Characterization of Mammary Tumors Arising from MMTV-PyVT Transgenic Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Transgenic Mice
2.2. Spontaneous Mammary Tumor
2.3. Histology and Whole-Mount Analysis
2.4. Whole-Exome Sequencing
2.5. Exome Data Analysis
3. Results
3.1. Spontaneous Tumor Formation
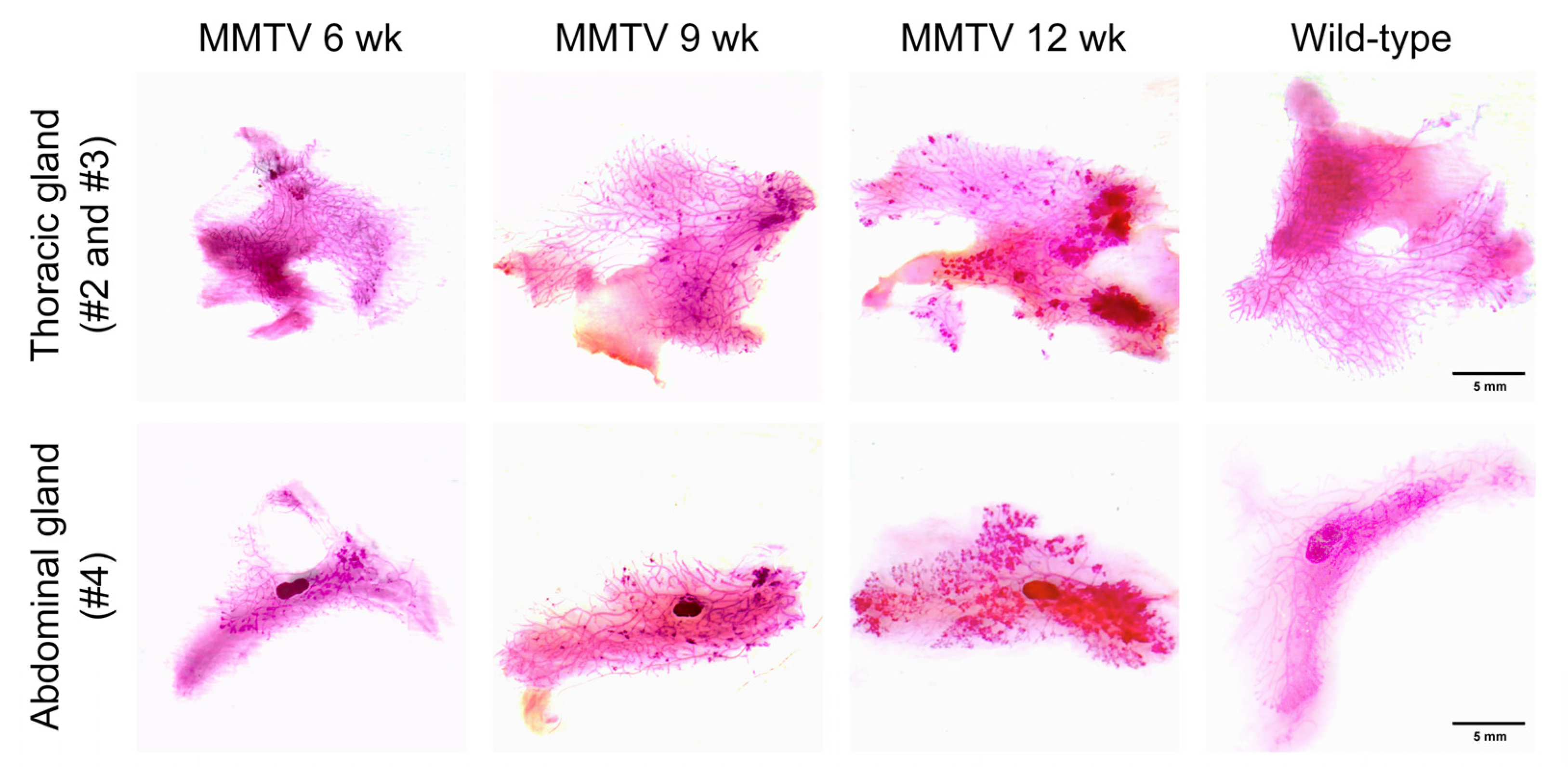
3.2. Histological Analysis
3.3. Mouse-Level Genetic Alterations
3.4. Tumor-Level Genetic Alterations
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Menezes, M.E.; Das, S.K.; Emdad, L.; Windle, J.J.; Wang, X.Y.; Sarkar, D.; Fisher, P.B. Genetically engineered mice as experimental tools to dissect the critical events in breast cancer. Adv. Cancer Res. 2014, 121, 331–382. [Google Scholar] [CrossRef] [PubMed]
- Mes, A.M.; Hassell, J.A. Polyoma viral middle T-antigen is required for transformation. J. Virol. 1982, 42, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Ha, G.; Khadka, P.; Jin, X.; Wong, B.; Franke, L.; Golub, T.R. The landscape of chromosomal aberrations in breast cancer mouse models reveals driver-specific routes to tumorigenesis. Nat. Commun. 2016, 7, 12160. [Google Scholar] [CrossRef] [PubMed]
- Attalla, S.; Taifour, T.; Bui, T.; Muller, W. Insights from transgenic mouse models of PyMT-induced breast cancer: Recapitulating human breast cancer progression in vivo. Oncogene 2021, 40, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Herschkowitz, J.I.; Simin, K.; Weigman, V.J.; Mikaelian, I.; Usary, J.; Hu, Z.; Rasmussen, K.E.; Jones, L.P.; Assefnia, S.; Chandrasekharan, S.; et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome. Biol. 2007, 8, R76. [Google Scholar] [CrossRef] [PubMed]
- Pfefferle, A.D.; Herschkowitz, J.I.; Usary, J.; Harrell, J.C.; Spike, B.T.; Adams, J.R.; Torres-Arzayus, M.I.; Brown, M.; Egan, S.E.; Wahl, G.M.; et al. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome. Biol. 2013, 14, R125. [Google Scholar] [CrossRef]
- Shishido, S.; Delahaye, A.; Beck, A.; Nguyen, T.A. The MMTV-PyVT transgenic mouse as a multistage model for mammary carcinoma and the efficacy of antineoplastic treatment. J. Cancer Ther. 2013, 4, 1187–1197. [Google Scholar] [CrossRef]
- Guy, C.T.; Cardiff, R.D.; Muller, W.J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol. Cell Biol. 1992, 12, 954–961. [Google Scholar] [CrossRef]
- Liu, C.L.; Cheng, S.P.; Chen, M.J.; Lin, C.H.; Chen, S.N.; Kuo, Y.H.; Chang, Y.C. Quinolinate phosphoribosyltransferase promotes invasiveness of breast cancer through myosin light chain phosphorylation. Front. Endocrinol. 2021, 11, 621944. [Google Scholar] [CrossRef]
- Liu, C.L.; Yang, P.S.; Wang, T.Y.; Huang, S.Y.; Kuo, Y.H.; Cheng, S.P. PGC1α downregulation and glycolytic phenotype in thyroid cancer. J. Cancer 2019, 10, 3819–3829. [Google Scholar] [CrossRef]
- Tolg, C.; Cowman, M.; Turley, E.A. Mouse mammary gland whole mount preparation and analysis. Bio. Protoc. 2018, 8, e2915. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Kallberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef]
- Geoffroy, V.; Herenger, Y.; Kress, A.; Stoetzel, C.; Piton, A.; Dollfus, H.; Muller, J. AnnotSV: An integrated tool for structural variations annotation. Bioinformatics 2018, 34, 3572–3574. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic. Acids. Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Sondka, Z.; Bamford, S.; Cole, C.G.; Ward, S.A.; Dunham, I.; Forbes, S.A. The COSMIC Cancer Gene Census: Describing genetic dysfunction across all human cancers. Nat. Rev. Cancer. 2018, 18, 696–705. [Google Scholar] [CrossRef]
- Rennhack, J.; To, B.; Wermuth, H.; Andrechek, E.R. Mouse models of breast cancer share amplification and deletion events with human breast cancer. J. Mammary Gland Biol. Neoplasia. 2017, 22, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Hollern, D.P.; Andrechek, E.R. A genomic analysis of mouse models of breast cancer reveals molecular features of mouse models and relationships to human breast cancer. Breast. Cancer Res. 2014, 16, R59. [Google Scholar] [CrossRef]
- Rennhack, J.P.; To, B.; Swiatnicki, M.; Dulak, C.; Ogrodzinski, M.P.; Zhang, Y.; Li, C.; Bylett, E.; Ross, C.; Szczepanek, K.; et al. Integrated analyses of murine breast cancer models reveal critical parallels with human disease. Nat. Commun. 2019, 10, 3261. [Google Scholar] [CrossRef]
- Ross, C.; Szczepanek, K.; Lee, M.; Yang, H.; Qiu, T.; Sanford, J.D.; Hunter, K. The genomic landscape of metastasis in treatment-naive breast cancer models. PLoS Genet. 2020, 16, e1008743. [Google Scholar] [CrossRef]
- Dilworth, S.M. Polyoma virus middle T antigen and its role in identifying cancer-related molecules. Nat. Rev. Cancer 2002, 2, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.T.; Muthuswamy, S.K.; Cardiff, R.D.; Soriano, P.; Muller, W.J. Activation of the c-Src tyrosine kinase is required for the induction of mammary tumors in transgenic mice. Genes. Dev. 1994, 8, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, C.A.; VanderVorst, K.; Free, S.; Rowson-Hodel, A.; Carraway, K.L. The role of membrane mucin MUC4 in breast cancer metastasis. Endocr. Relat. Cancer 2021, 29, R17–R32. [Google Scholar] [CrossRef] [PubMed]
- Price-Schiavi, S.A.; Jepson, S.; Li, P.; Arango, M.; Rudland, P.S.; Yee, L.; Carraway, K.L. Rat Muc4 (sialomucin complex) reduces binding of anti-ErbB2 antibodies to tumor cell surfaces, a potential mechanism for herceptin resistance. Int. J. Cancer 2002, 99, 783–791. [Google Scholar] [CrossRef]
- Rowson-Hodel, A.R.; Wald, J.H.; Hatakeyama, J.; O’Neal, W.K.; Stonebraker, J.R.; VanderVorst, K.; Saldana, M.J.; Borowsky, A.D.; Sweeney, C.; Carraway, K.L., 3rd. Membrane Mucin Muc4 promotes blood cell association with tumor cells and mediates efficient metastasis in a mouse model of breast cancer. Oncogene 2018, 37, 197–207. [Google Scholar] [CrossRef]
- Vargo-Gogola, T.; Rosen, J.M. Modelling breast cancer: One size does not fit all. Nat. Rev. Cancer 2007, 7, 659–672. [Google Scholar] [CrossRef]


| Location | Gene | Accession No. | Class |
|---|---|---|---|
| Chr1:88234242-88234312 | Mroh2a | NM_001281466 | DEL, INV |
| Chr1:88266278-88266329 | 6430706D22Rik | NR_040291 | DEL |
| Chr1:88270315-88271447 | A730008H23Rik | NM_172505 | DEL |
| Chr1:88270315-88271447 | Hjurp | NM_198652 | DEL |
| Chr1:171345007-171356531 | Nit1 | NM_012049 | DEL, BND |
| Chr1:171356616-171356702 | Pfdn2 | NM_001360825 | DEL |
| Chr6:116022400-116024804 | Tmcc1 | NM_001364577 | DEL |
| Chr7:24082235-24085425 | Zfp180 | NM_001045486 | DEL |
| Chr7:79651636-79665662 | Ticrr | NM_029835 | DEL |
| Chr7:84947480-84947640 | Vmn2r65 | NM_001105180 | DEL |
| Chr7:141638851-141639059 | Muc6 | NM_001368953 | DEL |
| Chr8:35482726-35491202 | Eri1 | NM_026067 | DEL |
| Chr9:56260762-56318663 | Peak1 | NM_172924 | DEL |
| Chr10:81178233-81178679 | Eef2 | NM_007907 | DEL |
| Chr11:3137135-3137136 | Sfi1 | NM_030207 | INS, BND |
| Chr12:106051004-106051759 | Vrk1 | NM_001029843 | DEL |
| Chr19:21598332-21598752 | 1110059E24Rik | NM_025423 | DEL |
| Location | Gene | Accession No. | Indel | Type |
|---|---|---|---|---|
| Chr1:139237087-139237087 | Crb1 | NM_133239 | c.3481delC, p.R1161Gfs*48 | frameshift deletion |
| Chr1:85094333-85094335 | A530032D15Rik | NM_213615 | c.392_394del, p.A131del | non-frameshift deletion |
| Chr1:85591567-85591567 | Sp110 | NM_030194 | c.538dupG, p.A180Gfs*26 | frameshift insertion |
| Chr1:88212372-88212372 | Ugt1a1 | NM_201645 | c.371delT, p.M124Sfs*3 | frameshift deletion |
| Chr1:88216161-88216162 | Ugt1a2 | NM_013701 | c.1108_1109del, p.I370Hfs*10 | frameshift deletion |
| Chr1:9546104-9546105 | Rrs1 | NM_021511 | c.581_582del, p.H197Qfs*18 | frameshift deletion |
| Chr3:130040795-130040797 | Sec24b | NM_207209 | c.751_753del, p.Q251del | non-frameshift deletion |
| Chr3:96683463-96683463 | Ankrd35 | NM_001081139 | c.1065dupA, p.N356Kfs*9 | frameshift insertion |
| Chr4:63171423-63171423 | Kif12 | NM_010616 | c.179_180insGCCGGGTGGAGGCCC, p.P60_D61insPGGGP | non-frameshift insertion |
| Chr5:32737988-32737988 | Pisd | NM_177298 | c.C994T, p.R332X | stopgain |
| Chr5:93637618-93637618 | Pramel34 | NM_001164284 | c.C802T, p.Q268X | stopgain |
| Chr8:104182034-104182034 | Bean1 | NM_001141922 | c.42delC, p.Q15Kfs*29 | frameshift deletion |
| Chr8:26160857-26160858 | Thap1 | NM_199042 | c.154_155del, p.S52Hfs*12 | frameshift deletion |
| Chr9:103355194-103355204 | Cdv3 | NM_175833 | c.805_815del, p.V269Sfs*16 | frameshift deletion |
| Chr9:39484333-39484333 | Or8g20 | NM_146830 | c.919delA, p.I307* | stopgain |
| Chr9:65280131-65280131 | Cilp | NM_173385 | c.3507delG, p.G1170Afs*16 | frameshift deletion |
| Chr13:64921972-64921972 | Spata31 | NM_030047 | c.C1933T, p.R645X | stopgain |
| Chr16:32752550-32752550 | Muc4 | NM_080457 | c.2427_2428insCA, p.Q810Hfs*30 | frameshift insertion |
| Chr16:44496429-44496431 | Boc | NM_172506 | c.1348_1350del, p.S450del | non-frameshift deletion |
| Chr16:45729926-45729926 | Abhd10 | NM_001272070 | c.870delC, p.D291Tfs*15 | frameshift deletion |
| Chr17:23291424-23291424 | Vmn2r114 | NM_001102584 | c.C2081G, p.S694X | stopgain |
| Chr17:23475353-23475353 | Vmn2r117 | NM_001104581 | c.G1519T, p.G507X | stopgain |
| Location | Gene | Accession No. | SNV |
|---|---|---|---|
| Chr1:12839899-12839899 | Sulf1 | NM_172294 | c.G1918A, p.D640N |
| Chr1:177773679-177773679 | Adss | NM_007422 | c.C686T, p.P229L |
| Chr1:26687400-26687400 | 4931408C20Rik | NM_001033764 | c.T27A, p.N9K |
| Chr1:59847167-59847167 | Bmpr2 | NM_007561 | c.G962A, p.R321Q |
| Chr1:75486756-75486756 | Obsl1 | NM_178884 | c.C5291T, p.T1764M |
| Chr1:85246950-85246950 | C130026I21Rik | NM_175219 | c.C863T, p.S288L |
| Chr1:85615212-85615212 | Sp140 | NM_001013817 | c.C443G, p.T148R |
| Chr2:131936461-131936461 | Prnp | NM_011170 | c.T32A, p.L11H |
| Chr2:84872044-84872044 | Rtn4rl2 | NM_199223 | c.G1165C, p.A389P |
| Chr3:144691910-144691910 | Sh3glb1 | NM_019464 | c.T980A, p.L327Q |
| Chr3:15548939-15548939 | Sirpb1b | NM_001173460 | c.A82G, p.M28V |
| Chr3:95734876-95734876 | Ecm1 | NM_007899 | c.A1396G, p.I466V |
| Chr3:96854557-96854557 | Pdzk1 | NM_021517 | c.A484G, p.N162D |
| Chr4:138221673-138221673 | Hp1bp3 | NM_010470 | c.C46T, p.L16F |
| Chr4:140798123-140798123 | Padi3 | NM_011060 | c.T578C, p.L193P |
| Chr4:147510785-147510785 | Zfp982 | NM_001039209 | c.A63C, p.E21D |
| Chr4:147581328-147581328 | Zfp985 | NM_001014397 | c.T117G, p.I39M |
| Chr4:147613775-147613775 | Zfp979 | NM_145078 | c.C476T, p.T159I |
| Chr4:148944359-148944359 | Casz1 | NM_027195 | c.T3260C, p.L1087P |
| Chr4:21873684-21873684 | Pnisr | NM_025669 | c.C1426G, p.R476G |
| Chr4:3184971-3184971 | Vmn1r3 | NM_001167535 | c.C335T, p.T112I |
| Chr5:112762721-112762721 | Myo18b | NM_028901 | c.G5804A, p.R1935H |
| Chr5:114398443-114398443 | Ube3b | NM_054093 | c.T749G, p.M250R |
| Chr5:13570208-13570208 | Sema3a | NM_009152 | c.A1423G, p.I475V |
| Chr5:26035024-26035024 | Speer4a | NM_029376 | c.A727C, p.T243P |
| Chr5:27501274-27501274 | Speer4b | NM_028561 | c.C94T, p.P32S |
| Chr5:38300085-38300085 | Otop1 | NM_172709 | c.G1187C, p.G396A |
| Chr5:89775351-89775351 | Adamts3 | NM_177872 | c.G595A, p.V199I |
| Chr5:96758142-96758142 | Fras1 | NM_175473 | c.T9404C, p.L3135P |
| Chr6:39400456-39400456 | Mkrn1 | NM_018810 | c.A1036T, p.N346Y |
| Chr7:102973309-102973309 | Or51g2 | NM_147109 | c.G682A, p.V228I |
| Chr7:105434593-105434593 | Cckbr | NM_007627 | c.C727G, p.R243G |
| Chr7:108465371-108465371 | Or5p73 | NM_146307 | c.T46A, p.F16I |
| Chr7:120135179-120135179 | Zp2 | NM_011775 | c.C1646T, p.A549V |
| Chr7:122167650-122167650 | Plk1 | NM_011121 | c.C1090T, p.R364W |
| Chr7:131065072-131065072 | Dmbt1 | NM_007769 | c.C1342A, p.P448T |
| Chr7:13801414-13801414 | Sult2a1 | NM_001111296 | c.A713G, p.Q238R |
| Chr7:141858623-141858623 | Muc5b | NM_028801 | c.T5305C, p.Y1769H |
| Chr7:3222537-3222537 | Nlrp12 | NM_001033431 | c.A3101G, p.K1034R |
| Chr7:43187290-43187290 | Zfp936 | NM_001034893 | c.G124A, p.A42T |
| Chr7:56131292-56131292 | Herc2 | NM_010418 | c.G3704A, p.G1235D |
| Chr7:79111354-79111354 | Acan | NM_007424 | c.A5813C, p.H1938P |
| Chr7:92858589-92858589 | Ddias | NM_001080995 | c.C2117T, p.P706L |
| Chr8:122890181-122890181 | Ankrd11 | NM_001081379 | c.G6868C, p.V2290L |
| Chr9:109145537-109145537 | Fbxw21 | NM_177069 | c.A914G, p.H305R |
| Chr9:120016907-120016907 | Xirp1 | NM_011724 | c.A2909C, p.Q970P |
| Chr9:25130622-25130622 | Herpud2 | NM_020586 | c.G253C, p.V85L |
| Chr9:38581513-38581513 | Or8b48 | NM_146810 | c.C235T, p.P79S |
| Chr9:44249891-44249891 | Pdzd3 | NM_133226 | c.T472C, p.C158R |
| Chr9:44942695-44942695 | Ube4a | NM_145400 | c.A1747T, p.N583Y |
| Chr10:58231344-58231344 | Dux | NM_001081954 | c.G1332C, p.L444F |
| Chr10:67238174-67238174 | Jmjd1c | NM_001242396 | c.T5144C, p.L1715P |
| Chr10:79169477-79169477 | Vmn2r80 | NM_001103368 | c.A947G, p.N316S |
| Chr10:88091833-88091833 | Pmch | NM_029971 | c.T395C, p.I132T |
| Chr11:46222615-46222615 | Cyfip2 | NM_133769 | c.C2903T, p.S968F |
| Chr11:90480671-90480671 | Stxbp4 | NM_011505 | c.G1603A, p.A535T |
| Chr13:100161909-100161909 | Naip2 | NM_010872 | c.T1618A, p.Y540N |
| Chr13:21468303-21468303 | Nkapl | NM_025719 | c.G139C, p.G47R |
| Chr13:27272475-27272475 | Prl3a1 | NM_025896 | c.C311T, p.T104I |
| Chr13:53117204-53117204 | Ror2 | NM_013846 | c.G1114A, p.V372M |
| Chr13:93063579-93063579 | Cmya5 | NM_023821 | c.G10240C, p.A3414P |
| Chr14:51413192-51413192 | Vmn2r88 | NM_001368932 | c.A361G, p.T121A |
| Chr14:70586204-70586204 | Fhip2b | NM_194345 | c.C1725A, p.S575R |
| Chr15:77638007-77638007 | Apol11b | NM_001143686 | c.T89G, p.I30R |
| Chr16:35291544-35291544 | Adcy5 | NM_001012765 | c.G2770A, p.V924M |
| Chr16:36772445-36772445 | Slc15a2 | NM_021301 | c.T629C, p.M210T |
| Chr16:38828345-38828345 | Tex55 | NM_029042 | c.C401G, p.T134S |
| Chr16:39024953-39024953 | Igsf11 | NM_170599 | c.G1045C, p.A349P |
| Chr16:43939116-43939116 | Ccdc191 | NM_027801 | c.G1279A, p.V427I |
| Chr16:44299802-44299802 | Sidt1 | NM_198034 | c.C515G, p.P172R |
| Chr16:44379308-44379308 | Spice1 | NM_144550 | c.C2122A, p.R708S |
| Chr16:44789572-44789572 | Cd200r1 | NM_021325 | c.A153G, p.I51M |
| Chr16:44820915-44820915 | Cd200r4 | NM_207244 | c.T20C, p.I7T |
| Chr16:45094982-45094982 | Ccdc80 | NM_026439 | c.A100G, p.T34A |
| Chr16:45239239-45239239 | Btla | NM_177584 | c.A305G, p.Q102R |
| Chr16:45392332-45392332 | Cd200 | NM_010818 | c.A751G, p.I251V |
| Chr16:45539592-45539592 | Slc9c1 | NM_198106 | c.T8C, p.M3T |
| Chr16:45664252-45664252 | Tmprss7 | NM_172455 | c.G1544T, p.S515I |
| Chr16:46049747-46049747 | Cd96 | NM_032465 | c.T1358C, p.F453S |
| Chr16:48817255-48817255 | Retnlb | NM_023881 | c.C43T, p.L15F |
| Chr17:35425194-35425194 | H2-Q6 | NM_207648 | c.A151G, p.N51D |
| Chr17:35440154-35440154 | H2-Q7 | NM_010394 | c.C580G, p.Q194E |
| Chr17:45517174-45517174 | Aars2 | NM_198608 | c.C1810T, p.R604C |
| Chr17:47400410-47400410 | Guca1a | NM_008189 | c.A10G, p.I4V |
| Chr17:67752883-67752883 | Lama1 | NM_008480 | c.G1966A, p.D656N |
| Chr18:24017781-24017781 | Zfp24 | NM_021559 | c.A307T, p.I103F |
| ChrX:124127783-124127783 | Vmn2r121 | NM_001100616 | c.A2539T, p.N847Y |
| Location | Gene | Accession No. | Amino Acid Change | Type |
|---|---|---|---|---|
| Chr3:15411378-15411378 | Sirpb1a | NM_001002898 | c.G559A, p.D187N | nonsynonymous SNV |
| Chr5:145803665-145803665 | Cyp3a44 | NM_177380 | c.C164A, p.T55K | nonsynonymous SNV |
| Chr5:94535811-94535811 | Pramel42 | NM_001243937 | c.T299A, p.L100H | nonsynonymous SNV |
| Chr6:29441097-29441102 | Flnc | NM_001081185 | c.1051_1054del, p.V351Pfs*16 | frameshift deletion |
| Chr7:35409643-35409645 | Cep89 | NM_028120 | c.546_548del, p.S190del | non-frameshift deletion |
| Chr8:122478985-122478987 | Ctu2 | NM_153775 | c.546_548del, p.Q190del | non-frameshift deletion |
| Chr8:22935648-22935650 | Kat6a | NM_001081149 | c.3208_3210del, p.E1077del | non-frameshift deletion |
| Chr9:99583673-99583675 | Dbr1 | NM_031403 | c.1303_1305del, p.E444del | non-frameshift deletion |
| Chr12:8728945-8728947 | Pum2 | NM_030723 | c.1516_1518del, p.Q513del | non-frameshift deletion |
| Chr14:98168891-98168893 | Dach1 | NM_007826 | c.417_419del, p.S156del | non-frameshift deletion |
| Chr15:101433138-101433138 | Krt87 | NM_001003668 | c.T1226C, p.I409T | nonsynonymous SNV |
| Chr15:98846446-98846448 | Kmt2d | NM_001033276 | c.10830_10832del, p.Q3610del | non-frameshift deletion |
| Chr17:23348034-23348034 | Vmn2r115 | NM_001104579 | c.G1519T, p.G507X | stopgain |
| Chr17:35873852-35873854 | Ppp1r18 | NM_175242 | c.1678_1680del, p.E570del | non-frameshift deletion |
| Chr17:46412515-46412517 | Zfp318 | NM_207671 | c.5443_5445del, p.E1823del | non-frameshift deletion |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.-L.; Huang, W.-C.; Cheng, S.-P.; Chen, M.-J.; Lin, C.-H.; Chang, S.-C.; Chang, Y.-C. Characterization of Mammary Tumors Arising from MMTV-PyVT Transgenic Mice. Curr. Issues Mol. Biol. 2023, 45, 4518-4528. https://doi.org/10.3390/cimb45060286
Liu C-L, Huang W-C, Cheng S-P, Chen M-J, Lin C-H, Chang S-C, Chang Y-C. Characterization of Mammary Tumors Arising from MMTV-PyVT Transgenic Mice. Current Issues in Molecular Biology. 2023; 45(6):4518-4528. https://doi.org/10.3390/cimb45060286
Chicago/Turabian StyleLiu, Chien-Liang, Wen-Chien Huang, Shih-Ping Cheng, Ming-Jen Chen, Chi-Hsin Lin, Shao-Chiang Chang, and Yuan-Ching Chang. 2023. "Characterization of Mammary Tumors Arising from MMTV-PyVT Transgenic Mice" Current Issues in Molecular Biology 45, no. 6: 4518-4528. https://doi.org/10.3390/cimb45060286
APA StyleLiu, C.-L., Huang, W.-C., Cheng, S.-P., Chen, M.-J., Lin, C.-H., Chang, S.-C., & Chang, Y.-C. (2023). Characterization of Mammary Tumors Arising from MMTV-PyVT Transgenic Mice. Current Issues in Molecular Biology, 45(6), 4518-4528. https://doi.org/10.3390/cimb45060286







