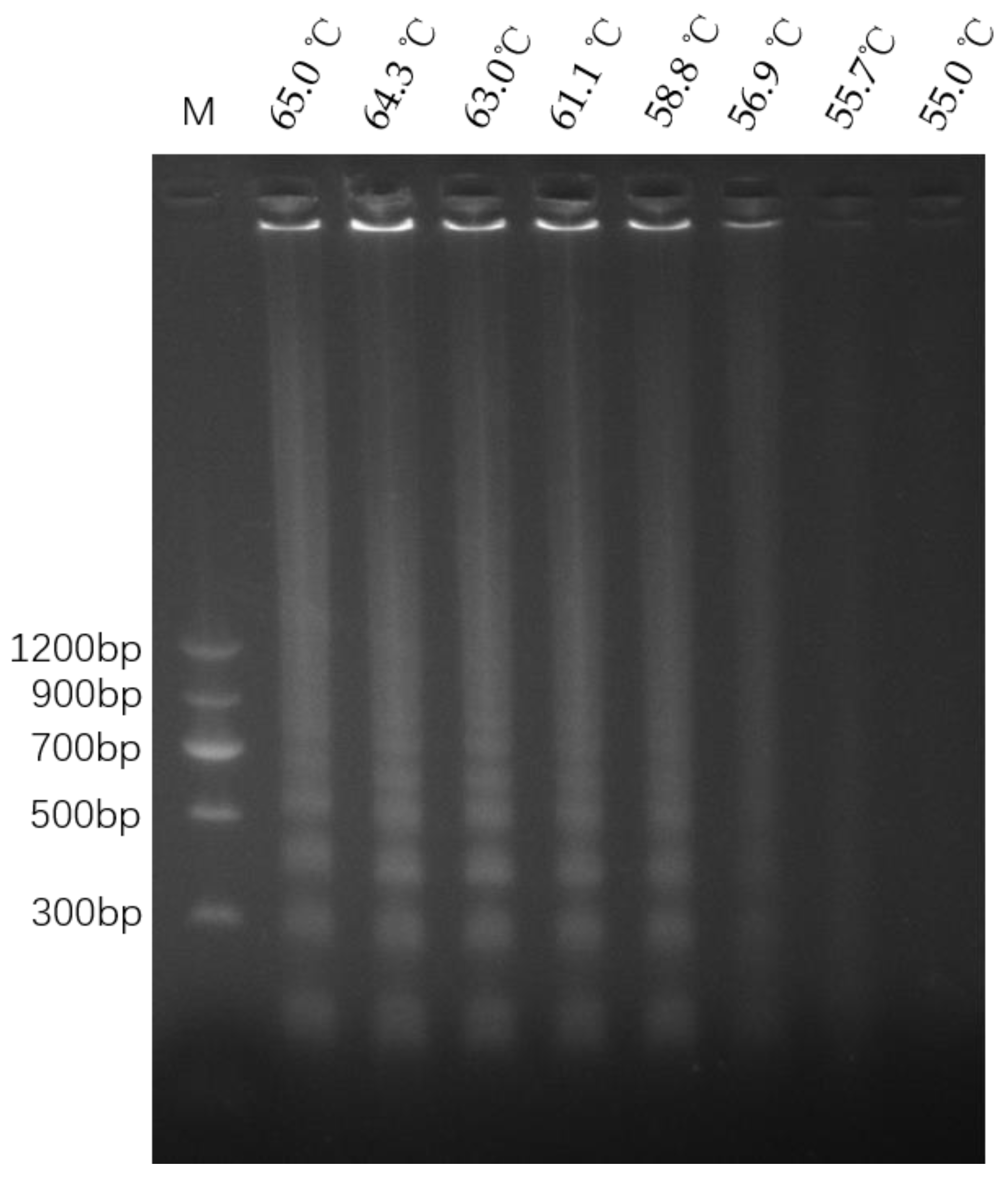
3.3. Optimization of LAMP Reaction Temperature
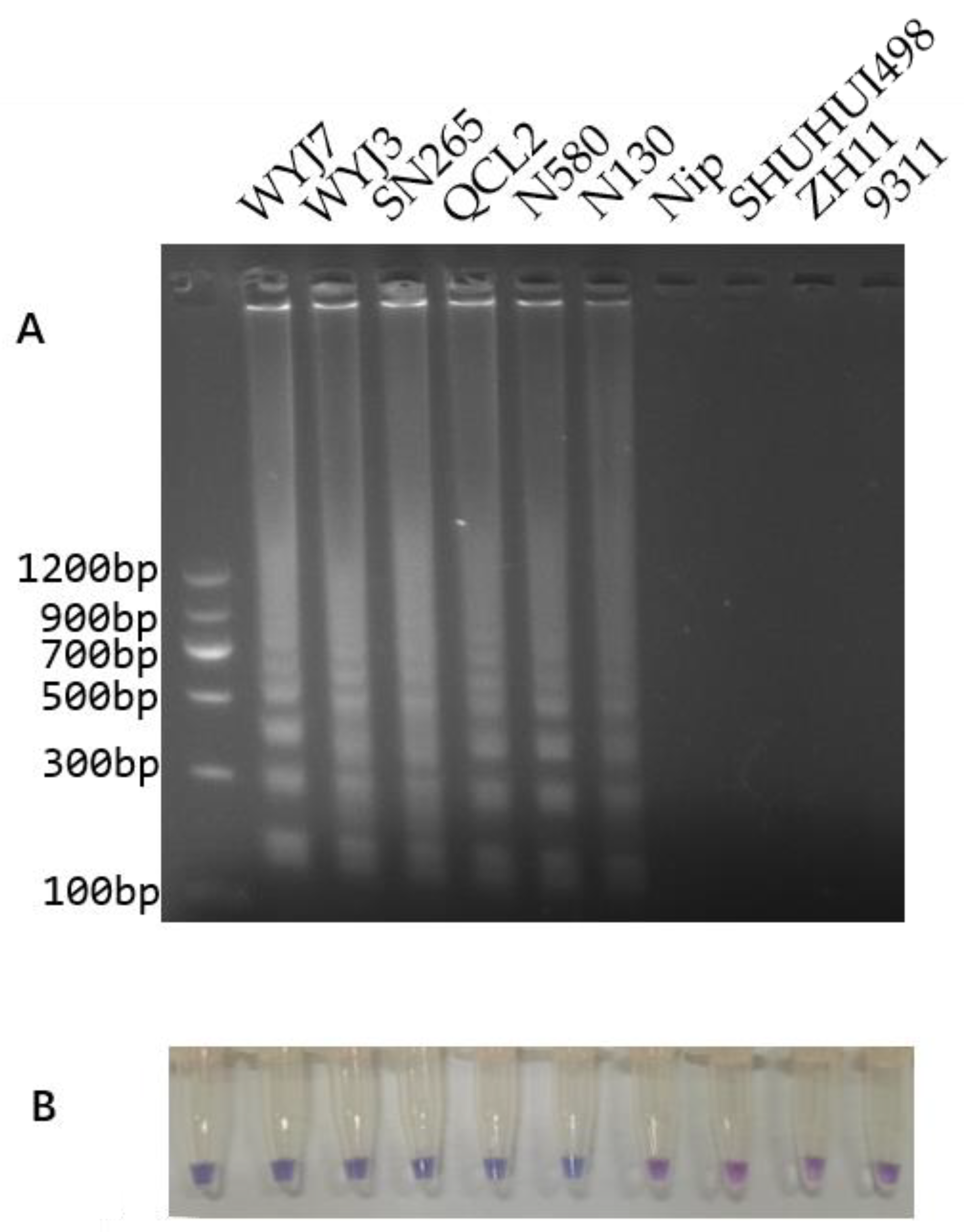
3.4. LAMP Reaction and Colorimetric LAMP Assay
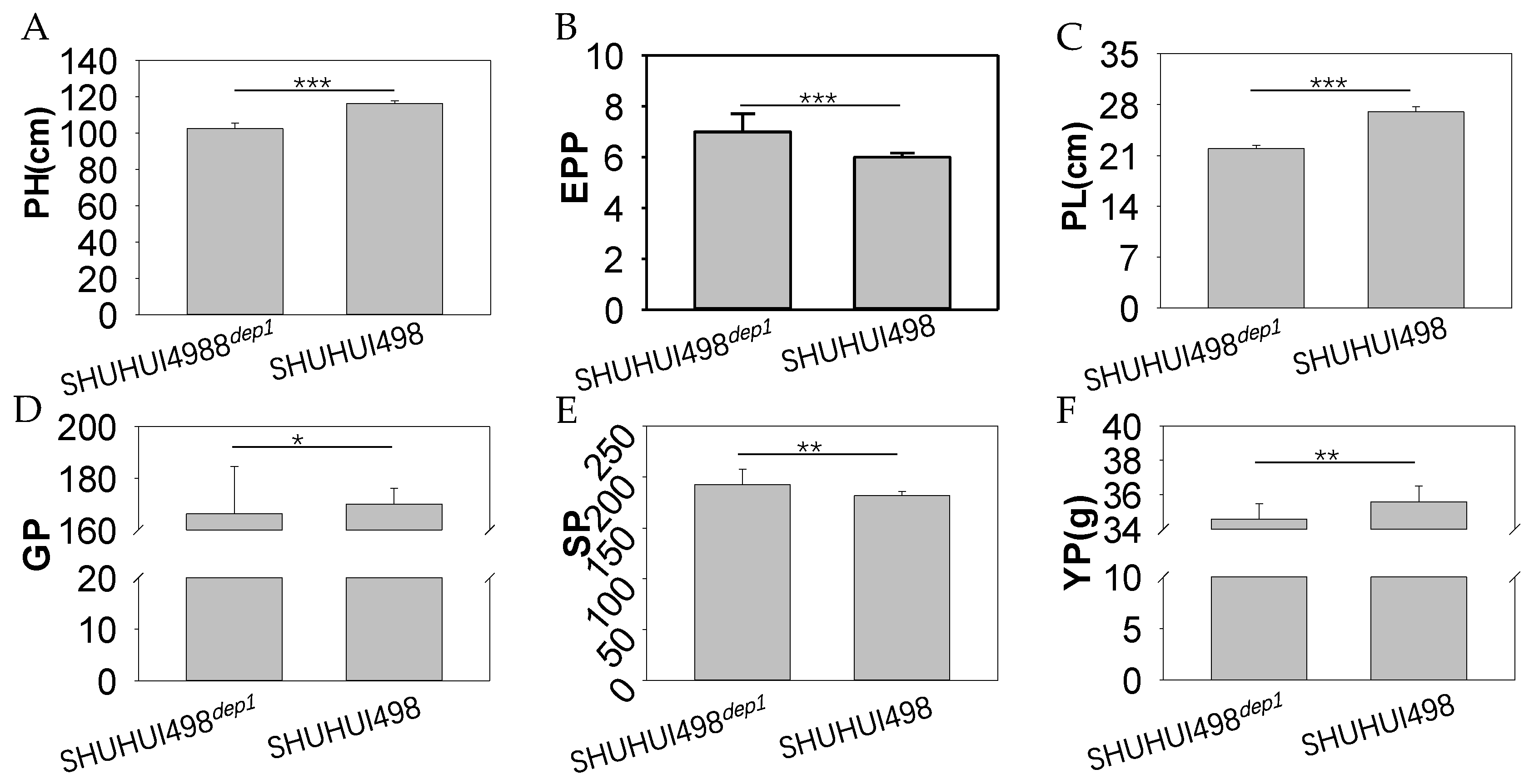
3.5. Application of the Colorimetric LAMP Assay in Breeding an Improved Heavy-Panicle Elite Hybrid Rice Restorer Line
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, K.; Ma, Y.; Liu, T.; Shen, M. The breeding of subspecific heavy ear hybrid rice-exploration about super-high yield breeding of hybrid rice. J. Sichuan Agric. Univ. 1995, 13, 403–407. [Google Scholar] [CrossRef]
- Zhang, A.; Wang, F.; Kong, D.; Hou, D.; Huang, L.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; Liu, G.; et al. Mutation of DEP1 facilitates the improvement of plant architecture in Xian rice (Oryza sativa). Plant Breed. 2023, 142, 338–344. [Google Scholar] [CrossRef]
- Huang, H.; Ye, Y.; Song, W.; Li, Q.; Han, R.; Wu, C.; Wang, S.; Yu, J.; Liu, X.; Fu, X.; et al. Modulating the C-terminus of DEP1 synergistically enhances grain quality and yield in rice. J. Genet. Genom. 2022, 49, 506–509. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Mei, Q.; Xue, C.; Wang, Z.; Li, D.; Zhang, Y.; Xuan, Y. Mutation of G-protein γ subunit DEP1 increases planting density and resistance to sheath blight disease in rice. Plant Biotechnol. J. 2020, 19, 418–420. [Google Scholar] [CrossRef]
- Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Li, J.; Fu, X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhao, M.; Zhang, Q.; Xu, Z.; Xu, Q. The DENSE AND ERECT PANICLE 1 (DEP1) gene offering the potential in the breeding of high-yielding rice. Breed. Sci. 2016, 66, 659–667. [Google Scholar] [CrossRef]
- Qian, Q.; Guo, L.; Smith, S.M.; Li, J. Breeding high-yield superior-quality hybrid super-rice by rational design. Natl. Sci. Rev. 2016, 3, 283–294. [Google Scholar] [CrossRef]
- Cheng, F.; Yu, J.; Xu, Z.; Xu, Q. Erect panicle architecture contributes to increased rice production through the improvement of canopy structure. Mol. Breed. 2019, 39, 128. [Google Scholar] [CrossRef]
- Li, Y.; Lv, X.; Rui, M.; Hu, J.; Volkov, V.S.; Zeng, D.; Wang, Y. Rice dep1 variety maintains larger stomatal conductance to enhance photosynthesis under low nitrogen conditions. Crop Des. 2023, 2, 100025. [Google Scholar] [CrossRef]
- Sun, H.; Qian, Q.; Wu, K.; Luo, J.; Wang, S.; Zhang, C.; Ma, Y.; Liu, Q.; Huang, X.; Yuan, Q.; et al. Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 2014, 46, 652–656. [Google Scholar] [CrossRef]
- Kunihiro, S.; Saito, T.; Matsuda, T.; Inoue, M.; Kuramata, M.; Taguchi-Shiobara, F.; Youssefian, S.; Berberich, T.; Kusano, T. Rice DEP1, encoding a highly cysteine-rich G protein γ subunit, confers cadmium tolerance on yeast cells and plants. J. Exp. Bot. 2013, 64, 4517–4527. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Jiang, N.; Xu, Z.; Xu, Q. Heterotrimeric G protein are involved in the regulation of multiple agronomic traits and stress tolerance in rice. BMC Plant Biol. 2020, 20, 90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhou, Y.; Yin, J.; Yan, X.; Lin, S.; Xu, W.; Baluška, F.; Wang, Y.; Xia, Y.; Liang, G.; et al. Rice G-protein subunits qPE9-1 and RGB1 play distinct roles in abscisic acid responses and drought adaptation. J. Exp. Bot. 2015, 66, 6371–6384. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhao, Y.; Yu, J.; Wu, C.; Wang, Q.; Liu, X.; Gao, X.; Wu, K.; Fu, X.; Liu, Q. Heterotrimeric G protein γ subunit DEP1 synergistically regulates grain quality and yield by modulating the TTP (TON1-TRM-PP2A) complex in rice. J. Genet. Genom. 2023, 50, 528–531. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, Q.; Sun, J.; Xu, Z.; Chen, W. Research progess of rice erect panicle gene DEP1. Sci. Sin. Vitae 2017, 47, 1036–1042. [Google Scholar] [CrossRef][Green Version]
- Xu, H.; Sun, J.; Xu, Q.; Pan, G.; Zhou, G.; Zhang, Z.; Sun, Y.; Xu, Z.; Chen, W. Research progress on optimizing panicle characters and quality improvement of indica pedigree in northern japonica rice. Chin. Sci. Bull. Kexue Tongbao 2022, 67, 135–142. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, J.; Yang, J.; Fan, F.; Yang, J.; Zhong, W. Development and application for a functional marker for erect panicle gene qPE9-1 of rice. Mol. Plant Breed. 2012, 10, 583–588. [Google Scholar]
- Pulindala, S.; Bommisetty, R.; Withanawasam, D.M.; Somagutta, S.; Kommana, M.; Kalluru, S.; Keerthi, I.; Chintala, S.; Vemireddy, L.R. Enhancement of yield through marker-assisted pyramiding of yield contributing genes in rice (Oryza sativa L.) var. BPT5204. Euphytica 2022, 218, 148. [Google Scholar] [CrossRef]
- Li, P.; Li, Z.; Liu, X.; Zhang, H.; Wang, Q.; Li, N.; Ding, H.; Yao, F. Development and application of intragenic markers for 14 nitrogen-use efficiency genes in rice (Oryza sativa L.). Front. Plant Sci. 2022, 13, 891860. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, N.; Chen, H.; Wang, F.; Huang, Y.; Jia, B.; Wang, S.; Wang, Y.; Xu, Z. Effects of DEP1 on grain yield and grain quality in the background of two japonica rice (Oryza sativa) cultivars. Plant Breed. 2020, 139, 608–617. [Google Scholar] [CrossRef]
- Soroka, M.; Wasowicz, B.; Rymaszewska, A. Loop-mediated isothermal amplification (LAMP): The better sibling of PCR? Cells 2021, 10, 1931. [Google Scholar] [CrossRef] [PubMed]
- Becherer, L.; Borst, N.; Bakheit, M.; Frischmann, S.; Zengerle, R.; von Stetten, F. Loop-mediated isothermal amplification (LAMP)–review and classification of methods for sequence-specific detection. Anal. Methods 2020, 12, 717–746. [Google Scholar] [CrossRef]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Cuadros, J.; Pérez-Tanoira, R.; Prieto-Pérez, L.; Martin-Martin, I.; Berzosa, P.; González, V.; Tisiano, G.; Balcha, S.; Ramos, J.M.; Górgolas, M. Field evaluation of malaria microscopy, rapid malaria tests and loop-mediated isothermal amplification in a rural hospital in south western Ethiopia. PLoS ONE 2015, 10, e0142842. [Google Scholar] [CrossRef] [PubMed]
- Narushima, J.; Kimata, S.; Soga, K.; Sugano, Y.; Kishine, M.; Takabatake, R.; Mano, J.; Kitta, K.; Kanamaru, S.; Shirakawa, N.; et al. Rapid DNA template preparation directly from a rice sample without purification for loop-mediated isothermal amplification (LAMP) of rice genes. Biosci. Biotechnol. Biochem. 2019, 84, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Prasannakumar, M.K.; Parivallal, B.P.; Manjunatha, C.; Pramesh, D.; Narayan, K.S.; Venkatesh, G.; Banakar, S.N.; Mahesh, H.B.; Vemanna, R.S.; Rangaswamy, K.T. Rapid genotyping of bacterial leaf blight resistant genes of rice using loop-mediated isothermal amplification assay. Mol. Biol. Rep. 2021, 48, 467–474. [Google Scholar] [CrossRef]
- Kishine, M.; Okunishi, T. Rapid identification of rice cultivar “Koshihikari” using loop-mediated isothermal amplification (LAMP). J. Jpn. Soc. Food Sci. Technol. 2011, 58, 591–596. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Report. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Kawahara, Y.; Bastide, M.d.l.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Goto, M.; Honda, E.; Ogura, A.; Nomoto, A.; Hanaki, K.-I. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 2009, 46, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Han, R.; Wu, K.; Zhang, J.; Ye, Y.; Wang, S.; Chen, J.; Pan, Y.; Li, Q.; Xu, X.; et al. G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 2018, 9, 852. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yu, Y.; Ma, Y.; Gao, Q.; Cao, Y.; Chen, Z.; Ma, B.; Qi, M.; Li, Y.; Zhao, X.; et al. Sequencing and de novo assembly of a near complete indica rice genome. Nat. Commun. 2017, 8, 15324. [Google Scholar] [CrossRef] [PubMed]
- Babu, N.N.; Krishnan, S.G.; Vinod, K.K.; Krishnamurthy, S.L.; Singh, V.K.; Singh, M.P.; Singh, R.; Ellur, R.K.; Rai, V.; Bollinedi, H.; et al. Marker aided incorporation of Saltol, a major QTL associated with seedling stage salt tolerance, into Oryza sativa ‘Pusa Basmati 1121’. Front. Plant Sci. 2017, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Berdrejo, V.J.; Magulama, E.E. Efficiency of five emasculation techniques in rice. USM CA Res. J. 1994, 5, 9–12. [Google Scholar]
- Plant Genetics and Breeding Program, Biology Department, Wuhan University. Development and Flowering of Rice Panicles. In Panicle Development, Flowering and Fruiting in Rice; Hubei People’s Publishing House: Wuhan, China, 1975; pp. 42–45. [Google Scholar]
- Srividya, A.; Maiti, B.; Chakraborty, A.; Chakraborty, G. Loop mediated isothermal amplification: A promising tool for screening genetic mutations. Mol. Diagn. Ther. 2019, 23, 723–733. [Google Scholar] [CrossRef]
- Gouda, G.; Gupta, M.K.; Donde, R.; Mohapatra, T.; Vadde, R.; Behera, L. Marker-assisted selection for grain number and yield-related traits of rice (Oryza sativa L.). Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2020, 26, 885–898. [Google Scholar] [CrossRef]
- Muhammad Gul, A.; Hameed, G. Application techniques of molecular marker and achievement of marker assisted selection (MAS) in three major crops rice, wheat and maize. Int. J. Res. Appl. Sci. Biotechnol. 2021, 8, 82–93. [Google Scholar] [CrossRef]
- Gu, Z.; Gong, J.; Zhu, Z.; Li, Z.; Feng, Q.; Wang, C.; Zhao, Y.; Zhan, Q.; Zhou, C.; Wang, A.; et al. Structure and function of rice hybrid genomes reveal genetic basis and optimal performance of heterosis. Nat. Genet. 2023, 55, 1745–1756. [Google Scholar] [CrossRef]
- Wei, S.; Jiang, J.; Liu, W.; Li, X.; Luo, J. Decade of crop science:Funding, outcomes and future prospects of national natural science foundation of China. Bull. Natl. Nat. Sci. Found. China 2022, 36, 972–981. [Google Scholar] [CrossRef]
- Amiteye, S. Basic concepts and methodologies of DNA marker systems in plant molecular breeding. Heliyon 2021, 7, e08093. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Chen, S.; Chen, L.; Sun, K.; Huang, C.; Zhou, D.; Huang, Y.; Wang, J.; Liu, Y.; Wang, H.; et al. Development of a core SNP arrays based on the KASP method for molecular breeding of rice. Rice 2019, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Ba, Y.; Song, Y.; Cui, J.; Zhang, X.; Zhang, D.; Yuan, Z.; Yang, L. Rapid and sensitive screening and identification of CRISPR/Cas9 edited rice plants using quantitative real-time PCR coupled with high resolution melting analysis. Food Control 2020, 112, 107088. [Google Scholar] [CrossRef]
- Glökler, J.; Lim, T.S.; Ida, J.; Frohme, M. Isothermal amplifications—A comprehensive review on current methods. Crit. Rev. Biochem. Mol. 2021, 56, 543–586. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.; Johnstone, C.; Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991, 19, 1349. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Zhao, X.; Pittelkow, C.M.; Fan, M.; Zhang, X.; Yan, X. Optimal nitrogen rate strategy for sustainable rice production in China. Nature 2023, 615, 73–79. [Google Scholar] [CrossRef]
- Zhang, Q. Strategies for developing Green Super Rice. Proc. Natl. Acad. Sci. USA 2007, 104, 16402–16409. [Google Scholar] [CrossRef]
- Yu, S.; Ali, J.; Zhou, S.; Ren, G.; Xie, H.; Xu, J.; Yu, X.; Zhou, F.; Peng, S.; Ma, L.; et al. From Green Super Rice to green agriculture: Reaping the promise of functional genomics research. Mol. Plant 2021, 15, 9–26. [Google Scholar] [CrossRef]
- Li, P.; Chang, T.; Chang, S.; Ouyang, X.; Qu, M.; Song, Q.; Xiao, L.; Xia, S.; Deng, Q.; Zhu, X.-G. Systems model-guided rice yield improvements based on genes controlling source, sink, and flow. J. Integr. Plant Biol. 2018, 60, 1154–1180. [Google Scholar] [CrossRef]
- Singh, B.K.; Delgado-Baquerizo, M.; Egidi, E.; Guirado, E.; Leach, J.E.; Liu, H.; Trivedi, P. Climate change impacts on plant pathogens, food security and paths forward. Nat. Rev. Microbiol. 2023, 21, 640–656. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Feng, T.; Zhang, C.; Zhang, F.; Li, H.; Chen, Y.; Liang, L.; Zhang, C.; Zeng, W.; Liu, E.; et al. Genetic dissection of salt tolerance and yield traits of geng (japonica) rice by selective subspecific introgression. Curr. Issues Mol. Biol. 2023, 45, 4796–4813. [Google Scholar] [CrossRef] [PubMed]
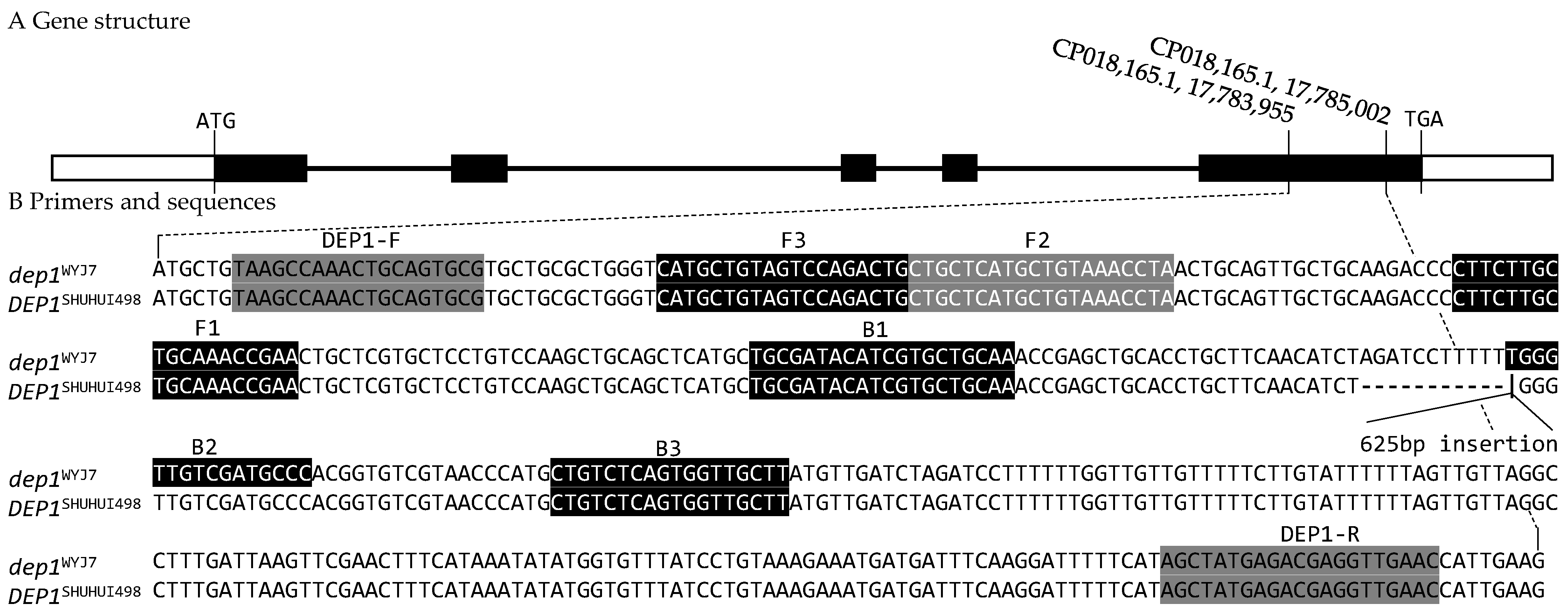


| Primers | Sequences (5′-3′) | Usage | Product Size |
|---|---|---|---|
| DEP1-F | TAAGCCAAACTGCAGTGCG | Forward Primer for InDel Marker | dep1 (409 bp), DEP1 (1034 bp) |
| DEP1-R | GTTCAACCTCGTCTCATAGCT | Reverse Primer for InDel Marker | |
| F3 | CATGCTGTAGTCCAGACTG | Forward Outer Primer for LAMP Marker | |
| B3 | AAGCAACCACTGAGACAG | Backward Outer Primer for LAMP Marker | |
| FIP (F1C-F2) | TTCGGTTTGCAGCAAGAAGG-CTGCTCATGCTGTAAACCTA | Forward Inner Primer for LAMP Marker | |
| BIP (B1C-B2) | TGCGATACATCGTGCTGCAA-GGGCATCGACAACCCA | Backward Inner Primer for LAMP Marker | |
| InDel-E5-F [17] | TCCAGGGATGTAATCATCTTTGTT | Forward Primer for InDel Marker InDel-E5 | dep1 (733 bp), DEP1 (1357 bp) |
| InDel-E5-R [17] | GGCTCCATATCTTCACGGTCTA | Forward Primer for InDel Marker InDel-E5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Y.; Chen, X.; Xu, P.; Wang, Y.; Wu, X.; Wu, K.; Fu, X.; Chin, Y.; Liao, Y. Rapid Visual Detection of Elite Erect Panicle Dense and Erect Panicle 1 Allele for Marker-Assisted Improvement in Rice (Oryza sativa L.) Using the Loop-Mediated Isothermal Amplification Method. Curr. Issues Mol. Biol. 2024, 46, 498-512. https://doi.org/10.3390/cimb46010032
Tian Y, Chen X, Xu P, Wang Y, Wu X, Wu K, Fu X, Chin Y, Liao Y. Rapid Visual Detection of Elite Erect Panicle Dense and Erect Panicle 1 Allele for Marker-Assisted Improvement in Rice (Oryza sativa L.) Using the Loop-Mediated Isothermal Amplification Method. Current Issues in Molecular Biology. 2024; 46(1):498-512. https://doi.org/10.3390/cimb46010032
Chicago/Turabian StyleTian, Yonghang, Xiyi Chen, Peizhou Xu, Yuping Wang, Xianjun Wu, Kun Wu, Xiangdong Fu, Yaoxian Chin, and Yongxiang Liao. 2024. "Rapid Visual Detection of Elite Erect Panicle Dense and Erect Panicle 1 Allele for Marker-Assisted Improvement in Rice (Oryza sativa L.) Using the Loop-Mediated Isothermal Amplification Method" Current Issues in Molecular Biology 46, no. 1: 498-512. https://doi.org/10.3390/cimb46010032
APA StyleTian, Y., Chen, X., Xu, P., Wang, Y., Wu, X., Wu, K., Fu, X., Chin, Y., & Liao, Y. (2024). Rapid Visual Detection of Elite Erect Panicle Dense and Erect Panicle 1 Allele for Marker-Assisted Improvement in Rice (Oryza sativa L.) Using the Loop-Mediated Isothermal Amplification Method. Current Issues in Molecular Biology, 46(1), 498-512. https://doi.org/10.3390/cimb46010032






