Anti-Staphylococcal, Anti-Candida, and Free-Radical Scavenging Potential of Soil Fungal Metabolites: A Study Supported by Phenolic Characterization and Molecular Docking Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Fungal Isolation
2.2. Identification of Fungal Isolates
2.3. Fermentation and Extraction of Fungal Metabolites
2.4. Antimicrobial Susceptibility Testing
2.4.1. Test Microorganisms
2.4.2. Agar Well-Diffusion Method
2.4.3. Determination of Minimum Inhibitory Concentration
2.5. Free-Radical-Scavenging Assay
2.6. Brine-Shrimp Lethality Assay
2.7. Phylogenetic Analysis of the Most Potent Isolates
2.7.1. Isolation of Genomic DNA
2.7.2. PCR Amplification and Nucleotide Sequence Analysis
2.8. Determination of Total Phenolic and Flavonoid Content
2.9. High-Performance Liquid Chromatography Analysis
2.10. Molecular Docking Simulations with Target Proteins
2.11. Data Analysis
3. Results
3.1. Identification of Fungal Isolates
3.2. Antimicrobial Activity
Determination of the Minimum Inhibitory Concentration (MIC) of Extracts
3.3. Antioxidative and Cytotoxic Activities of Extracts
3.4. Molecular Identification of the of Most Potent Isolates
3.5. Evaluation of Total Phenolics and Flavonoids
3.6. Flavonoid and Phenolic HPLC Profile of Extracts
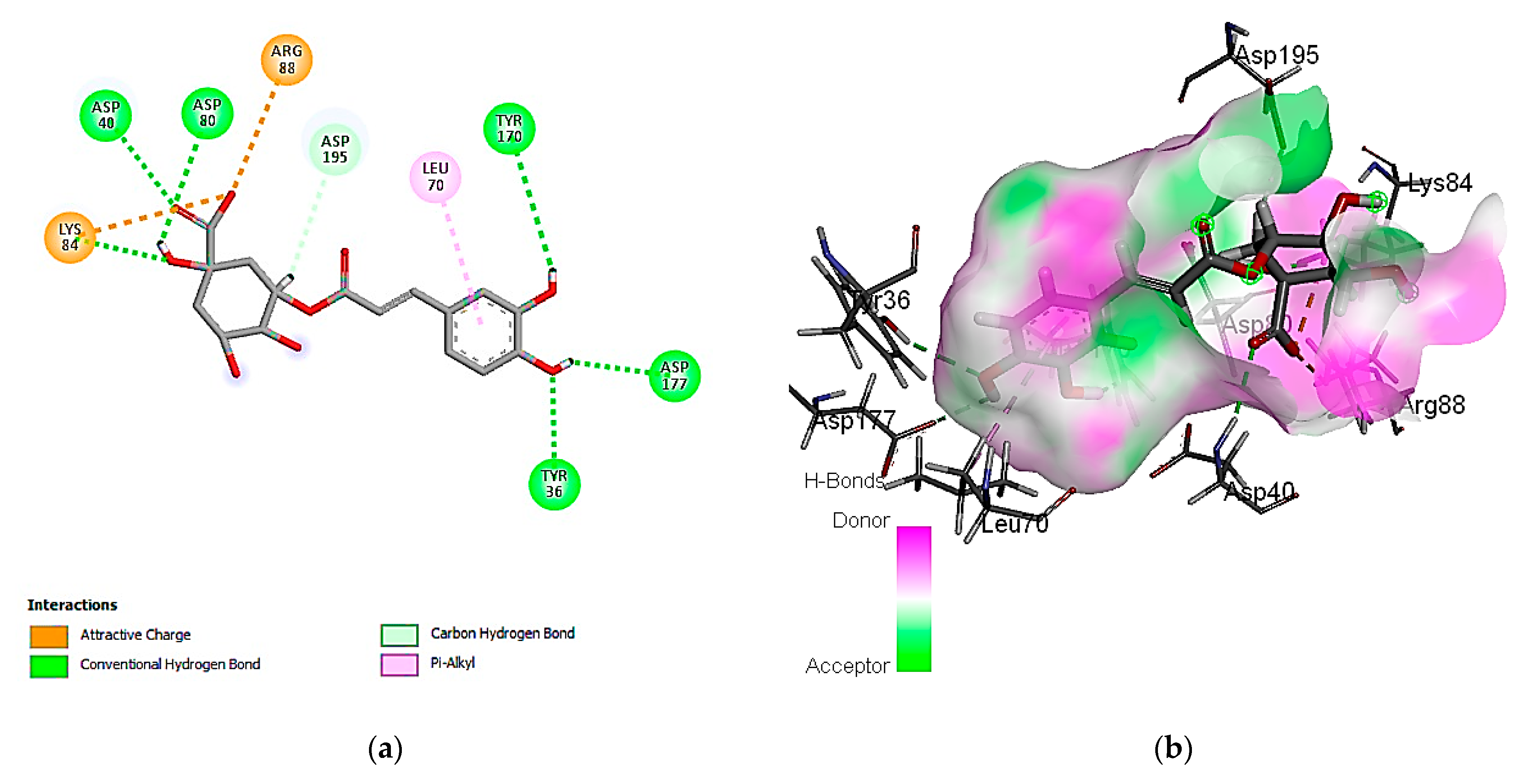
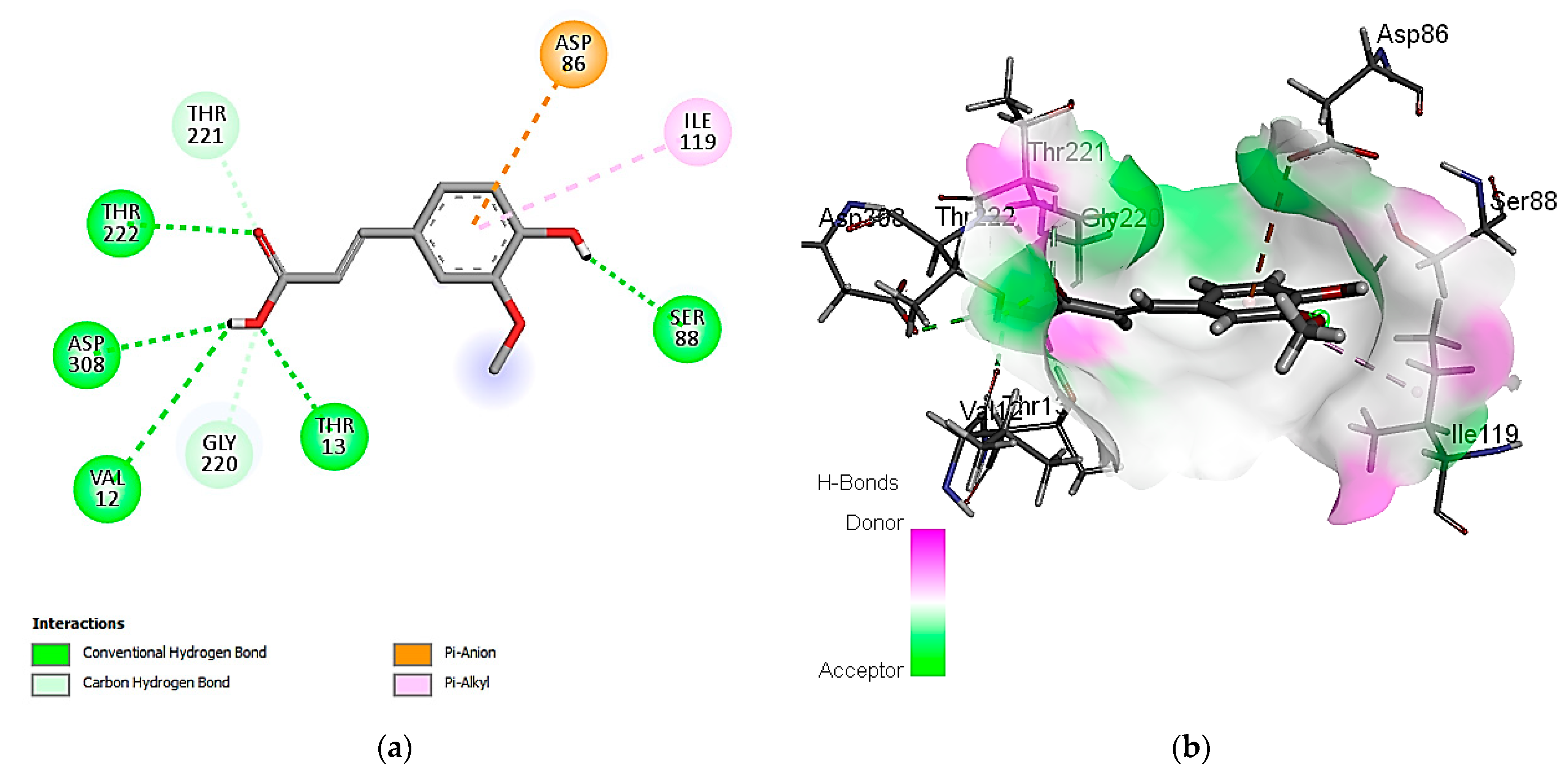
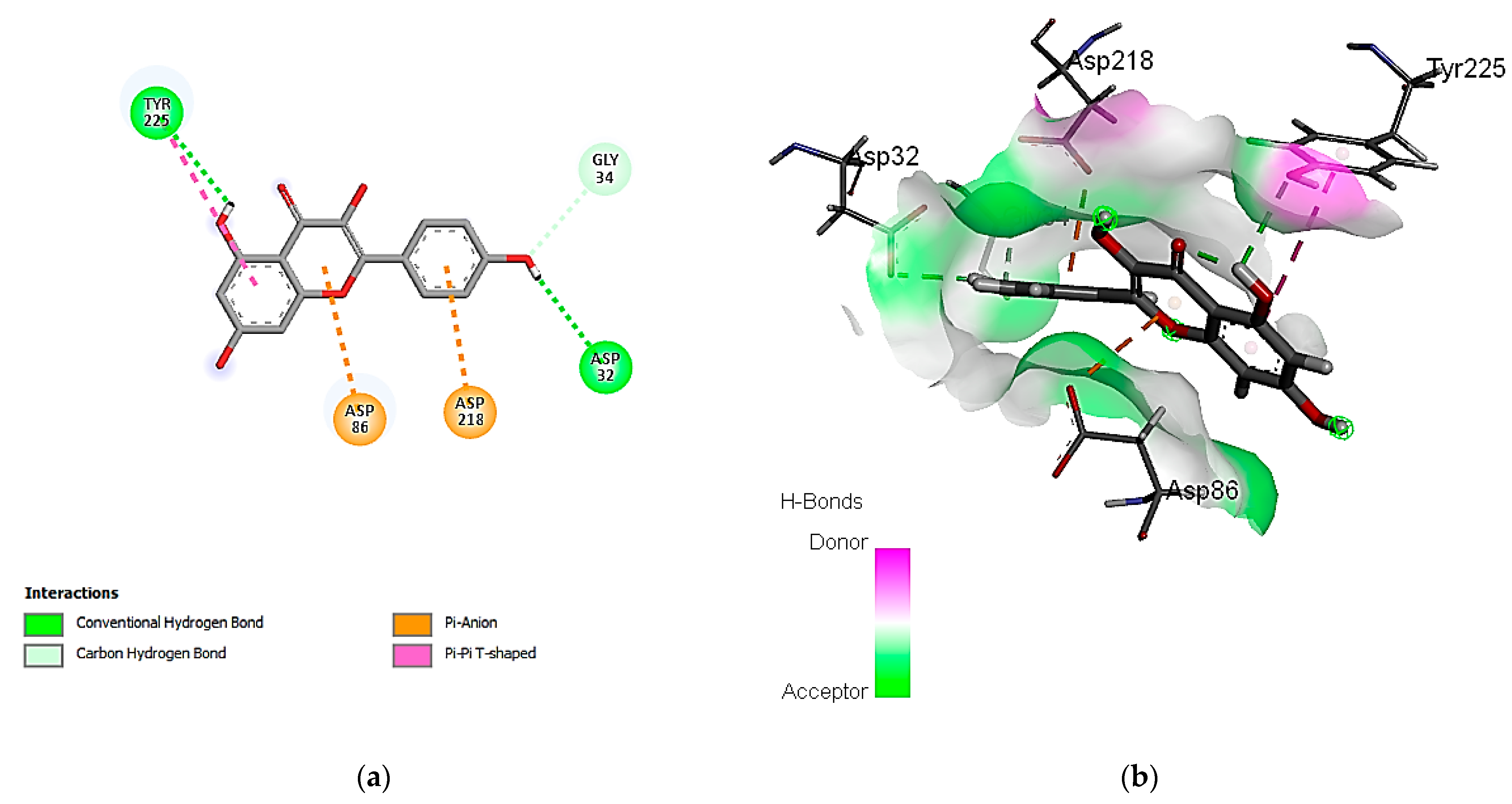
3.7. In Silico Molecular Docking of Identified Compounds
3.7.1. Molecular Docking Simulation against S. aureus Tyrosyl-tRNA Synthetase
3.7.2. Molecular Docking Simulation with the C. albicans Secreted Aspartic Protease 2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mohammed, A.E.; Sonbol, H.; Alwakeel, S.S.; Alotaibi, M.O.; Alotaibi, S.; Alothman, N.; Suliman, R.S.; Ahmedah, H.T.; Ali, R. Investigation of biological activity of soil fungal extracts and LC/MS-QTOF based metabolite profiling. Sci. Rep. 2021, 11, 4760. [Google Scholar] [CrossRef] [PubMed]
- Higginbotham, S.J.; Arnold, A.E.; Ibanez, A.; Spadafora, C.; Coley, P.D.; Kursar, T.A. Bioactivity of fungal endophytes as a function of endophyte taxonomy and the taxonomy and distribution of their host plants. PLoS ONE 2013, 8, e73192. [Google Scholar] [CrossRef] [PubMed]
- Giavasis, I. Bioactive fungal polysaccharides as potential functional ingredients in food and nutraceuticals. Curr. Opin. Biotechnol. 2014, 26, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Al Mousa, A.A.; Abo-Dahab, N.F.; Hassane, A.M.A.; Gomaa, A.F.; Aljuriss, J.A.; Dahmash, N.D. Harnessing Mucor spp. for xylanase production: Statistical optimization in submerged fermentation using agro-industrial wastes. BioMed Res. Int. 2022, 2022, 3816010. [Google Scholar] [CrossRef] [PubMed]
- Al Mousa, A.A.; Hassane, A.M.A.; Gomaa, A.F.; Aljuriss, J.A.; Dahmash, N.D.; Abo-Dahab, N.F. Response-surface statistical optimization of submerged fermentation for pectinase and cellulase production by Mucor circinelloides and M. hiemalis. Fermentation 2022, 8, 205. [Google Scholar] [CrossRef]
- Mohamed, H.; Awad, M.F.; Shah, A.M.; Nazir, Y.; Naz, T.; Hassane, A.; Nosheen, S.; Song, Y. Evaluation of different standard amino acids to enhance the biomass, lipid, fatty acid, and γ -linolenic acid production in Rhizomucor pusillus and Mucor circinelloides. Front. Nutr. 2022, 9, 876817. [Google Scholar] [CrossRef]
- Rajamanikyam, M.; Vadlapudi, V.; Amanchy, R.; Upadhyayula, S.M. Endophytic fungi as novel resources of natural therapeutics. Braz. Arch. Biol. Technol. 2017, 60, e17160542. [Google Scholar] [CrossRef]
- Pimentel, M.R.; Molina, G.; Dionisio, A.P.; Maróstica, M.R.; Pastore, G.M. Use of endophytes to obtain bioactive compounds and their application in biotransformation process. Biotechnol. Res. Int. 2011, 2011, 576286. [Google Scholar] [CrossRef]
- Al Mousa, A.A.; Abouelela, M.E.; Hassane, A.M.A.; Al-Khattaf, F.S.; Hatamleh, A.A.; Alabdulhadi, H.S.; Dahmash, N.D.; Abo-Dahab, N.F. Cytotoxic potential of Alternaria tenuissima AUMC14342 mycoendophyte extract: A study combined with LC-MS/MS metabolic profiling and molecular docking simulation. Curr. Issues Mol. Biol. 2022, 44, 5067–5085. [Google Scholar] [CrossRef]
- Hassane, A.M.A.; Taha, T.M.; Awad, M.F.; Mohamed, H.; Melebari, M. Radical scavenging potency, HPLC profiling and phylogenetic analysis of endophytic fungi isolated from selected medicinal plants of Saudi Arabia. Electron. J. Biotechnol. 2022, 58, 37–45. [Google Scholar] [CrossRef]
- Hassane, A.M.A.; Hussien, S.M.; Abouelela, M.E.; Taha, T.M.; Awad, M.F.; Mohamed, H.; Hassan, M.M.; Hassan, M.H.A.; Abo-Dahab, N.F.; El-Shanawany, A.A. In vitro and in silico antioxidant efficiency of bio-potent secondary metabolites from different taxa of black seed-producing plants and their derived mycoendophytes. Front. Bioeng. Biotechnol. 2022, 10, 930161. [Google Scholar] [CrossRef] [PubMed]
- Alkhulaifi, M.M.; Awaad, A.S.; AL-Mudhayyif, H.A.; Alothman, M.R.; Alqasoumi, S.I.; Zain, S.M. Evaluation of antimicrobial activity of secondary metabolites of fungi isolated from Sultanate Oman soil. Saudi Pharm. J. 2019, 27, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahem, M.M.M.; Hassane, A.M.A.; Abouelela, M.E.; Abo-Dahab, N.F. Comparative bioactivity and metabolites produced by fungal co-culture system against myco-phytopathogens. J. Environ. Stud. 2023, 31, 1–15. [Google Scholar] [CrossRef]
- Ujam, N.T.; Ajaghaku, D.L.; Okoye, F.B.C.; Esimone, C.O. Antioxidant and immunosuppressive activities of extracts of endophytic fungi isolated from Psidium guajava and Newbouldia laevis. Phytomed. Plus 2021, 1, 100028. [Google Scholar] [CrossRef]
- Tawfike, A.F.; Romli, M.; Clements, C.; Abbott, G.; Young, L.; Schumacher, M.; Diederich, M.; Farag, M.; Edrada-Ebel, R. Isolation of anticancer and anti-trypanosome secondary metabolites from the endophytic fungus Aspergillus flocculus via bioactivity guided isolation and MS based metabolomics. J. Chromatogr. B 2019, 1106–1107, 71–83. [Google Scholar] [CrossRef] [PubMed]
- El-Kassem, L.A.; Hawas, U.W.; El-Souda, S.; Ahmed, E.F.; El-Khateeb, W.; Fayad, W. Anti-HCV protease potential of endophytic fungi and cytotoxic activity. Biocatal. Agric. Biotechnol. 2019, 19, 101170. [Google Scholar] [CrossRef]
- Al-Fakih, A.A.; Almaqtri, W.Q.A. Overview on antibacterial metabolites from terrestrial Aspergillus spp. Mycology 2019, 10, 191–209. [Google Scholar] [CrossRef]
- Saber, S.M.; Youssef, M.S.; Arafa, R.F.; Hassane, A.M.A. Mycotoxins production by Aspergillus ostianus Wehmer and using phytochemicals as control agent. J. Sci. Eng. Res. 2016, 3, 198–213. [Google Scholar]
- Hassane, A.M.A.; El-Shanawany, A.A.; Abo-Dahab, N.F.; Abdel-Hadi, A.M.; Abdul-Raouf, U.M.; Mwanza, M. Cultural and analytical assays for aflatoxin B production by Aspergillus flavus isolates. J. Nat. Prod. Chem. 2017, 1, 17–23. [Google Scholar] [CrossRef]
- Zhong, J.J.; Xiao, J.H. Secondary metabolites from higher fungi: Discovery, bioactivity, and bioproduction. Adv. Biochem. Eng. Biotechnol. 2009, 113, 79–150. [Google Scholar] [CrossRef]
- Oliver, S.; Vittorio, O.; Cirillo, G.; Boyer, C. Enhancing the therapeutic effects of polyphenols with macromolecules. Polym. Chem. 2016, 7, 1529–1544. [Google Scholar] [CrossRef]
- Dantas, S.B.S.; Moraes, G.K.A.; Araujo, A.R.; Chapla, V.M. Phenolic compounds and bioactive extract produced by endophytic fungus Coriolopsis rigida. Nat. Prod. Res. 2023, 37, 2037–2042. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Maldonado, A.F.; Schieber, A.; Gänzle, M.G. Structure–function relationships of the antibacterial activity of phenolic acids and their metabolism by lactic acid bacteria. J. Appl. Microbiol. 2011, 111, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Keman, D.; Soyer, F. Antibiotic-resistant Staphylococcus aureus does not develop resistance to vanillic acid and 2-hydroxycinnamic acid after continuous exposure in vitro. ACS Omega 2019, 4, 15393–15400. [Google Scholar] [CrossRef] [PubMed]
- Ecevit, K.; Barros, A.A.; Silva, J.M.; Reis, R.L. Preventing microbial infections with natural phenolic compounds. Future Pharmacol. 2022, 2, 460–498. [Google Scholar] [CrossRef]
- Wu, S.-C.; Liu, F.; Zhu, K.; Shen, J.-Z. Natural products that target virulence factors in antibiotic-resistant Staphylococcus aureus. J. Agric. Food Chem. 2019, 67, 13195–13211. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Johnson, A.D. Candida albicans biofilms and human disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef]
- Patil, S.; Rao, R.S.; Majumdar, B.; Anil, S. Clinical appearance of oral candida infection and therapeutic strategies. Front. Microbiol. 2015, 6, 1391. [Google Scholar] [CrossRef]
- Silva, S.; Negri, M.; Henriques, M.; Oliveira, R.; Williams, D.W.; Azeredo, J. Candida glabrata, Candida parapsilosis and Candida tropicalis: Biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbial. Rev. 2012, 36, 288–305. [Google Scholar] [CrossRef]
- Ng, K.P.; Kuan, C.S.; Kaur, H.; Na, S.L.; Atiya, N.; Velayuthan, R.D. Candida species epidemiology 2000-2013: A laboratory-based report. Trop. Med. Int. Health 2015, 20, 1447–1453. [Google Scholar] [CrossRef]
- De Bernardis, F.; Graziani, S.; Tirelli, F.; Antonopoulou, S. Candida vaginitis: Virulence, host response and vaccine prospects. Med. Mycol. 2018, 56, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Meenambiga, S.S.; Venkataraghavan, R.; Biswal, A. In silico analysis of plant phytochemicals against secreted aspartic proteinase enzyme of Candida albicans. J. Appl. Pharm. Sci. 2018, 8, 140–150. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and mechanisms of antifungal resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- Harriott, M.M.; Noverr, M.C. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: Effects on antimicrobial resistance. Antimicrob. Agents Chemother. 2009, 53, 3914–3922. [Google Scholar] [CrossRef] [PubMed]
- Harriott, M.M.; Noverr, M.C. Ability of Candida albicans mutants to induce Staphylococcus aureus vancomycin resistance during polymicrobial biofilm formation. Antimicrob. Agents Chemother. 2010, 54, 3746–3755. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Stark, L. Staphylococcus aureus: Aspects of Pathogenesis and Molecular Epidemiology. Ph.D. Thesis, Linköping University Electronic Press, Linköping, Sweden, 2013; p. 1371. [Google Scholar]
- Saxena, S.; Gomber, C. Surmounting antimicrobial resistance in the Millennium Superbug: Staphylococcus aureus. Open Med. 2010, 5, 12–29. [Google Scholar] [CrossRef]
- Klotz, S.A.; Chasin, B.S.; Powell, B.; Gaur, N.K.; Lipke, P.N. Polymicrobial bloodstream infections involving Candida species: Analysis of patients and review of the literature. Diagn. Microbiol. Infect. Dis. 2007, 59, 401–406. [Google Scholar] [CrossRef]
- Carolus, H.; Van Dyck, K.; Van Dijck, P. Candida albicans and Staphylococcus species: A threatening twosome. Front. Microbiol. 2019, 10, 2162. [Google Scholar] [CrossRef]
- Budzynska, A.; Rozalska, S.; Sadowska, B.; Rozalska, B. Candida albicans/Staphylococcus aureus dual-species biofilm as a target for the combination of essential oils and fluconazole or mupirocin. Mycopathologia 2017, 182, 989–995. [Google Scholar] [CrossRef]
- Sanguinetti, M.; Posteraro, B.; Lass-Florl, C. Antifungal drug resistance among Candida species: Mechanisms and clinical impact. Mycoses 2015, 58, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Guignard, B.; Entenza, J.M.; Moreillon, P. β-Lactams against methicillin-resistant Staphylococcus aureus. Curr. Opin. Pharmacol. 2005, 5, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Shevelev, A.B.; La Porta, N.; Isakova, E.P.; Martens, S.; Biryukova, Y.K.; Belous, A.S.; Sivokhin, D.A.; Trubnikova, E.V.; Zylkova, M.V.; Belyakova, A.V.; et al. In vivo antimicrobial and wound-healing activity of resveratrol, dihydroquercetin, and dihydromyricetin against Staphylococcus aureus, Pseudomonas aeruginosa, and Candida albicans. Pathogens 2020, 9, 296. [Google Scholar] [CrossRef] [PubMed]
- Schaller, M.; Borelli, C.; Korting, H.C.; Hube, B. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 2005, 48, 365–377. [Google Scholar] [CrossRef]
- Selmecki, A.; Forche, A.; Berman, J. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot. Cell 2010, 9, 991–1008. [Google Scholar] [CrossRef]
- Naglik, J.; Albrecht, A.; Bader, O.; Hube, B. Candida albicans proteinases and host/pathogen interactions. Cell. Microbiol. 2004, 6, 915–926. [Google Scholar] [CrossRef]
- Talapko, J.; Juzbašić, M.; Matijević, T.; Pustijanac, E.; Bekić, S.; Kotris, I.; Škrlec, I. Candida albicans-the virulence factors and clinical manifestations of infection. J. Fungi 2021, 7, 79. [Google Scholar] [CrossRef]
- Mseddi, K.; Alimi, F.; Noumi, E.; Veettil, V.N.; Deshpande, S.; Adnan, M.; Hamdi, A.; Elkahoui, S.; Ahmed, A.; Kadri, A.; et al. Thymus musilii Velen. as a promising source of potent bioactive compounds with its pharmacological properties: In vitro and in silico analysis. Arab. J. Chem. 2020, 13, 6782–6801. [Google Scholar] [CrossRef]
- Pohlmann, J.; Brotz-Oesterhelt, H. New aminoacyltRNA synthetase inhibitors as antibacterial agents. Curr. Drug Targets Infect. Disord. 2004, 4, 261–272. [Google Scholar] [CrossRef]
- Farshadfar, C.; Mollica, A.; Rafii, F.; Noorbakhsh, A.; Nikzad, M.; Seyedi, S.H.; Abdi, F.; Verki, S.A.; Mirzaie, S. Novel potential inhibitor discovery against tyrosyl-tRNA synthetase from Staphylococcus aureus by virtual screening, molecular dynamics, MMPBSA and QMMM simulations. Mol. Simul. 2020, 46, 507–520. [Google Scholar] [CrossRef]
- Pisano, M.B.; Kumar, A.; Medda, R.; Gatto, G.; Pal, R.; Fais, A.; Era, B.; Cosentino, S.; Uriarte, E.; Santana, L.; et al. Antibacterial activity and molecular docking studies of a selected series of hydroxy-3-arylcoumarins. Molecules 2019, 24, 2815. [Google Scholar] [CrossRef] [PubMed]
- Vallavan, V.; Krishnasamy, G.; Zin, N.M.; Abdul Latif, M. A review on antistaphylococcal secondary metabolites from Basidiomycetes. Molecules 2020, 25, 5848. [Google Scholar] [CrossRef] [PubMed]
- Chopade, A.R.; Sayyad, F.J.; Pore, Y.V. Molecular docking studies of phytocompounds fom Phyllanthus species as potential chronic pain modulators. Sci. Pharm. 2015, 83, 243–267. [Google Scholar] [CrossRef] [PubMed]
- Othman, I.M.M.; Gad-Elkareem, M.A.M.; Anouar, E.; Aouadi, K.; Snoussi, M.; Kadri, A. New substituted pyrazolones and dipyrazolotriazines as promising tyrosyl-tRNA synthetase and peroxiredoxin-5 inhibitors: Design, synthesis, molecular docking and structure-activity relationship (SAR) analysis. Bioorg. Chem. 2021, 109, 104704. [Google Scholar] [CrossRef] [PubMed]
- Moubasher, A.H.; El-Naghy, M.A.; Abdel-Hafez, S.I.I. Studies on the fungus flora of three grains in Egypt. Mycopathol. Mycol. Appl. 1972, 47, 261–274. [Google Scholar] [CrossRef]
- Raper, K.B.; Fennell, D.I. The Genus Aspergillus; Williams & Wilkins Company: Baltimore, MD, USA, 1965. [Google Scholar]
- Moubasher, A.H. Soil Fungi in Qatar and Other Arab Countries; University of Qatar, Center of Scientific and Applied Research: Doha, Qatar, 1993; p. 566. [Google Scholar]
- Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of Soil Fungi, 2nd ed.; IHW Verlag: Eching, Germany, 2007; p. 672. [Google Scholar]
- Al Mousa, A.A.; Mohamed, H.; Hassane, A.M.; Abo-Dahab, N.F. Antimicrobial and cytotoxic potential of an endophytic fungus Alternaria tenuissima AUMC14342 isolated from Artemisia judaica L. growing in Saudi Arabia. J. King Saud Univ. Sci. 2021, 33, 101462. [Google Scholar] [CrossRef]
- Mohamed, H.; Hassane, A.; Atta, O.; Song, Y. Deep learning strategies for active secondary metabolites biosynthesis from fungi: Harnessing artificial manipulation and application. Biocatal. Agric. Biotechnol. 2021, 38, 102195. [Google Scholar] [CrossRef]
- Pretsch, A.; Nagl, M.; Schwendinger, K.; Kreiseder, B.; Wiederstein, M.; Pretsch, D.; Genov, M.; Hollaus, R.; Zinssmeister, D.; Debbab, A.; et al. Antimicrobial and anti-inflammatory activities of endophytic fungi Talaromyces wortmannii extracts against acne-inducing bacteria. PLoS ONE 2014, 9, e97929. [Google Scholar] [CrossRef]
- Baz, A.A.; Bakhiet, E.K.; Abdul-Raouf, U.; Abdelkhalek, A. Prevalence of enterotoxin genes (SEA to SEE) and antibacterial resistant pattern of Staphylococcus aureus isolated from clinical specimens in Assiut city of Egypt. Egypt. J. Med. Hum. Genet. 2021, 22, 84. [Google Scholar] [CrossRef]
- Mohamed, H.; Hassane, A.; Rawway, M.; El-Sayed, M.; Gomaa, A.; Abdul-Raouf, U.; Shah, A.M.; Abdelmotaal, H.; Song, Y. Antibacterial and cytotoxic potency of thermophilic Streptomyces werraensis MI-S.24-3 isolated from an Egyptian extreme environment. Arch. Microbiol. 2021, 203, 4961–4972. [Google Scholar] [CrossRef]
- Al Halteet, S.; Abdel-Hadi, A.; Hassan, M.; Awad, M. Prevalence and antifungal susceptibility profile of clinically relevant Candida species in postmenopausal women with diabetes. BioMed Res. Int. 2020, 2020, 7042490. [Google Scholar] [CrossRef] [PubMed]
- Jahangirian, H.; Haron, M.; Ismail, M.H.S.; Rafiee-Moghaddam, R.; Afsah-Hejri, L.; Abdollahi, Y.; Rezayi, M.; Vafaei, N. Well diffusion method for evaluation of antibacterial activity of copper phenyl fatty hydroxamate synthesized from canola and palm kernel oils. Dig. J. Nanomater. Biostructures 2013, 8, 1263–1270. [Google Scholar]
- Lall, N.; Henley-Smith, C.J.; De Canha, M.N.; Oosthuizen, C.B.; Berrington, D. Viability reagent, PrestoBlue, in comparison with other available reagents, utilized in cytotoxicity and antimicrobial assays. Inter. J. Microbiol. 2013, 2013, 420601. [Google Scholar] [CrossRef] [PubMed]
- John, C.N.; Abrantes, P.M.D.S.; Prusty, B.K.; Ablashi, D.V.; Africa, C.W.J. K21 compound, a potent antifungal agent: Implications for the treatment of fluconazole-resistant HIV-associated Candida species. Front. Microbiol. 2019, 10, 1021. [Google Scholar] [CrossRef] [PubMed]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Moshi, M.J.; Van den Beukel, C.J.; Hamza, O.J.M.; Mbwambo, Z.H.; Nondo, R.O.S.; Masimba, P.J.; Matee, M.I.; Kapingu, M.C.; Mikx, F.; Verweije, P.J. Brine shrimp toxicity evaluation of some Tanzanian plants used traditionally for the treatment of fungal infections. Afr. J. Tradit. Complement. Altern. Med. 2007, 4, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.M.; Farid, M.A.; Gaber, A. Rapid identification of Trichoderma koningiopsis and Trichoderma longibrachiatum using sequence characterized amplified region markers. Egypt. J. Biol. Pest Control 2019, 29, 13. [Google Scholar] [CrossRef]
- Mohamed, H.; El-Shanawany, A.; Shah, A.M.; Nazir, Y.; Naz, T.; Ullah, S.; Mustafa, K.; Song, Y. Comparative analysis of different isolated oleaginous Mucoromycota fungi for their γ-linolenic acid and carotenoid production. BioMed Res. Int. 2020, 2020, 3621543. [Google Scholar] [CrossRef]
- Suleria, H.A.; Butt, M.S.; Anjum, F.M.; Saeed, F.; Khalid, N. Onion: Nature protection against physiological threats. Crit. Rev. Food Sci. Nutr. 2015, 55, 50–66. [Google Scholar] [CrossRef]
- Quettier-Deleu, C.; Gressier, B.; Vasseur, J.; Dine, T.; Brunet, C.; Luyckx, M.; Cazin, M.; Cazin, J.; Bailleul, F.; Trotin, F. Phenolic compounds and antioxidant activities of buckwheat (Fagopyrum esculentum Moench) hulls and flour. J. Ethnopharm. 2002, 72, 35–42. [Google Scholar] [CrossRef]
- Akpotu, M.O.; Eze, P.M.; Abba, C.C.; Umeokoli, B.O.; Nwachukwu, C.U.; Okoye, F.B.C.; Esimone, C.O. Antimicrobial activities of secondary metabolites of endophytic fungi isolated from Catharanthus roseus. J. Health Sci. 2017, 7, 15–22. [Google Scholar] [CrossRef]
- Orabi, M.A.A.; Alshahrani, M.M.; Sayed, A.M.; Abouelela, M.E.; Shaaban, K.A.; Abdel-Sattar, E.-S. Identification of potential Leishmania N-myristoyltransferase inhibitors from Withania somnifera (L.) Dunal: A molecular docking and molecular dynamics investigation. Metabolites 2023, 13, 93. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Kader, N.S.; Moustafa, H.; El-Ansary, A.L.; Farghaly, A.M. Theoretical calculations for new coumarin Schiff base complexes as candidates for in vitro and in silico biological applications. Appl. Organomet. Chem. 2022, 36, e6840. [Google Scholar] [CrossRef]
- Bhairavi, V.A.; Vidya, S.L.; Sathishkumar, R. Identification of effective plant extracts against candidiasis: An in silico and in vitro approach. Future J. Pharm. Sci. 2023, 9, 38. [Google Scholar] [CrossRef]
- Sharma, S.; Sharma, A.; Gupta, U. Molecular docking studies on the anti-fungal activity of Allium sativum (garlic) against mucormycosis (black fungus) by BIOVIA discovery studio visualizer 21.1.0. Ann. Antivir. Antiretrovir. 2021, 5, 28–32. [Google Scholar]
- Chugh, A.; Sehgal, I.; Khurana, N.; Verma, K.; Rolta, R.; Vats, P.; Salaria, D.; Fadare, O.A.; Awofisayo, O.; Verma, A.; et al. Comparative docking studies of drugs and phytocompounds for emerging variants of SARS-CoV-2. 3 Biotech 2023, 13, 36. [Google Scholar] [CrossRef]
- Nisa, H.; Kamili, A.N.; Nawchoo, I.A.; Shafi, S.; Shameem, N.; Bandh, S.A. Fungal endophytes as prolific source of phytochemicals and other bioactive natural products: A review. Microb. Pathog. 2015, 82, 50–59. [Google Scholar] [CrossRef]
- Ibrahim, D.; Nurhaida; Hong, L.S. Anti-candidal activity of Aspergillus flavus IBRL-C8, an endophytic fungus isolated from Cassia siamea Lamk leaf. J. App. Pharm. Sci. 2018, 8, 83–87. [Google Scholar] [CrossRef]
- Nath, A.; Joshi, S. Anti-candidal effect of endophytic fungi isolated from Calotropis gigantean. Rev. Biol. Trop. 2017, 65, 1437–1447. [Google Scholar] [CrossRef][Green Version]
- Jawaid, K.; Shafique, M.; Versiani, A.; Muhammed, H.; Naz, S.A.; Jabeen, N. Antimicrobial potential of newly isolated Aspergillus terreus MK-1: An approach towards new antibiotics. J. Pak. Med. Assoc. 2019, 69, 18. [Google Scholar][Green Version]
- Phupiewkham, W.; Sirithorn, P.; Saksirirat, W.; Thammasirirak, S. Antibacterial agents from Trichoderma harzianum strain T9 against pathogenic bacteria. Chiang Mai J. Sci. 2015, 42, 304–316. [Google Scholar][Green Version]
- Bekoe, E.O.; Wiafe-Kwagyan, M.; Gaysi, J. Antibacterial activity of the metabolites of Aspergillus chevalieri and Trichoderma harzianum. Int. J. Pharm. Pharm. Sci. 2021, 13, 67–70. [Google Scholar] [CrossRef]
- Leelavathi, M.S.; Vani, L.; Reena, P. Antimicrobial activity of Trichoderma harzianum against bacteria and fungi. Int. J. Curr. Microbiol. App. Sci. 2014, 3, 96–103. [Google Scholar]
- Meenambiga, S.S.; Rajagopal, K. Antibiofilm activity and molecular docking studies of bioactive secondary metabolites from endophytic fungus Aspergillus nidulans on oral Candida albicans. J. Appl. Pharm. Sci. 2018, 8, 037–045. [Google Scholar] [CrossRef]
- Perumal, S.; Pillai, S.; Cai, L.W.; Mahmud, R.; Ramanathan, S. Determination of minimum inhibitory concentration of Euphorbia hirta (L) extracts by tetrazolium microplate assay. J. Nat. Prod. 2012, 5, 68–76. [Google Scholar]
- Chukwujekwu, J.C.; van Staden, J. In vitro antibacterial activity of Combretum edwardsii, Combretum krausii, and Maytenus nemorosa and their synergistic effects in combination with antibiotics. Front. Pharmacol. 2016, 7, 208. [Google Scholar] [CrossRef]
- Pauli, G.F.; Case, R.J.; Inui, T.; Wang, Y.; Cho, S.; Fischer, N.H.; Franzblau, S.G. New perspectives on natural products in TB drug research. Life Sci. 2005, 78, 485–494. [Google Scholar] [CrossRef]
- Valgas, C.; de Souza, S.M.; Smania, E.F.A.; Junior, A.S. Screening methods to determine antibacterial activity of natural products. Braz. J. Microbiol. 2007, 38, 369–380. [Google Scholar] [CrossRef]
- Dewi, R.T.; Tachibana, S.; Itoh, K.; Ilyas, M. Isolation of antioxidant compounds from Aspergillus terreus LSJ. Microbial Biochem. Technol. 2012, 4, 1. [Google Scholar] [CrossRef]
- Saleh, A.M.; El-Refai, H.A.; Hashem, A.M.; El-Menoufy, H.A.; Mansour, N.M.; El-Beih, A.A. Optimization studies and chemical investigations of Aspergillus terreus-18 showing antioxidant activity. Egypt. J. Chem. 2019, 62, 215–230. [Google Scholar] [CrossRef]
- Chandra, P.; Arora, D.S. Production of antioxidant bioactive phenolic compounds by solid-state fermentation on agro-residues using various fungi isolated from soil. Asian J. Biotechnol. 2016, 8, 8–15. [Google Scholar] [CrossRef]
- Qiu, M.; Xie, R.; Shi, Y.; Zhang, H.; Chen, H. Isolation and identification of two flavonoid-producing endophytic fungi from Ginkgo biloba L. Ann. Microbiol. 2010, 60, 143–150. [Google Scholar] [CrossRef]
- Goncalves, R.C.R.; Pombeiro-Sponchiado, S.R. Antioxidant activity of the melanin pigment extracted from Aspergillus nidulans. Biol. Pharm. Bull. 2005, 28, 1129–1131. [Google Scholar] [CrossRef] [PubMed]
- Al-Askar, A.A. Bioactive compounds produced by Trichoderma harzianum 1-SSR for controlling Fusarium verticillioides (Sacc.) Nirenberg and growth promotion of Sorghum vulgare. Egypt. J. Biol. Pest Control 2016, 26, 379–386. [Google Scholar]
- Tavares, D.G.; Barbosa, B.V.L.; Ferreira, R.L.; Duarte, W.F.; Cardoso, P.G. Antioxidant activity and phenolic compounds of the extract from pigment producing fungi isolated from Brazilian caves. Biocatal. Agric. Biotechnol. 2018, 16, 148–154. [Google Scholar] [CrossRef]
- Ahmed, N.S.; Al-Shamary, E.I. Optimization of phenolic compound production by local Aspergillus niger B1b isolate. IOP Conf. Ser.: Earth Environ. Sci. 2021, 761, 012119. [Google Scholar] [CrossRef]
- Barakat, K.M.; Gohar, Y.M. Antimicrobial agents produced by marine Aspergillus terreus var. africanus against some virulent fish pathogens. Indian J. Microbiol. 2012, 52, 366–372. [Google Scholar] [CrossRef]
- Mohamed, H.; Awad, M.F.; Shah, A.M.; Sadaqat, B.; Nazir, Y.; Naz, T.; Yang, W.; Song, Y. Coculturing of Mucor plumbeus and Bacillus subtilis bacterium as an efficient fermentation strategy to enhance fungal lipid and gamma-linolenic acid (GLA) production. Sci. Rep. 2022, 12, 13111. [Google Scholar] [CrossRef]
- Mazrou, Y.S.; Makhlouf, A.H.; Elbealy, E.R.; Salem, M.A.; Awad, M.F.; Hassan, M.M.; Ismail, M. Molecular characterization of phosphate solubilizing fungi Aspergillus niger and its correlation to sustainable agriculture. J. Environ. Biol. 2020, 41, 592–599. [Google Scholar] [CrossRef]
- Abdel-Wareth, M.T.A.; Ghareeb, M.A. Bioprospecting certain freshwater-derived fungi for phenolic compounds with special emphasis on antimicrobial and larvicidal activity of methyl gallate and p-coumaric acid. Egypt. J. Chem. 2018, 61, 773–784. [Google Scholar] [CrossRef]
- Abdel-Aty, A.M.; Barakat, A.Z.; Bassuiny, R.I.; Mohamed, S.A. Improved production of antioxidant-phenolic compounds and certain fungal phenolic-associated enzymes under solid-state fermentation of chia seeds with Trichoderma reesei: Response surface methodology-based optimization. J. Food Meas. Charact. 2022, 16, 3488–3500. [Google Scholar] [CrossRef]
- Xing, C.; Wu, J.; Xia, J.; Fan, S.; Yang, X. Steroids and anthraquinones from the deep-sea-derived fungus Aspergillus nidulans MCCC 3ABiochem. Syst. Ecol. 2019, 83, 103–105. [Google Scholar] [CrossRef]
- Kepa, M.; Miklasinska-Majdanik, M.; Wojtyczka, R.D.; Idzik, D.; Korzeniowski, K.; Smolen-Dzirba, J.; Wasik, T.J. Antimicrobial potential of caffeic acid against Staphylococcus aureus clinical strains. BioMed Res. Int. 2018, 2018, e7413504. [Google Scholar] [CrossRef] [PubMed]
- Imana, S.N.; Ningsih, E.G.; Tambunan, U.S.F. In silico identification of peptide as epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer treatment. Pak. J. Biol. Sci. 2020, 23, 567–574. [Google Scholar] [CrossRef][Green Version]
- Deepika, M.S.; Thangam, R.; Sakthidhasan, P.; Arun, S.; Sivasubramanian, S.; Thirumurugan, R. Combined effect of a natural flavonoid rutin from Citrus sinensis and conventional antibiotic gentamicin on Pseudomonas aeruginosa biofilm formation. Food Control 2018, 90, 282–294. [Google Scholar] [CrossRef]
- Bautista-Baños, S.; Hernández-Lauzardo, A.N.; Velázquez-del Valle, M.G.; Hernández-López, M.; Ait Barka, E.; Bosquez-Molina, E.; Wilson, C.L. Chitosan as a potential natural compound to control pre and postharvest diseases of horticultural commodities. Crop Prot. 2006, 25, 108–118. [Google Scholar] [CrossRef]
- Chew, J.; Peh, S.-C.; Sin Yeang, T. Non-microbial natural products that inhibit drug-resistant Staphylococcus aureus. In Staphylococcus aureus; Hemeg, H., Ozbak, H., Afrin, F., Eds.; IntechOpen: London, UK, 2019; pp. 1–30. [Google Scholar] [CrossRef]
- Campos, F.M.; Couto, J.A.; Figueiredo, A.R.; Tóth, I.V.; Rangel, A.O.S.S.; Hogg, T.A. Cell membrane damage induced by phenolic acids on wine lactic acid bacteria. Int. J. Food Microbiol. 2009, 135, 144–151. [Google Scholar] [CrossRef]
- Bouarab-Chibane, L.; Forquet, V.; Lantéri, P.; Clément, Y.; Léonard-Akkari, L.; Oulahal, N.; Degraeve, P.; Bordes, C. Antibacterial properties of polyphenols: Characterization and QSAR (Quantitative Structure–Activity Relationship) models. Front. Microbiol. 2019, 10, 829. [Google Scholar] [CrossRef]
- Kumar, N.; Goel, N. Phenolic acids: Natural versatile molecules with promising therapeutic applications. Biotechnol. Rep. 2019, 24, e00370. [Google Scholar] [CrossRef]
- Lou, Z.; Wang, H.; Rao, S.; Sun, J.; Ma, C.; Li, J. p-Coumaric acid kills bacteria through dual damage mechanisms. Food Control 2012, 25, 550–554. [Google Scholar] [CrossRef]
- Cushnie, T.P.T.; Lamb, A.J. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 2005, 26, 343–356. [Google Scholar] [CrossRef] [PubMed]
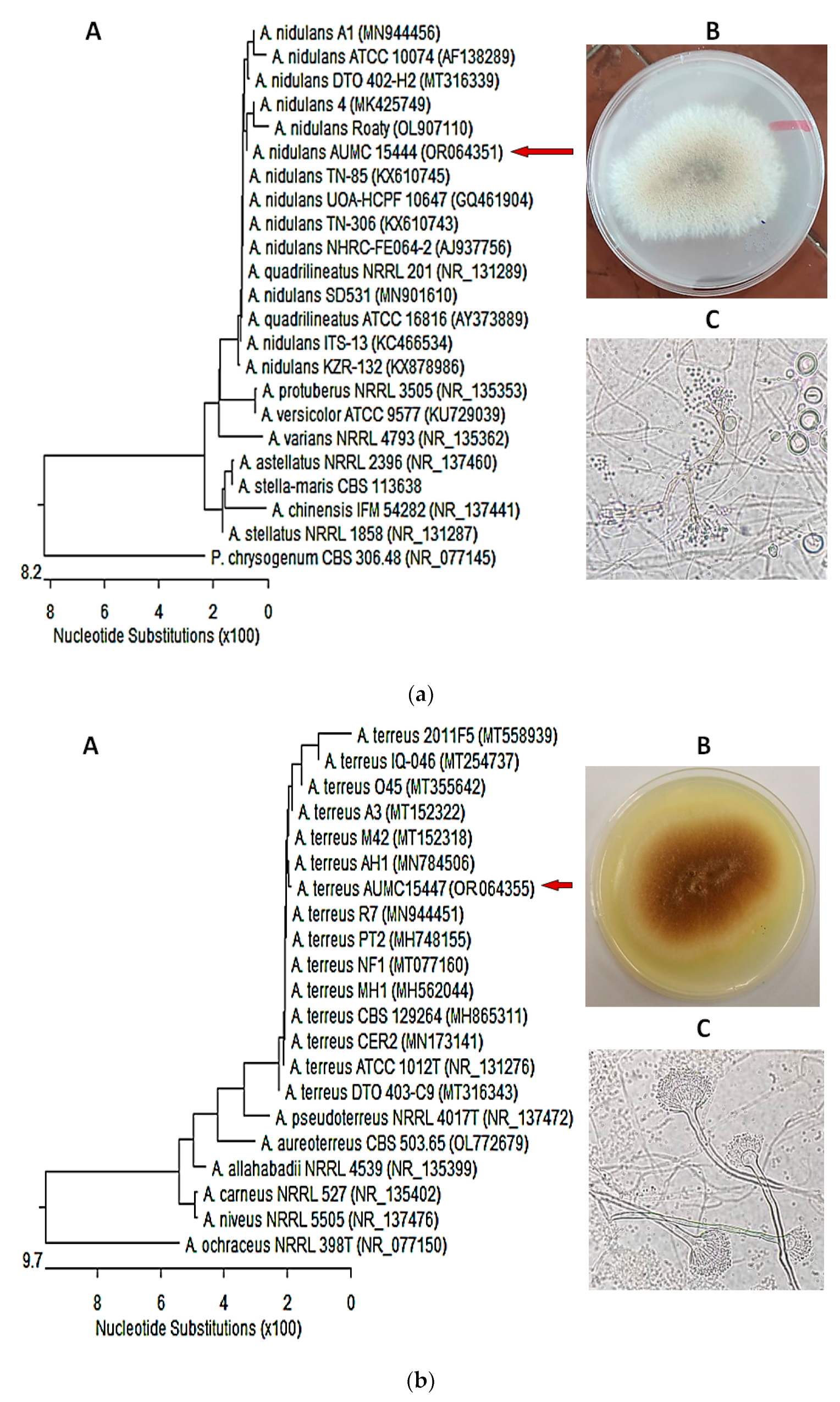



| AUMC No. | Fungal Isolate Identification | Fungal Phylum; Class; Order |
|---|---|---|
| 15440 | Trichoderma harzianum Rifai | Ascomycota; Sordariomycetes; Hypocreales |
| 15443 | T. harzianum Rifai | |
| 15441 | Aspergillus aureolatus Munt.-Cvetk. & Bata | Ascomycota; Eurotiomycetes; Eurotiales |
| 15446 | A. aureolatus Munt.-Cvetk. & Bata | |
| 15444 | A. nidulans (Eidam) Wint. | |
| 15447 | A. terreus Thom | |
| 15448 | A. terreus Thom | |
| 15445 | Penicillium crustosum Thom | |
| 15442 | P. novae-zeelandiae Van Beyma |
| Fungal Extract AUMC No. | Diameter of Inhibition Zone (mm) | ||||
|---|---|---|---|---|---|
| S. aureus ATCC 6538 | S. aureus AZHAR2 | S. aureus BLBM 112 | S. aureus AUHL 123 | S. aureus MLBM 117 | |
| 15440 | 19.67 ± 0.33 c | 11.67 ± 0.33 c | 13.67 ± 0.33 e | 17.33 ± 0.33 b,c | 16.33 ± 0.88 c,d |
| 15443 | 11.00 ± 0.58 d | 14.33 ± 0.67 b,c | 17.33 ± 0.67 b,c,d | 14.33 ± 0.33 c | 14.00 ± 0.58 d |
| 15441 | R | R | R | 10.00 ± 0.58 d | R |
| 15446 | 10.00 ± 0.00 d | 14.00 ± 0.58 b,c | 14.67 ± 0.33 d,e | 15.00 ± 0.00 c | 12.67 ± 0.67 e |
| 15444 | 23.33 ± 0.33 b | 16.33 ± 0.88 b | 18.00 ± 0.58 b | 19.67 ± 0.88 b | 20.33 ± 0.33 b |
| 15447 | 22.33 ± 0.67 b | 16.33 ± 0.33 b | 17.67 ± 0.67 b,c | 24.67 ± 0.33 a | 23.00 ± 0.58 a,b |
| 15448 | 17.67 ± 0.33 c | 14.67 ± 0.33 b | 15.00 ± 0.00 c,d,e | 19.33 ± 0.88 b | 15.33 ± 0.33 c,d |
| 15445 | 9.67 ± 0.33 d | 13.67 ± 0.88 b,c | 12.67 ± 0.33 e | 16.67 ± 0.88 b,c | 17.00 ± 0.00 c |
| 15442 | R | R | R | R | R |
| Chloramphenicol | 26.00 ± 0.58 a | 27.33 ± 0.33 a | 27.67 ± 1.20 a | 26.00 ± 1.53 a | 25.33 ± 0.88 a |
| Fungal Extract AUMC No. | Diameter of Inhibition Zone (mm) | ||||
|---|---|---|---|---|---|
| C. albicans ATCC 10231 | C. albicans MLBM 73 | C. albicans C-13 | C. glabrata C-4 | C. krusei C-30 | |
| 15440 | 20.33 ± 0.88 b | 19.33 ± 0.33 b | 13.67 ± 0.33 b | 13.00 ± 0.58 b | R |
| 15443 | 9.67 ± 0.33 d,e | R | R | R | R |
| 15441 | 9.00 ± 0.00 e | 9.33 ± 0.33 c | R | 10.00 ± 0.00 c | R |
| 15446 | 12.00 ± 0.00 c,d | R | R | R | R |
| 15444 | 20.67 ± 0.67 b | 19.67 ± 0.33 b | 22.33 ± 0.88 a | 20.00 ± 0.58 a | 24.00 ± 0.58 a |
| 15447 | 13.00 ± 0.00 c | R | 12.00 ± 0.58 b | R | R |
| 15448 | 13.33 ± 0.33 c | R | R | R | 11.00 ± 0.58 b |
| 15445 | R | R | R | R | R |
| 15442 | 12.33 ± 0.33 c,d | 11.00 ± 0.58 c | 10.67 ± 0.33 b | 10.00 ± 0.58 c | R |
| Fluconazole | 24.33 ± 1.20 a | 26.00 ± 0.58 a | 24.33 ± 0.88 a | 21.67 ± 0.88 a | 22.67 ± 0.33 a |
| Fungal Extract AUMC No. | MIC and MBC (mg/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ATCC 6538 | AZHAR2 | BLBM 112 | AUHL 123 | MLBM 117 | ||||||
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| 15440 | 3.125 | 6.25 | 50 | 100 | 25 | 50 | 3.125 | 6.25 | 3.125 | 6.25 |
| 15443 | 100 | R | 12.5 | 25 | 3.125 | 6.25 | 25 | 50 | 25 | 50 |
| 15441 | R | R | R | R | R | R | 100 | R | R | R |
| 15446 | 100 | R | 25 | 50 | 25 | 50 | 25 | 50 | 50 | 100 |
| 15444 | 0.78 | 1.56 | 25 | 50 | 12.5 | 25 | 0.78 | 1.56 | 0.78 | 1.56 |
| 15447 | 0.78 | 1.56 | 12.5 | 25 | 6.25 | 12.5 | 0.39 | 0.78 | 0.39 | 0.78 |
| 15448 | 6.25 | 12.5 | 12.5 | 25 | 12.5 | 25 | 3.125 | 6.25 | 12.5 | 25 |
| 15445 | 100 | R | 25 | 50 | 25 | 50 | 12.5 | 25 | 6.25 | 12.5 |
| 15442 | R | R | R | R | R | R | R | R | R | R |
| Chloramphenicol | 0.39 | 0.78 | 0.78 | 1.56 | 0.78 | 1.56 | 0.195 | 0.39 | 0.195 | 0.39 |
| Fungal Extract AUMC No. | MIC and MBC (mg/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ATCC 10231 | MLBM 73 | C-13 | C-4 | C-30 | ||||||
| MIC | MFC | MIC | MFC | MIC | MFC | MIC | MFC | MIC | MFC | |
| 15440 | 1.56 | 3.125 | 3.125 | 6.25 | 6.25 | 12.5 | 12.5 | 25 | R | R |
| 15443 | 100 | R | R | R | R | R | R | R | R | R |
| 15441 | 100 | R | 100 | R | R | R | 100 | R | R | R |
| 15446 | 50 | 100 | R | R | R | R | R | R | R | R |
| 15444 | 1.56 | 3.125 | 1.56 | 3.125 | 0.78 | 1.56 | 3.125 | 6.25 | 0.39 | 0.78 |
| 15447 | 25 | 50 | R | R | 12.5 | 25 | R | R | R | R |
| 15448 | 12.5 | 25 | R | R | R | R | R | R | 50 | 100 |
| 15445 | R | R | R | R | R | R | R | R | R | R |
| 15442 | 25 | 50 | 100 | R | 100 | R | 100 | R | R | R |
| Fluconazole | 0.78 | 1.56 | 1.56 | 3.125 | 0.78 | 1.56 | 0.39 | 0.78 | 0.78 | 1.56 |
| Fungal Isolate AUMC No. | IC50 Value (mg/mL) | LC50 Value (mg/mL) |
|---|---|---|
| 15440 | 3.51 ± 0.02 b | 1.18 |
| 15443 | 42.87 ± 1.73 f | 2.01 |
| 15441 | 17.69 ± 0.07 e | 1.45 |
| 15446 | 9.73 ± 0.01 d | 1.34 |
| 15444 | 6.06 ± 0.10 c | 1.39 |
| 15447 | 0.47 ± 0.00 a | 2.00 |
| 15448 | 0.58 ± 0.01 a | 1.29 |
| 15445 | 11.09 ± 0.01 d | 1.56 |
| 15442 | 17.12 ± 0.19 e | 1.71 |
| BHT | 3.38 ± 0.04 b | - |
| Fungal Isolate AUMC No. | Total Phenolics Gallic Acid Equivalent (mg/g) | Total Flavonoids Quercetin Equivalent (mg/g) |
|---|---|---|
| 15440 | 38.95 ± 1.45 e | 32.14 ± 0.40 c |
| 15443 | 25.81 ± 0.52 g | 22.94 ± 0.65 e |
| 15441 | 31.87 ± 0.70 f | 17.43 ± 0.40 g |
| 15446 | 103.11 ± 0.71 c | 29.09 ± 0.50 d |
| 15444 | 51.86 ± 1.01 d | 21.66 ± 0.39 e,f |
| 15447 | 138.30 ± 1.14 a | 72.09 ± 0.71 a |
| 15448 | 116.54 ± 1.04 b | 53.60 ± 0.49 b |
| 15445 | 28.04 ± 0.85 g | 20.42 ± 0.42 f |
| 15442 | 20.60 ± 0.26 h | 18.48 ± 0.17 g |
| Peak | RT (min) | Name | Fungal isolate extracts | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUMC 15440 | AUMC 15443 | AUMC 15441 | AUMC 15446 | AUMC 15444 | AUMC 15447 | AUMC 15448 | AUMC 15445 | AUMC 15442 | ||||||||||||
| Area % | Conc. (µg/g) | Area % | Conc. (µg/g) | Area % | Conc. (µg/g) | Area % | Conc. (µg/g) | Area % | Conc. (µg/g) | Area % | Conc. (µg/g) | Area % | Conc. (µg/g) | Area % | Conc. (µg/g) | Area % | Conc. (µg/g) | |||
| 1 | 3.49 | Gallic acid | 2.07 | 410.40 | 3.62 | 142.75 | 0.80 | 115.12 | 0.56 | 635.79 | 0.46 | 42.13 | 0.24 | 140.54 | 2.18 | 680.12 | 1.31 | 85.8 | 1.76 | 108.74 |
| 2 | 4.38 | Chlorogenic acid | 6.26 | 1937.16 | 27.44 | 1685.53 | 66.39 | 14,915.62 | 23.36 | 41,284.66 | 3.64 | 522.12 | 0.36 | 331.10 | 9.49 | 4614.26 | 40.99 | 4167.3 | 4.50 | 432.88 |
| 3 | 4.74 | Catechin | 0.00 | ND | 0.26 | 17.90 | 0.00 | ND | 0.00 | ND | 0.15 | 24.37 | 0.00 | ND | 0.00 | ND | 0.00 | ND | 0.00 | ND |
| 4 | 5.68 | Methyl gallate | 13.90 | 372.12 | 19.89 | 105.73 | 0.23 | 4.49 | 0.14 | 20.71 | 3.40 | 42.19 | 0.26 | 20.58 | 0.25 | 10.68 | 0.43 | 3.8 | 0.24 | 1.99 |
| 5 | 6.17 | Caffeic acid | 13.26 | 1969.90 | 11.10 | 327.34 | 1.11 | 119.96 | 2.78 | 2359.24 | 15.43 | 1062.60 | 0.03 | 15.07 | 3.02 | 704.88 | 10.18 | 497.0 | 1.27 | 58.73 |
| 6 | 6.66 | Syringic acid | 0.08 | 10.23 | 1.54 | 39.77 | 9.20 | 867.33 | 14.49 | 10,746.51 | 2.85 | 171.22 | 0.05 | 20.85 | 3.74 | 762.56 | 0.82 | 34.9 | 1.43 | 57.79 |
| 7 | 8.21 | Rutin | 0.03 | 10.64 | 8.17 | 553.19 | 0.78 | 194.25 | 5.11 | 9946.40 | 1.88 | 297.56 | 0.33 | 338.60 | 3.64 | 1949.47 | 4.52 | 506.1 | 0.47 | 49.32 |
| 8 | 9.15 | Ellagic acid | 12.34 | 3586.82 | 4.17 | 240.92 | 2.31 | 486.81 | 2.40 | 3977.62 | 1.26 | 169.33 | 0.10 | 86.16 | 1.60 | 731.22 | 1.07 | 102.5 | 0.19 | 17.20 |
| 9 | 9.24 | p-Coumaric acid | 1.02 | 56.50 | 2.73 | 30.13 | 0.41 | 16.41 | 0.02 | 6.56 | 1.08 | 27.73 | 0.04 | 6.18 | 0.64 | 55.79 | 0.86 | 15.7 | 0.37 | 6.31 |
| 10 | 9.76 | Vanillin | 0.91 | 69.33 | 0.24 | 3.59 | 0.47 | 25.87 | 0.45 | 196.28 | 0.10 | 3.51 | 0.00 | ND | 0.52 | 62.81 | 0.09 | 2.4 | 0.42 | 9.92 |
| 11 | 10.32 | Ferulic acid | 11.37 | 1544.83 | 5.14 | 138.74 | 2.25 | 222.14 | 2.27 | 1763.41 | 3.89 | 245.23 | 1.47 | 596.68 | 14.19 | 3029.11 | 1.96 | 87.6 | 3.15 | 133.19 |
| 12 | 10.58 | Naringenin | 0.73 | 167.46 | 1.39 | 63.46 | 1.04 | 173.17 | 1.63 | 2145.88 | 1.19 | 127.38 | 2.16 | 1490.00 | 7.50 | 2713.25 | 0.39 | 29.7 | 2.73 | 195.45 |
| 13 | 12.33 | Daidzein | 2.12 | 226.28 | 4.52 | 96.05 | 6.96 | 540.72 | 28.24 | 17,248.31 | 8.05 | 398.89 | 23.18 | 7426.39 | 5.66 | 950.92 | 1.83 | 64.3 | 7.36 | 244.70 |
| 14 | 12.81 | Quercetin | 0.72 | 121.88 | 0.65 | 22.05 | 0.79 | 97.93 | 3.10 | 3010.94 | 14.78 | 1165.77 | 26.30 | 13,411.96 | 1.12 | 300.74 | 0.27 | 15.2 | 3.54 | 187.42 |
| 15 | 14.03 | Cinnamic acid | 0.34 | 11.14 | 5.13 | 33.21 | 1.68 | 39.68 | 0.37 | 69.20 | 1.49 | 22.51 | 41.65 | 4070.05 | 36.58 | 1874.54 | 0.81 | 8.7 | 64.83 | 657.33 |
| 16 | 14.54 | Apigenin | 15.30 | 2678.13 | 0.53 | 18.57 | 4.18 | 531.59 | 8.67 | 8669.62 | 10.41 | 844.85 | 3.38 | 1770.67 | 4.06 | 1116.32 | 13.52 | 777.5 | 0.00 | ND |
| 17 | 15.03 | Kaempferol | 0.04 | 6.85 | 1.61 | 59.46 | 0.00 | ND | 0.00 | ND | 0.69 | 59.40 | 0.28 | 153.69 | 1.24 | 360.20 | 9.34 | 569.5 | 0.00 | ND |
| 18 | 15.55 | Hesperetin | 0.38 | 31.35 | 1.85 | 30.25 | 1.40 | 83.56 | 6.41 | 3014.19 | 29.24 | 1116.63 | 0.19 | 47.99 | 4.56 | 590.43 | 11.60 | 314.0 | 7.74 | 198.38 |
| Seq. | Compound | S. aureus Tyrosyl-tRNA Synthetase | C. albicans Aspartic Protease 2 | ||
|---|---|---|---|---|---|
| Score (kcal/mol) | RMSD (Å) | Score (kcal/mol) | RMSD (Å) | ||
| 1 | Caffeic acid | −12.57 | 0.58 | −7.71 | 1.12 |
| 2 | Catechin | −11.10 | 0.96 | −9.93 | 1.03 |
| 3 | Chlorogenic acid | −13.94 | 1.40 | −9.57 | 1.34 |
| 4 | Cinnamic acid | −7.30 | 0.98 | −9.08 | 0.84 |
| 5 | Ellagic acid | −13.89 | 0.55 | −9.09 | 0.67 |
| 6 | Ferulic acid | −9.04 | 0.60 | −11.15 | 0.60 |
| 7 | Gallic acid | −10.06 | 0.70 | −9.64 | 0.52 |
| 8 | Hesperetin | −11.17 | 0.80 | −8.47 | 0.92 |
| 9 | Kaempferol | −11.76 | 0.89 | −10.97 | 0.72 |
| 10 | Methyl gallate | −11.47 | 0.66 | −10.76 | 0.77 |
| 11 | Naringenin | −11.45 | 0.68 | −9.21 | 0.67 |
| 12 | p-Coumaric acid | −7.97 | 1.00 | −7.82 | 1.03 |
| 13 | Querectin | −12.76 | 0.78 | −9.30 | 1.03 |
| 14 | Rutin | −16.43 | 1.32 | −12.35 | 1.05 |
| 15 | Syringic acid | −10.08 | 0.63 | −8.91 | 1.25 |
| 16 | Vanillin | −8.03 | 0.65 | −6.33 | 1.06 |
| 17 | Apigenin | −12.44 | 0.85 | −8.71 | 0.90 |
| 18 | Daidzein | −10.53 | 0.64 | −7.86 | 1.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Mousa, A.A.; Abouelela, M.E.; Al Ghamidi, N.S.; Abo-Dahab, Y.; Mohamed, H.; Abo-Dahab, N.F.; Hassane, A.M.A. Anti-Staphylococcal, Anti-Candida, and Free-Radical Scavenging Potential of Soil Fungal Metabolites: A Study Supported by Phenolic Characterization and Molecular Docking Analysis. Curr. Issues Mol. Biol. 2024, 46, 221-243. https://doi.org/10.3390/cimb46010016
Al Mousa AA, Abouelela ME, Al Ghamidi NS, Abo-Dahab Y, Mohamed H, Abo-Dahab NF, Hassane AMA. Anti-Staphylococcal, Anti-Candida, and Free-Radical Scavenging Potential of Soil Fungal Metabolites: A Study Supported by Phenolic Characterization and Molecular Docking Analysis. Current Issues in Molecular Biology. 2024; 46(1):221-243. https://doi.org/10.3390/cimb46010016
Chicago/Turabian StyleAl Mousa, Amal A., Mohamed E. Abouelela, Nadaa S. Al Ghamidi, Youssef Abo-Dahab, Hassan Mohamed, Nageh F. Abo-Dahab, and Abdallah M. A. Hassane. 2024. "Anti-Staphylococcal, Anti-Candida, and Free-Radical Scavenging Potential of Soil Fungal Metabolites: A Study Supported by Phenolic Characterization and Molecular Docking Analysis" Current Issues in Molecular Biology 46, no. 1: 221-243. https://doi.org/10.3390/cimb46010016
APA StyleAl Mousa, A. A., Abouelela, M. E., Al Ghamidi, N. S., Abo-Dahab, Y., Mohamed, H., Abo-Dahab, N. F., & Hassane, A. M. A. (2024). Anti-Staphylococcal, Anti-Candida, and Free-Radical Scavenging Potential of Soil Fungal Metabolites: A Study Supported by Phenolic Characterization and Molecular Docking Analysis. Current Issues in Molecular Biology, 46(1), 221-243. https://doi.org/10.3390/cimb46010016










