Experimental Dengue Virus Type 4 Infection Increases the Expression of MicroRNAs-15/16, Triggering a Caspase-Induced Apoptosis Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Viral Samples
2.3. Infection of Hepatic Human Cells
2.4. RNA Extraction
2.5. Quantification of Viral Load by RT-qPCR
2.6. Quantification of miRNA Levels by RT-qPCR
2.7. NS1 Quantification
2.8. Apoptosis Detection
2.9. NS1 Fluorescent Imaging
2.10. Statistical Analysis
3. Results
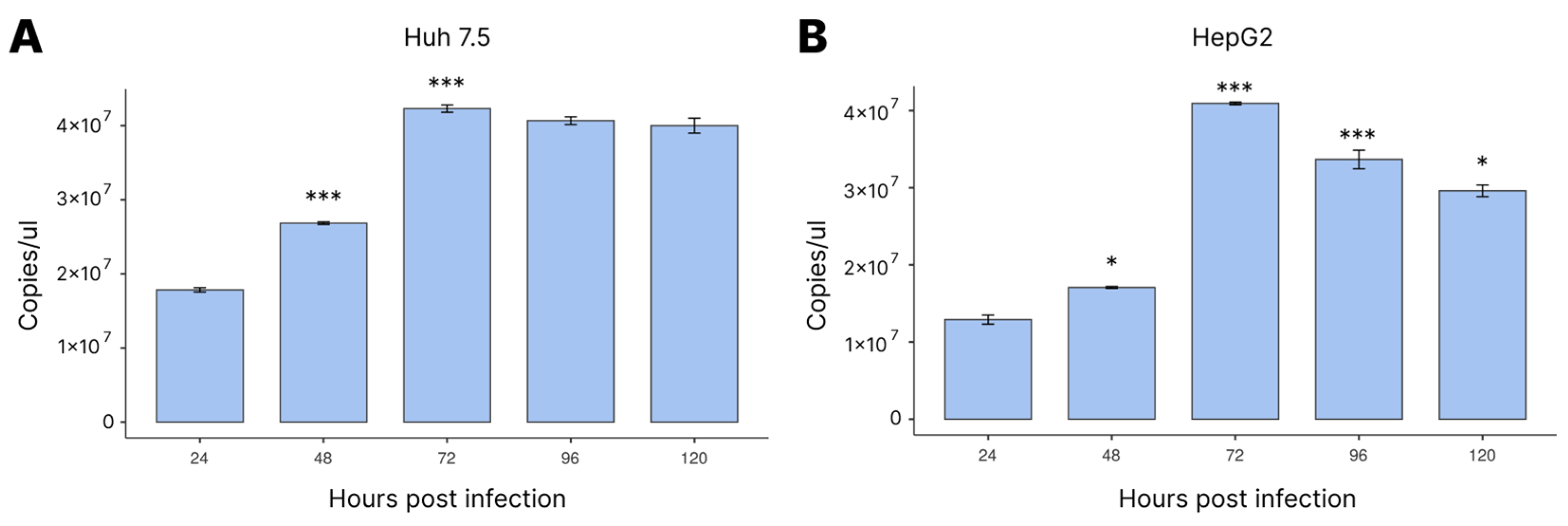
3.1. Viral Load Profile during Infection
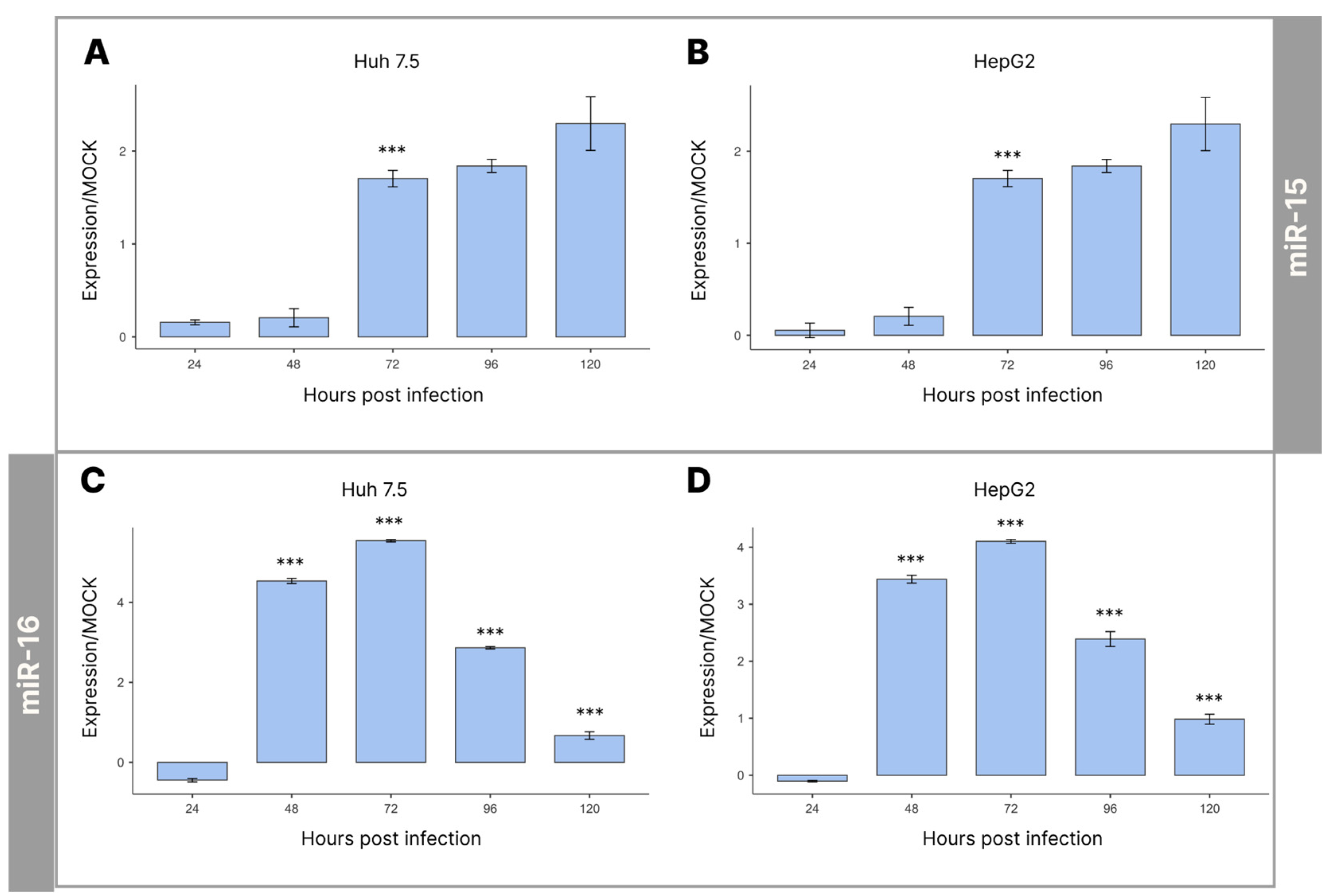
3.2. Expression Levels of miRNAs-15/16 during Infection
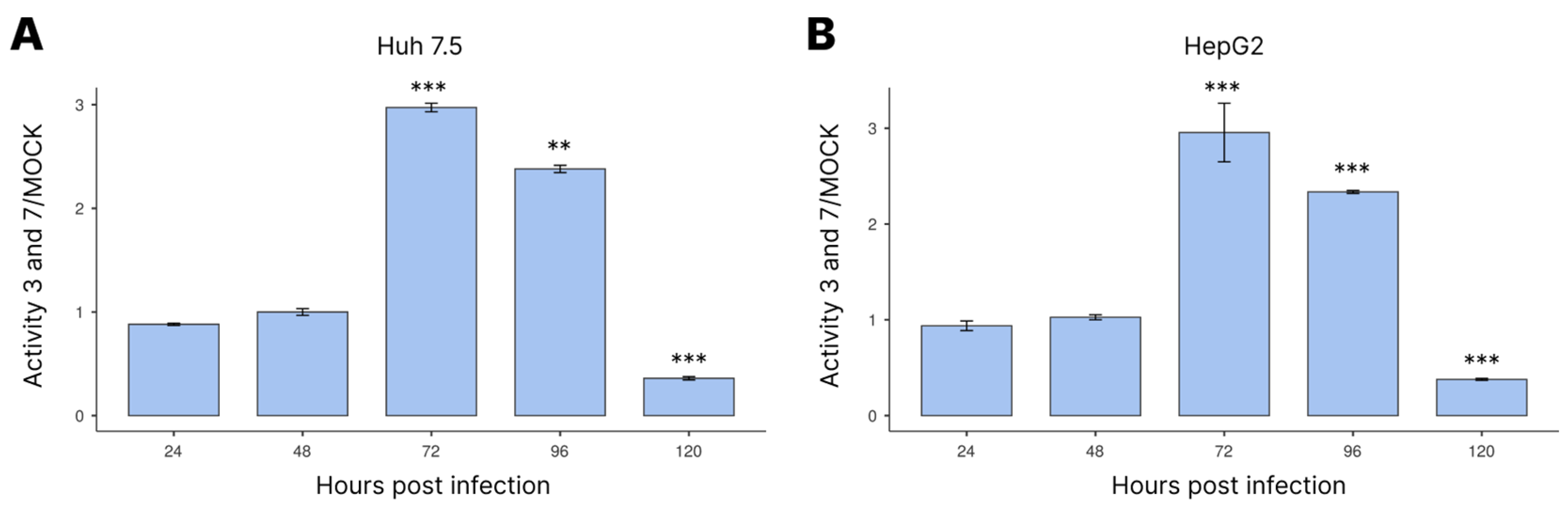
3.3. Activation of Caspases-3/7 during Infection
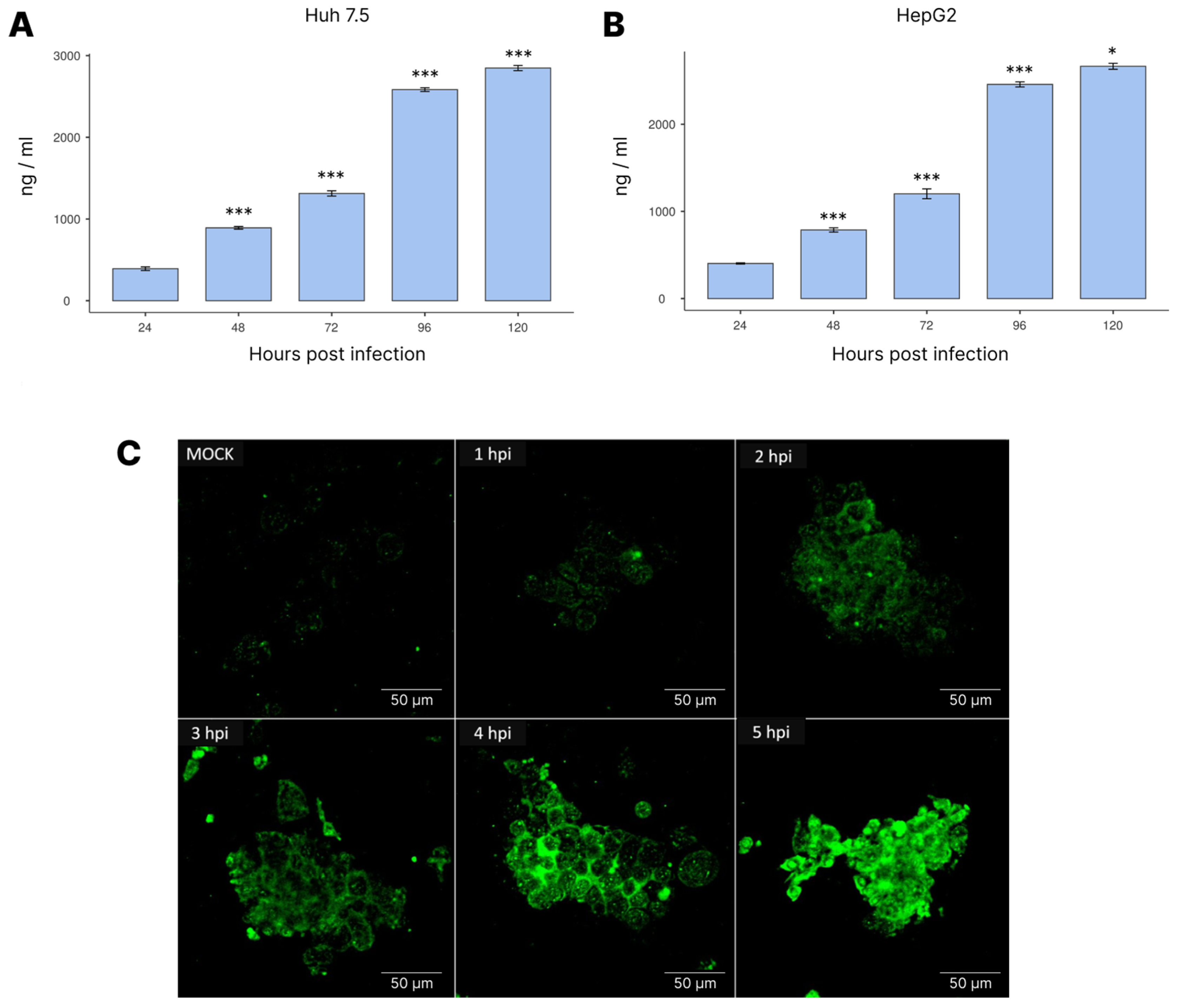
3.4. NS1 Expression during Infection
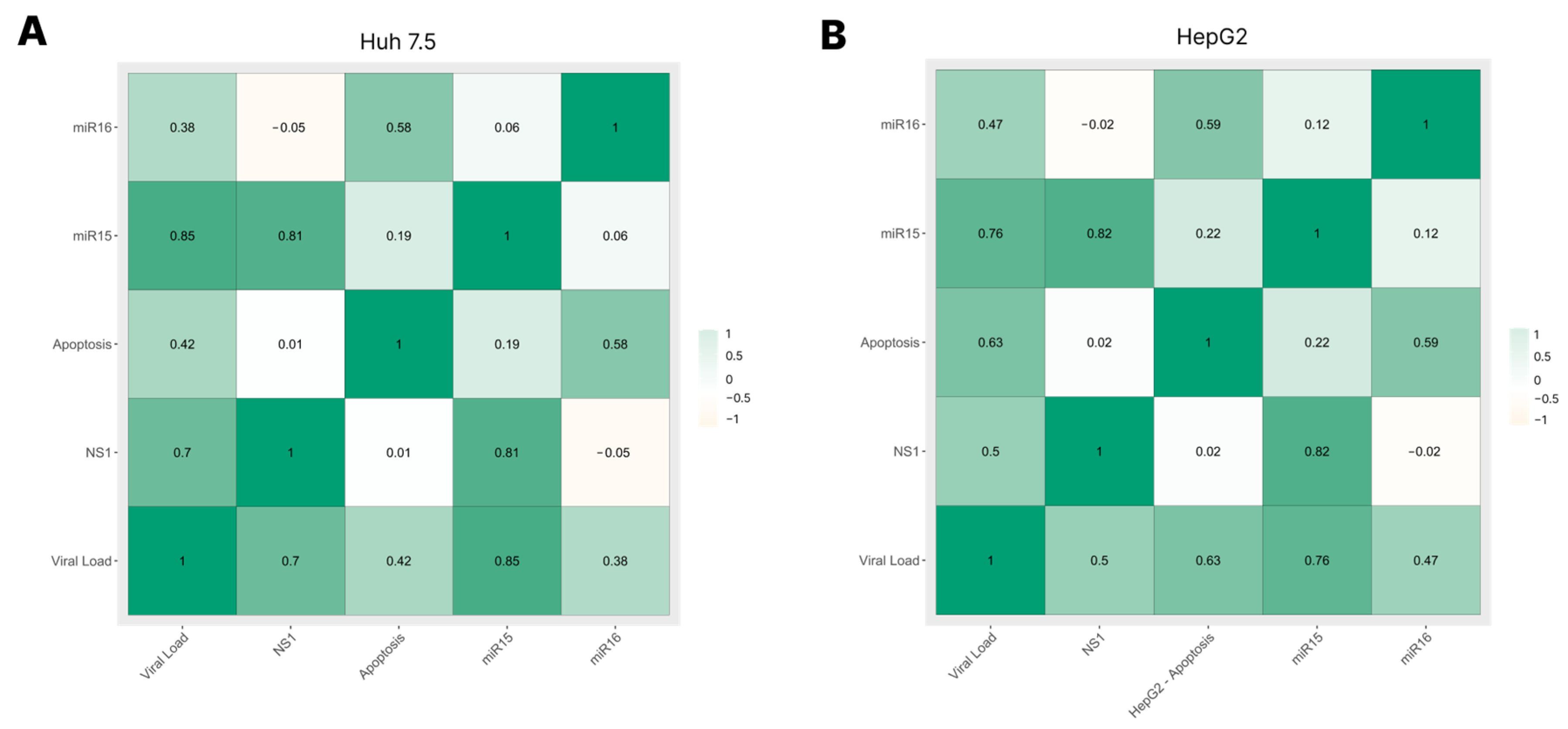
3.5. Correlation among Viral Load, NS1 Levels, miRNA-15/16 Expression, and Caspase-3/7 Activity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Horstick, O.; Tozan, Y.; Wilder-Smith, A. Reviewing dengue: Still a neglected tropical disease? PLoS Negl. Trop. Dis. 2015, 9, e0003632. [Google Scholar] [CrossRef]
- Li, G.; Khandekar, A.; Yin, T.; Hicks, S.C.; Guo, Q.; Takahashi, K.; Lipovsky, C.E.; Brumback, B.D.; Rao, P.K.; Weinheimer, C.J.; et al. Differential Wnt-mediated programming and arrhythmogenesis in right versus left ventricles. J. Mol. Cell. Cardiol. 2018, 123, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.B.; Martelli, C.M.T.; Coelho, G.E.; Simplicio, A.C.d.R.; Hatch, D.L. Dengue and dengue hemorrhagic fever, Brazil, 1981-2002. Emerg. Infect. Dis. 2005, 11, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Azeredo, E.L.; Dos Santos, F.B.; Barbosa, L.S.; Souza, T.M.A.; Badolato-Corrêa, J.; Sánchez-Arcila, J.C.; Nunes, P.C.G.; de-Oliveira-Pinto, L.M.; de Filippis, A.M.; Dal Fabbro, M.; et al. Clinical and Laboratory Profile of Zika and Dengue Infected Patients: Lessons Learned from the Co-circulation of Dengue, Zika and Chikungunya in Brazil. PLoS Curr. Influenza 2018, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Díaz, D.A.; Gutiérrez-Díaz, A.A.; Orozco-García, E.; Puerta-González, A.; Bermúdez-Santana, C.I.; Gallego-Gómez, J.C. Dengue virus potentially promotes migratory responses on endothelial cells by enhancing pro-migratory soluble factors and miRNAs. Virus Res. 2019, 259, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Hafirassou, M.L.; Meertens, L.; Umaña-Diaz, C.; Labeau, A.; Dejarnac, O.; Bonnet-Madin, L.; Kümmerer, B.M.; Delaugerre, C.; Roingeard, P.; Vidalain, P.-O.; et al. A global interactome map of the dengue virus NS1 identifies virus restriction and dependency host factors. Cell. Rep. 2017, 21, 3900–3913. [Google Scholar] [CrossRef]
- Halstead, S. Recent advances in understanding dengue. [version 1; peer review: 2 approved]. F1000Res 2019, 8, 1279. [Google Scholar] [CrossRef]
- Halstead, S.B.; Russell, P.K.; Brandt, W.E. NS1, Dengue’s Dagger. J. Infect. Dis. 2020, 221, 857–860. [Google Scholar] [CrossRef]
- Yen, P.-S.; Chen, C.-H.; Sreenu, V.; Kohl, A.; Failloux, A.-B. Assessing the potential interactions between cellular miRNA and arboviral genomic RNA in the yellow fever mosquito, Aedes aegypti. Viruses 2019, 11, 540. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microrna biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Huang, Y.; Shen, X.J.; Zou, Q.; Wang, S.P.; Tang, S.M.; Zhang, G.Z. Biological functions of microRNAs: A review. J. Physiol. Biochem. 2011, 67, 129–139. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Dou, Y.; Chen, L.; Wang, J.; Jiang, N.; Guo, C.; Yao, Q.; Wang, C.; Liu, L.; et al. Degradation of unmethylated miRNA/miRNA*s by a DEDDy-type 3’ to 5’ exoribonuclease Atrimmer 2 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E6659–E6667. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Jayathilaka, D.; Gomes, L.; Jeewandara, C.; Jayarathna, G.S.B.; Herath, D.; Perera, P.A.; Fernando, S.; Wijewickrama, A.; Hardman, C.S.; Ogg, G.S.; et al. Role of NS1 antibodies in the pathogenesis of acute secondary dengue infection. Nat. Commun. 2018, 9, 5242. [Google Scholar] [CrossRef] [PubMed]
- Glasner, D.R.; Puerta-Guardo, H.; Beatty, P.R.; Harris, E. The good, the bad, and the shocking: The multiple roles of dengue virus nonstructural protein 1 in protection and pathogenesis. Annu. Rev. Virol. 2018, 5, 227–253. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-R.; Yeh, S.-F.; Ruan, X.-M.; Zhang, H.; Hsu, S.-D.; Huang, H.-D.; Hsieh, C.-C.; Lin, Y.-S.; Yeh, T.-M.; Liu, H.-S.; et al. Honeysuckle aqueous extract and induced let-7a suppress dengue virus type 2 replication and pathogenesis. J. Ethnopharmacol. 2017, 198, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.W.; Russell, B.J.; Lanciotti, R.S. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J. Clin. Microbiol. 2005, 43, 4977–4983. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Casseb, S.M.M.; Simith, D.B.; Melo, K.F.L.; Mendonça, M.H.; Santos, A.C.M.; Carvalho, V.L.; Cruz, A.C.R.; Vasconcelos, P.F.C. Drosha, DGCR8, and Dicer mRNAs are down-regulated in human cells infected with dengue virus 4, and play a role in viral pathogenesis. Genet. Mol. Res. 2016, 15, 1–8. [Google Scholar] [CrossRef]
- Castillo, J.A.; Urcuqui-Inchima, S. Letter to the Editor “Drosha, DGCR8, and Dicer mRNAs are downregulated in human cells infected with dengue virus 4” - Genet. Mol. Res. 15 (2): Gmr.15027891—Drosha, Dicer, and TRBP mRNA are downregulated in Vero cells with the 3’UTR of Dengue virus. Genet. Mol. Res. 2016, 15, 1–4. [Google Scholar] [CrossRef]
- Holanda, G.M.; Casseb, S.M.M.; Mello, K.F.L.; Vasconcelos, P.F.C.; Cruz, A.C.R. Yellow Fever Virus Modulates the Expression of Key Proteins Related to the microRNA Pathway in the Human Hepatocarcinoma Cell Line HepG2. Viral Immunol. 2017, 30, 336–341. [Google Scholar] [CrossRef]
- Murphy Schafer, A.R.; Smith, J.L.; Pryke, K.M.; DeFilippis, V.R.; Hirsch, A.J. The E3 ubiquitin ligase SIAH1 targets myd88 for proteasomal degradation during dengue virus infection. Front. Microbiol. 2020, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, J.D.; Kageyama, R.; Shehata, H.M.; Fassett, M.S.; Mar, D.J.; Wigton, E.J.; Johansson, K.; Litterman, A.J.; Odorizzi, P.; Simeonov, D.; et al. miR-15/16 Restrain Memory T Cell Differentiation, Cell Cycle, and Survival. Cell. Rep. 2019, 28, 2169–2181. [Google Scholar] [CrossRef]
- Mishra, R.; Kumar, A.; Ingle, H.; Kumar, H. The Interplay Between Viral-Derived miRNAs and Host Immunity During Infection. Front. Immunol. 2019, 10, 3079. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Ampudia, Y.; Monsalve, D.M.; Castillo-Medina, L.F.; Rodríguez, Y.; Pacheco, Y.; Halstead, S.; Willison, H.J.; Anaya, J.-M.; Ramírez-Santana, C. Autoimmune neurological conditions associated with zika virus infection. Front. Mol. Neurosci. 2018, 11, 116. [Google Scholar] [CrossRef]
- Modhiran, N.; Watterson, D.; Muller, D.A.; Panetta, A.K.; Sester, D.P.; Liu, L.; Hume, D.A.; Stacey, K.J.; Young, P.R. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci. Transl. Med. 2015, 7, 304ra142. [Google Scholar] [CrossRef] [PubMed]
- Alhoot, M.A.; Wang, S.M.; Sekaran, S.D. RNA interference mediated inhibition of dengue virus multiplication and entry in HepG2 cells. PLoS ONE 2012, 7, e34060. [Google Scholar] [CrossRef]
- Aqeilan, R.I.; Calin, G.A.; Croce, C.M. miR-15a and miR-16-1 in cancer: Discovery, function and future perspectives. Cell Death Differ. 2010, 17, 215–220. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, L.F.; de Andrade, A.A.S.; Pagliari, C.; de Carvalho, L.V.; Silveira, T.S.; Cardoso, J.F.; Silva, A.L.T.E.; de Vasconcelos, J.M.; Moreira-Nunes, C.A.; Burbano, R.M.R.; et al. Differential expression analysis and profiling of hepatic miRNA and isomiRNA in dengue hemorrhagic fever. Sci. Rep. 2021, 11, 5554. [Google Scholar] [CrossRef]
- Chen, C.-L.; Lin, C.-F.; Wan, S.-W.; Wei, L.-S.; Chen, M.-C.; Yeh, T.-M.; Liu, H.-S.; Anderson, R.; Lin, Y.-S. Anti-dengue virus nonstructural protein 1 antibodies cause NO-mediated endothelial cell apoptosis via ceramide-regulated glycogen synthase kinase-3β and NF-κB activation. J. Immunol. 2013, 191, 1744–1752. [Google Scholar] [CrossRef]
- Lin, J.-C.; Lin, S.-C.; Chen, W.-Y.; Yen, Y.-T.; Lai, C.-W.; Tao, M.-H.; Lin, Y.-L.; Miaw, S.-C.; Wu-Hsieh, B.A. Dengue viral protease interaction with NF-κB inhibitor α/β results in endothelial cell apoptosis and hemorrhage development. J. Immunol. 2014, 193, 1258–1267. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, X.; Jiang, X.; Gu, D.; Zhang, Y.; Kong, S.K.; Jiang, C.; Xie, W. Dysregulated Serum MiRNA Profile and Promising Biomarkers in Dengue-infected Patients. Int. J. Med. Sci. 2016, 13, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Kayesh, M.E.H.; Tsukiyama-Kohara, K. Mammalian animal models for dengue virus infection: A recent overview. Arch. Virol. 2021, 167, 31–44. [Google Scholar] [CrossRef]
- Nanaware, N.; Banerjee, A.; Mullick Bagchi, S.; Bagchi, P.; Mukherjee, A. Dengue virus infection: A tale of viral exploitations and host responses. Viruses 2021, 13, 1967. [Google Scholar] [CrossRef]
- Wong, R.R.; Abd-Aziz, N.; Affendi, S.; Poh, C.L. Role of microRNAs in antiviral responses to dengue infection. J. Biomed. Sci. 2020, 27, 4. [Google Scholar] [CrossRef]
- Linsley, P.S.; Schelter, J.; Burchard, J.; Kibukawa, M.; Martin, M.M.; Bartz, S.R.; Johnson, J.M.; Cummins, J.M.; Raymond, C.K.; Dai, H.; et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell. Biol. 2007, 27, 2240–2252. [Google Scholar] [CrossRef] [PubMed]
- El-Abd, N.E.; Fawzy, N.A.; El-Sheikh, S.M.; Soliman, M.E. Circulating miRNA-122, miRNA-199a, and miRNA-16 as Biomarkers for Early Detection of Hepatocellular Carcinoma in Egyptian Patients with Chronic Hepatitis C Virus Infection. Mol. Diagn. Ther. 2015, 19, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Pagliari, C.; Quaresma, J.A.S.; Fernandes, E.R.; Stegun, F.W.; Brasil, R.A.; de Andrade, H.F.; Barros, V.; Vasconcelos, P.F.C.; Duarte, M.I.S. Immunopathogenesis of dengue hemorrhagic fever: Contribution to the study of human liver lesions. J. Med. Virol. 2014, 86, 1193–1197. [Google Scholar] [CrossRef]
- Acharya, B.; Gyeltshen, S.; Chaijaroenkul, W.; Na-Bangchang, K. Significance of autophagy in dengue virus infection: A brief review. Am. J. Trop. Med. Hyg. 2019, 100, 783–790. [Google Scholar] [CrossRef]
- Lu, Z.-Y.; Cheng, M.-H.; Yu, C.-Y.; Lin, Y.-S.; Yeh, T.-M.; Chen, C.-L.; Chen, C.-C.; Wan, S.-W.; Chang, C.-P. Dengue Nonstructural Protein 1 Maintains Autophagy through Retarding Caspase-Mediated Cleavage of Beclin-1. Int. J. Mol. Sci. 2020, 21, 9702. [Google Scholar] [CrossRef] [PubMed]
- Arboleda, J.F.; Fernandez, G.J.; Urcuqui-Inchima, S. Vitamin D-mediated attenuation of miR-155 in human macrophages infected with dengue virus: Implications for the cytokine response. Infect. Genet. Evol. 2019, 69, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Silva, B.M.; Sousa, L.P.; Gomes-Ruiz, A.C.; Leite, F.G.G.; Teixeira, M.M.; da Fonseca, F.G.; Pimenta, P.F.P.; Ferreira, P.C.P.; Kroon, E.G.; Bonjardim, C.A. The dengue virus nonstructural protein 1 (NS1) increases NF-κB transcriptional activity in HepG2 cells. Arch. Virol. 2011, 156, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Puerta-Guardo, H.; Glasner, D.R.; Espinosa, D.A.; Biering, S.B.; Patana, M.; Ratnasiri, K.; Wang, C.; Beatty, P.R.; Harris, E. Flavivirus NS1 Triggers Tissue-Specific Vascular Endothelial Dysfunction Reflecting Disease Tropism. Cell. Rep. 2019, 26, 1598–1613.e8. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Casseb, S.M.M.; Melo, K.F.L.d.; Carvalho, C.A.M.d.; Santos, C.R.d.; Franco, E.C.S.; Vasconcelos, P.F.d.C. Experimental Dengue Virus Type 4 Infection Increases the Expression of MicroRNAs-15/16, Triggering a Caspase-Induced Apoptosis Pathway. Curr. Issues Mol. Biol. 2023, 45, 4589-4599. https://doi.org/10.3390/cimb45060291
Casseb SMM, Melo KFLd, Carvalho CAMd, Santos CRd, Franco ECS, Vasconcelos PFdC. Experimental Dengue Virus Type 4 Infection Increases the Expression of MicroRNAs-15/16, Triggering a Caspase-Induced Apoptosis Pathway. Current Issues in Molecular Biology. 2023; 45(6):4589-4599. https://doi.org/10.3390/cimb45060291
Chicago/Turabian StyleCasseb, Samir Mansour Moraes, Karla Fabiane Lopes de Melo, Carlos Alberto Marques de Carvalho, Carolina Ramos dos Santos, Edna Cristina Santos Franco, and Pedro Fernando da Costa Vasconcelos. 2023. "Experimental Dengue Virus Type 4 Infection Increases the Expression of MicroRNAs-15/16, Triggering a Caspase-Induced Apoptosis Pathway" Current Issues in Molecular Biology 45, no. 6: 4589-4599. https://doi.org/10.3390/cimb45060291
APA StyleCasseb, S. M. M., Melo, K. F. L. d., Carvalho, C. A. M. d., Santos, C. R. d., Franco, E. C. S., & Vasconcelos, P. F. d. C. (2023). Experimental Dengue Virus Type 4 Infection Increases the Expression of MicroRNAs-15/16, Triggering a Caspase-Induced Apoptosis Pathway. Current Issues in Molecular Biology, 45(6), 4589-4599. https://doi.org/10.3390/cimb45060291






