Abstract
Osteosarcoma describes a tumor of mesenchymal origin with an annual incidence rate of four to five people per million. Even though chemotherapy treatment has shown success in non-metastatic osteosarcoma, metastatic disease still has a low survival rate of 20%. A targeted therapy approach is limited due to high heterogeneity of tumors, and different underlying mutations. In this review, we will summarize new advances obtained by new technologies, such as next generation sequencing and single-cell sequencing. These new techniques have enabled better assessment of cell populations within osteosarcoma, as well as an understanding of the molecular pathogenesis. We also discuss the presence and properties of osteosarcoma stem cells—the cell population within the tumor that is responsible for metastasis, recurrence, and drug resistance.
1. Introduction
Osteosarcoma describes a malignant mesenchymal tumor whose cells produce osteoid, a non-mineralized bone matrix. According to the classification of tumors of soft tissues and bones by the World Health Organization (WHO), there are several types of osteosarcomas, of different malignancy grades. The subject of this review article is the conventional (classical) osteosarcoma (OS), which, according to the WHO, also includes secondary osteosarcomas. OS is the most common primary non-hematopoietic malignant bone tumor, with a worldwide incidence of 3.4 per million [1], which accounts for about 35% of all primary malignant bone tumors. It is the most common primary malignant mesenchymal tumor in children and adolescents, and in that age group, it accounts for about 80% of all malignant bone tumors. Over the age of fifty, OS accounts for about 50% of all primary malignant bone tumors, with a higher share of secondary OS, and is in second place in the overall share of primary malignant bone tumors in this age group [2]. This tumor has a highly heterogeneous genetic profile, but advances were made in understanding its biology and revealing genetic aberrations that define possible patient subgroups. These developments were made feasible by the emergence of thoroughly annotated tissue banks, along with the advancement and broader availability of technologies for detailed molecular profiling [3]. This review aims to gather current knowledge in the field of osteosarcoma molecular biology.
2. Cell Populations in Osteosarcoma
OSs are very complex tumors, because they contain heterogeneous sets of cells that differ in morphology, phenotype, gene expression, metabolism, immunogenicity, and proliferative and metastatic potential. Osteosarcomas grow in very complex and dynamic bone microenvironments, made up of bone cells (osteoclasts, osteoblasts, and osteocytes), stromal cells (mesenchymal stem cells and fibroblasts), vascular cells (endothelial cells and pericytes), immune cells (macrophages and lymphocytes), and a mineralized extracellular matrix (ECM).
Precise intratumor cell populations could not be distinguished using conventional transcriptomics due to insufficient resolution of the bulk analysis. However, single-cell RNA sequencing (scRNA-seq) provided insight into the complex intratumor heterogeneity and tumor interaction with the microenvironment. Zhou et al. performed scRNA-seq on 7 primary, 2 recurrent, and 2 lung metastatic osteosarcomas and identified 11 cell clusters based on unbiased clustering of gene expression profiles and canonical markers [4]. Different cell clusters included osteoblastic cells, myeloid cells, osteoblastic proliferative cells, tumor-infiltrated lymphocytes (TILs), chondroblastic cells, endothelial cells, mesenchymal stem cells (MSCs), pericytes, fibroblasts, and myeloblasts [4,5]. In an analysis performed by Wu et al. [5], the main cell types were osteoblastic, myeloid, osteoblastic proliferative, and osteoclasts. However, the composition changed in metastatic OS, and included a large proportion of TILs, osteoblastic cells, and MSCs. In recurrent OS, there was a large proportion of chondroblastic cells. TILs were recruited to the tumor site, and several studies pointed out their importance for clinical outcome. The higher infiltration of immune cells was correlated with better overall survival and progression-free survival of OS patients [4].
A plethora of growth factors are released from the degraded bone matrix, as they establish a vicious cycle of bone remodeling processes. Pediatric osteosarcomas have a peak incidence around the time of the puberty, mostly located in the growth plate of the long bones. This suggests a connection between intensive bone growth and OS [6]. Bone remodeling relies on precise communication between various cell types crucial for bone homeostasis, including bone matrix-secreting osteoblasts and bone-degrading osteoclasts. MSCs are a part of the bone marrow niche, and they can differentiate into osteoblasts under the control of specific transcription factors. Growth factors, such as transforming growth factors (TGFs) and fibroblast growth factors (FGFs), must first engage a sophisticated signal transduction cascade in order to activate those transcription factors [7]. Most of these cytokines and growth factors are connected to the development of OS. The tumor microenvironment is rich in TGF-β, a factor that stimulates growth and metastasis. TGF-β-1 is mostly linked to the formation of OS during the growth of primary tumors or metastatic progression [8]. FGFs are crucial regulators of skeletal development, as FGF18 is required for the maturation of osteoblasts and FGF2 is essential for the growth and development of pre-osteoblasts [9]. Mostly, osteosarcomas are osteolytic, suggesting increased osteoclast activity [10]. Osteoclast activity is regulated by many cytokines and growth factors, such as macrophage-colony stimulating factor (M-CSF), insulin-like growth factors (IGFs), parathyroid hormone (PTH), RANK ligand (RANKL), and osteoprotegerin (OPG).
3. Cellular Theory of Osteosarcoma Tumorigenesis
3.1. Osteosarcoma Stem Cells
The tumorigenesis of OS is not fully explained; however, as for other malignant tumors, it is complemented by different theories, the latest being the theory about tumor stem cells. According to that theory, in the evolutionary progression of a malignant tumor or its recurrence after therapy, the main unit of selection is the cell, and, in particular, the one that has a high potential for a self-renewal—the cancer stem cell (CSC). The hypothesis of the CSC’s existence was created as part of experiments on the transplantation of leukemic cells, and although it was published as a general feature of all malignant tumors, it is still disputed [11,12,13]. This theory also applies to OS stem cells, OS-CSCs, which is supported by clinical cases describing the appearance of metastases after more than 20 years of remission [14].
OS-CSCs theoretically arise either by transformation of undifferentiated MSCs, or by transformation of directed, progenitor cells of the osteoblastic lineage [15]. MSCs are non-hematopoietic precursor cells found in numerous tissues, including bone marrow, peripheral blood, placenta, umbilical cord, and adipose tissue, which show the ability to self-renew and differentiate into various mesenchymal cell lines, such as osteocytes, chondrocytes, myocytes, and adipocytes, and have roles in wound healing [16]. According to this assumption, MSCs are recruited from the bone marrow (BM-MSCs) in response to genetic and epigenic changes and microenvironmental signals [17]. The histological type of mesenchymal tumors may be related to the type of oncogenic stress in MSCs. Thus, for example, it was found that a loss of p53 expression and a simultaneous loss of expression of the Rb and p53 genes led to the formation of leiomyosarcoma in vivo, regardless of the source of MSCs [16]. BM-MSCs (Rb -/- p53 -/-) differentiated into the osteogenic lineage and developed tumors in vivo with histological features consistent with OS [18,19]. A change in cell cycle regulators can also occur in osteoblast precursors, which indicates that they could also be a source of OS, orchestrated by signals from the tumor microenvironment and responses to genetic and epigenic changes within these cells [20].
OS-CSCs interact with local cells. Circulating normal MSCs that enter a tumor can support its growth, enhance invasiveness, and participate in the creation of metastatic niches in cooperation with tumor cells. Under the influence of MSCs, dedifferentiation of OS cells can occur so that they show stemness properties [21,22]. Among other ways, this intercellular communication occurs via vesicles. Researching the influence of the aforementioned vesicles secreted from OS cells, it was found that MSCs can internalize vesicles, after which such so-called tumor-directed MSCs play a role in the promotion of OS growth and metastasis [23]. Vesicles secreted from OS cells contain TGF-β (transforming growth factor beta), which is important for the development of the CSC’s phenotype [24].
CSCs play a significant role in achieving tumor heterogeneity, by constantly enriching the tumor mass with new mutated cells and dominant subclones, and by regulating the microenvironment of tumor cells [25]. Generally, the presence of stem cells defines the microenvironment in which they reside, and the microenvironment influences the properties of stem cells. It consists of CSCs, fibroblasts, inflammatory cells, capillaries, extracellular matrix, and chemokines, and cytokines [26]. This microenvironment facilitates the entry of CSCs into a state of rest, retention of stemness properties, and self-renewal, and provides them physical support. It strengthens the resistance of CSCs to stresses, such as hypoxia, the immune system, antitumor drugs, and radiotherapy [27,28]. The tumor can colonize tissue locally and remotely, and CSCs play a key role in this process, as they contribute to the creation and maintenance of the tumor niche [29].
Fibroblasts associated with a tumor (cancer-associated fibroblasts, CAFs) are activated fibroblasts that share similarities with normal fibroblasts, and are stimulated by inflammatory conditions associated with tumor development. They represent a significant component of the stroma in tumor tissue, surround tumor cells, and play an important role in mechanical support, proliferation, survival, angiogenesis, metastasis, and immunogenicity [30]. Stem properties can be enhanced by conditioned medium from CAFs, mediated by the presence of paracrine-secreted molecules that suppress tumor differentiation, in which, for example, FGF (fibroblast growth factor) plays an important role [31]. Poor differentiation of the human OS cell line MG-63 could be associated with overproduction of basic FGF (bFGF) and bFGF receptors [32].
OS-CSCs may represent a separate therapeutic target that complements conventional treatment, so elucidating the characteristics and role of CSCs in OS may improve its treatment [33]. The necessity to understand the complex osteosarcoma microenvironment and CSC niche led to the development of new, better research models—tissue-engineered 3D models [34]. Wang et al. (2022) used 3D printing technology to fabricate a PLLA-based biomimetic scaffold resembling the physiochemical environment of cortical bones. A comparison of the transcriptome data revealed that 3D models can recapitulate OS properties better than patient biopsy samples and better than spheroid cultures. KEGG pathway analysis confirmed 20 enriched pathways, especially metabolic, MAPK, PI3K, and TGFbeta/SMAD pathways, in engineered tumors in comparison to spheroid cultures [35].
3.2. Phenotypic Characteristics of Osteosarcoma Stem Cells
Gibbs et al. were the first to prove the existence of OS-CSCs by identifying a subpopulation of cells that were able to form sarcospheres (Figure 1) in serum-free conditions in OS tissue samples obtained from patients [36].
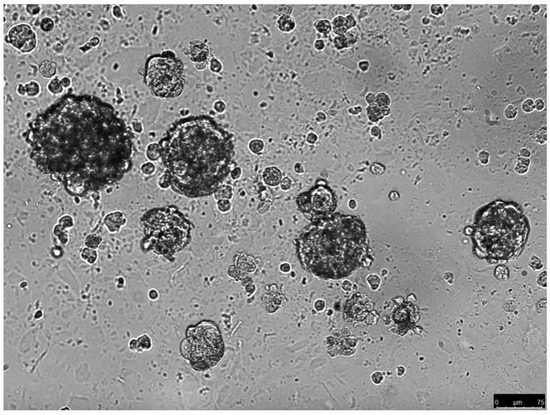
Figure 1.
Sarcospheres growing in serum-free conditions (unpublished data). The spheres were derived from the parental cell culture of an osteosarcoma biopsy. The spheres were cultured in methyl-cellulose with serum-free medium for 14 days.
It is believed that one of the mechanisms by which CSCs achieve drug resistance is by increasing the level and activity of transporters responsible for the elimination of drugs from the cell, such as MDR/ABC (multidrug resistant transporters/ATP-binding cassette) [37]. This feature is used to identify CSCs by measuring the ability of the cell to release a dye that binds to DNA (Hoechst 33342 or Rhodamine 123) and sorting the cells by measuring fluorescence [38]. Cells that express a high level of the mentioned transporters emit color during the analysis and are also labeled as side populations in the literature [39]. The presence of a cell population that eliminates Hoechst 33342 dye has been demonstrated in OS cell lines and in human primary OS [40]. Martins-Neves et al. have identified such subpopulations of cells in the MNNG/HOS tumor line that showed resistance to antitumor drugs and radiotherapy [41].
OS-CSCs are phenotypically similar to stem cells due to the expression of genes for the Oct-3 and Oct-4 proteins (transcription factors that bind octamer 3 and octamer 4, respectively), which play a role in the self-renewal of undifferentiated embryonic stem cells and are often used as markers for undifferentiated cells. In addition to them, OS-CSCs also express the Nanog protein, encoded by the Nanog gene, which is a transcription factor that helps embryonic stem cells (ESCs) to maintain pluripotency by suppressing differentiation cell factors. Therefore, Nanog gene deletion triggers ESC differentiation. Basu Roy et al. found cells that have an increased ability to grow in non-adherent conditions by forming spheres, express the stem cell markers Sox2 (SRY-box transcription factor 2) and Sca-1 (stem cell antigen 1) in lineage and non-lineage OS samples [42]. Loss of Sox2 gene function resulted in enhanced OS cell differentiation and decreased clonality, migration, and invasiveness [43].
In the last few decades, numerous studies have tried to identify specific markers and properties of OS-CSCs [44]. In addition to the expression of stem cell markers shared with ESCs, OS-CSCs have the ability to form cell spheres in vitro, which are highly tumorigenic in vivo and have a high level of aldehyde dehydrogenase-1 (ALDH-1) [45]. The ALDH-1 expression is associated with resistance to antitumor drugs and enhanced metastatic potential in tumor cells [46]. Adhikari et al. showed the presence of cells expressing CD117 (a protein that binds to stem cell factor, SCF) and Stro-1 (MSCs marker) in mouse and human OS cell lines. The double-positive cell fraction, compared to the double-negative cell fraction, showed a greater ability to form spheres, an increased resistance to doxorubicin treatment, greater metastatic potential, and overexpression of the ABCG2 drug efflux transporter (ATP-binding cassette super-family G member 2) [47]. CD44 is another marker of MSCs and plays a role in the migration and metastasis of OS-CSCs [48,49]. CD133, used as a marker for CSCs of various types of tumors, including OS, is strongly expressed in patients with lung metastases and a poor prognosis. Oct4+ OS cells that also expressed CD105 had a greater propensity to metastasize to the lungs [50,51]. CD105 is a cell membrane glycoprotein mainly expressed on endothelial cells and overexpressed in the tumor endothelium, and is a component of the receptor for TGF-β [52]. Furthermore, OS-CSCs express CD271, a low-affinity receptor for neural crest growth factor and a brain stem cell marker. CD271+ cells showed resistance to antitumor drugs and higher sphere-forming potential, as well as higher tumorigenic potential after injection into nude mice [53].
4. Driver Mutations and Epigenetic Changes of Osteosarcoma
4.1. Coding DNA Changes in Osteosarcoma
Osteosarcoma develops under the pressure of random mutational changes, and its growth is led by preferential clonal proliferation and epigenetic modifications [54]. Understanding the pathogenesis of osteosarcoma is complicated by the low tumor incidence and significant genetic differences between subtypes. For easier understanding, we will categorize OS into two groups based on their origin—hereditary syndromes, and sporadic OS that can be further categorized as pediatric OS or adult OS.
Several hereditary syndromes and genetic alterations have been linked to OS, such as Li–Fraumeni and retinoblastoma syndromes, as well as germline mutations in genes encoding DNA helicases of the RecQ family [55].
Li–Fraumeni syndrome (LFS), caused by a germline mutation in the TP53 gene, is the syndrome that most frequently predisposes children to sarcomas. TP53 encodes for p53, a master transcription factor, and about one-third of people with LFS develop OS. Therefore, it is no surprise that loss of the tumor suppressor function of p53 also happens in sporadic cases of OS [56]. Approximately 20% of OS cases have mutations in the TP53 gene [55]. Protein p53 is involved in many antitumor cellular processes regulating cell growth, cell division, apoptosis, and DNA repair. Mutations in OS may result in loss of function or gain of function. The majority of mutations are missense mutations localized in the DNA binding domain. Additionally, 74% of OS cases have structural variations (SV) and somatic nucleotide variations (SNV). Among those, translocations with a breakpoint in the first intron result in the loss of TP53 expression [57]. Other intron mutations result in splice site changes or a frameshift in sequence [58]. Amplification of p53 main negative regulator MDM2 is also found in 10% of OS cases as a part of a larger amplicon containing CDK4, MDM2, and SAS [59]. In total, 32 variants of the TP53 gene were found in tumor samples from 765 patients with LFS [60] as well as some heterozygous mutations of the CHK2 gene [61].
Another tumor suppressor gene linked with OS is Rb, which codes for pRb. The carriers of a germline mutation (autosomal dominant) of RB have an increased risk of various neoplasms, especially OS. The incidence of OS in hereditary retinoblastoma is 400 times higher than in the general population [62]. The Rb gene is often altered in 70% of sporadic OS cases. It is affected by point mutations or structural variations. The very common finding is a loss of heterozygosity at 13q [63]. The Rb protein is an important guardian of cell cycle entry, and loss of its function leads to uncontrolled cell division. Other upstream and downstream components of the Rb pathway can also be affected by mutations in OS cases. pRb is regulated by phosphorylation via complexes of cyclin-dependent kinases and their coactivator cyclins. Phosphorylation of pRb activates cell division. CDK4 and cyclin D1 are found upregulated in OS, while CDK inhibitors are commonly lost [59].
It has been shown that a loss of RecQ helicases, which unwind DNA prior to replication, also represents an increased risk of developing OS (Table 1). Germline mutations in the RecQ family of genes cause rare recessive autosomal cancer predisposition syndromes—Rothmund–Thompson II (RTS II), RAPALIDINO, and Bloom and Werner syndromes—which are associated with an increased frequency in OS development [55,64].

Table 1.
Osteosarcoma-predisposing syndromes caused by mutations in the RecQ family gene.
Another syndrome, named Diamond–Blackfan anemia, is caused by a mutation in the gene for ribosomal protein, and is classified as ribosomopathy. Two studies reported an increased risk for OS in Diamond–Blackfan anemia patients [65], and mutations were found in the genes RPS19, RPL5, RPL11, RPL35A, RPS24, RPS17, RPS7, RPS10, and RPS26.
The c-Myc gene is mutated in more than 10% of OS cases. It is a known oncogene, mainly affecting cell proliferation and migration by activating the MAP kinase pathway [56]. A high expression of Myc correlates with metastatic disease and poor prognosis [55].
In adult OS cases, 14% of tumors present IGF1 receptor amplification [66]. In addition to previously described genes, sequencing data revealed other driver mutations of Notch1, FOS, NF2, WIF1, BRCA2, APC, PTCH1, and PRKAR1A. Additionally identified were synergistic genes including Rb1, TWIST, PTEN, and JUN [67].
Two very distinguished mutational processes are involved in osteosarcoma genesis, causing a heterogenous intratumoral profile of OS. The first one is complex chromosomal rearrangement, also known as chromothripsis, present in about 77% of OS cases [68]. It is a single catastrophic event that results in several genomic rearrangements in one or more chromosomes, including amplifications of CDK4, MDM2, COPS3, gains of RICTOR, TERT, and losses of TP53 and NF1 [69]. The second process is kataegis—an accumulation of mutations at localized regions (50–85% of cases) [70]. Those localized hypermutated regions are common in the regions of the TP53 and ATRX genes [71].
4.2. Non-Coding RNA Changes in Osteosarcoma [55,56,60,61,62,63,64,65,66,67,68,69,70,71]
Aside from alterations in the protein-coding genes, it is evident that many non-coding RNA (ncRNA) genes are dysregulated in OS. ncRNAs are commonly used as tumor markers due to easy sample acquisition, and higher sensitivity and specificity than protein markers. Clinically relevant ncRNAs include microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA). miRNAs are often 18–25 nucleotides in length and regulate gene expression targeting specific mRNAs via complementary binding with 3′UTR of mRNAs [55,56,60,61,62,63,64,65,66,67,68,69,70,71]. lncRNAs have over 200 nucleotides and exert their functions mainly via sponging miRNAs and targeting specific substrates.
lncRNAs act as miRNA sponges, preventing miRNA from acting on mRNA, therefore enhancing the translation of target mRNA (Figure 2). Interestingly, miRNA can also negatively regulate the expression of lncRNA. In the context of osteosarcoma, the affected signaling pathways are Notch, PI3K/AKT, JNK, and Wnt [72], important for cell proliferation, cell survival, self-renewal, and drug resistance. One example is lncRNA SNGH12 inhibiting miR-195-5p, which inhibits Notch2. Very similar is the case of lncRNA SNGH7 that inhibits miR-34a, inhibiting Notch1, BCL-2, Ki-67, and CDK6. As a consequence, in osteosarcoma, Notch1 or 2 signaling is upregulated, promoting tumorigenesis and metastases [73,74]. JNK and P13/AKT signaling pathways are induced in osteosarcoma cells via axes lncRNA CAT104—miR-381—mRNA ZEB1 [75], lncRNA MEG3—miR 127—mRNA ZEB1 [76], and lncRNA ATB—miR-200—mRNA ZEB1 and ZEB2 [77]. Inhibition of apoptosis, and cell cycle arrest, is the result of axes lncRNA PVT1—miR-195, miR-497—BCL2, CCND1 and HK2 [78], lnc RNA SNHG20—miR-139—RUNX2 [79], lnc RNA TUG1—miR132-3p, and miR-212-3p—SOX4, FOXA1 [80].
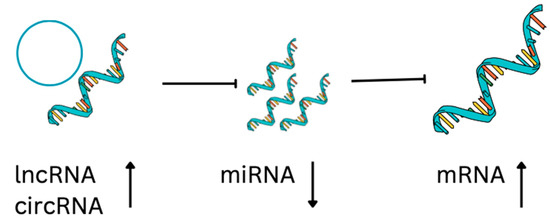
Figure 2.
Regulatory ncRNA-mRNA axis. lncRNA and circRNA can suppress the miRNA that sponges mRNA. As a result of their presence, gene expression is upregulated. As a result of their absence, gene expression is downregulated.
Additionally, lncRNAs can suppress osteosarcoma proliferation and metastasis if the result of miRNA sponging is the upregulation of tumor suppressors. The examples affecting osteosarcoma cells are axes lncRNA CASC2—miR-181a—RASSF6 [81], PTEN and ATM, lncRNA-p21—miR-130b and PTEN [82], lncRNA NBAT1—miR-21—PTEN, PDCD4, TPM1, and RECK [83]. The situation between ncRNAs is even more complex because miRNA can negatively regulate lncRNA, and there is also reciprocal regulation between lncRNA and miRNA [72] (Table 2).
CircRNAs are ssRNAs that form closed loops and modulate gene expression, splicing, and sponging of miRNAs, and interact with RNA binding proteins [84] (Figure 2). For example, Qiu et al. [85] identified 15 downregulated circRNAs, 136 upregulated miRNAs, and 52 downregulated mRNAs from an OS database, in which 14 circRNAs, 24 miRNAs, and 52 mRNAs formed a network affecting common cancer-related pathways [84]. For example, CircTVF25 promotes cell proliferation and migration, upregulating cyclin D1, CDK6, MMP2, MMP9, and vimentin. circTCF25 reduces miR-206-inducing pathways MEK/ERK and AKT/mTOR [86]. CircPVT1 increases metastatic potential of osteosarcoma cells by promoting epithelial to mesenchymal transition. CircPVT1 reduces miR-205-5p activation of c-FLIP [87]. CircMMP9 is present in advanced osteosarcoma, it sponges miR-1265 activating chitinase-3-like protein 1 (CHI3L1) circMMP [88] (Table 3).

Table 2.
lncRNA in osteosarcoma.
Table 2.
lncRNA in osteosarcoma.
| Signaling Patway | lncRNAs | References |
|---|---|---|
| Notch | SNHG 12 | [75] |
| SNGH 7 | [76] | |
| PIK3/AKT | CCAT1 | [89] |
| DANCR | [90] | |
| GAS5 | [91] | |
| AKT | lncRNA-p21 | [84] |
| JNK and Wnt | CAT104 | [77] |
| MEG3 | [78] | |
| ATB | [79] | |
| p38/ERK/AKT and Wnt/β-catenin | C2dat1 | [92] |
| PI3K/AKT, JAK/STAT, and Notch | TUG1 | [93] |
| NEAT1 | [94] | |
| PVT1 | [95] | |
| APTR | [88,96] |

Table 3.
circRNA in osteosarcoma.
Table 3.
circRNA in osteosarcoma.
| Signaling Patway | circRNA | References |
|---|---|---|
| MEK/ERK and AKT/mTOR | circTCF25 | [88] |
| Akt/PI3K and Bcl-2 | circ-0001785 | [97] |
| Atk/PTEN | circ_ORC2 | [98] |
| MEK and ERK | circ-ITCH | [99] |
| VEGF, CDK4 | Circ_001621 | [100] |
| CREB3 | CircTADA2A | [101] |
| SOCS3 and STAT3 | Circ_ANKIB1 | [102] |
| β-catenin | circMYO10 | [103] |
4.3. Osteosarcoma Extracellular Vesicles
Extracellular vesicles (EV) are released from the cells in the extracellular matrix. Vesicles are surrounded by membranes and they carry cargo collected within the cell, enabling communication within the microenvironment, as well as distant communication. Their cargo is composed of proteins, DNA (mitochondrial and fragmented genomic), RNA (mRNA and ncRNA), lipids, and metabolites [104]. EV derived from osteosarcoma cells and mesenchymal stem cells (MSCs) from the tumor microenvironment are important for osteosarcoma progression, invasion, and metastasis. Via EV, OS cells can establish crosstalk with surrounding cells (osteoblasts, osteoclasts, osteocytes, MSCs, fibroblasts, immune cells, endothelial cells, and pericytes), modulate the epigenetic status of MSCs, and prepare a premetastatic niche. Many of the previously mentioned lncRNAs, miRNAs, and circRNAs are found in the EVs secreted by OS cells affecting angiogenesis, matrix remodeling, proliferation, and migration [105]. Examples of miRNAs in the EVs that support OS metastasis are miR-21 [106], miR-1307 [107], miR-675 [108], and miRNAs that change the OS microenvironment are miR-148-3p and miR-148-5p [109]. OS EVs also contain proteins that help the immune scape, such as PD-L1 [110]. EVs play an important role in the multidrug resistance of osteosarcoma. EVs transfer MDR-1 mRNA [111] and miR-135b [112], as well as circRNA_103801 [113], spreading resistance to chemotherapy-sensitive cells.
EVs can be easily collected from biological fluids (blood, urine, saliva, breast milk, amniotic fluid, cerebrospinal fluid, pleural effusion, and bronchoalveolar lavage fluid), and used for prognosis, diagnosis, and evaluation of the progression of osteosarcoma [114]. Potential biomarkers are proteins PD-L1 and N-cadherin [115], and ncRNAs miR-675, miR-25-3p, miR-21-5, miR143-3p, miR-101, and has-circ-103801 [104,110,116,117]. The interesting potential application of EVs is the delivery of therapeutics, especially for delivering ncRNA that will induce tumor suppressor activity, as well as reduce metastatic potential and cell proliferation.
5. Therapy and Resistance
Current treatment of osteosarcoma includes surgical resection and systemic chemotherapy, resulting in the 5-year event-free survival of approximately 70% of patients with localized disease. Unfortunately, overall survival rates are less than 20% in patients with metastatic and recurrent disease [118]. The outcome for patients has not changed for decades, despite many new targeted therapies for cancer.
One reason for this is the complexity of osteosarcoma molecular mechanisms and the influence of the microenvironment and immune system. Therefore, transcriptome and proteome analyses could point out differentially expressed genes in osteosarcoma and provide information about new potential therapeutic targets. These data were used by Chaiyawat et al. (2017) for generation of the list of therapeutic targets for already-approved drugs. Upregulated proteins with FDA-approved antineoplastic drugs were categorized as epigenetic regulators (DNMT1, HDAC1, and HDAC2), kinases (ERBB2, FGFR1, KIT, MET, MTOR, and PDGFRalpha), proteasomes (PSMC5 and PSMC6) and other drugs (GSR and PARP1). The analysis was also performed in order to identify proteins that can be targeted with non-antineoplastic drugs approved for other diseases. Among immunosuppressants for rheumatic arthritis, leflunomide, the inhibitor of DHODH, can be repurposed for osteosarcoma. Digoxin, a drug in the group of cardiac glycosides, can also be repurposed for osteosarcoma [119]. Additionally, patient-derived 2D and 3D cultures of giant cell tumors of bone and desmoplastic fibroma showed an upregulation of genes involved in bone vicious-cycle-related genes (RANK-L, RANK, OPN, CXCR4, RUNX2, FLT1), suggesting the therapeutic combination of denosumab and lenvatinib. The clinical data confirmed the usefulness of molecular and pharmacological results [120].
The second reason for poor outcome is that an osteosarcoma is considered a drug-resistant tumor. Chemotherapy resistance can be linked to drug import and efflux, intracellular detoxification, resistance to apoptosis, DNA damage repair, tumor microenvironment, immunity, and aquired mutations of the drug target. In order to achieve a better drug response, it is necessary to unravel the underlying mechanisms of osteosarcoma resistance to treatment. The main causes for methotrexat resistance are inefficient drug delivery due to poor vascularisation, an acidic tumor microenvironment that inactivates the drug [121], and impaired cellular influx [122]. Drug efflux is mediated by a membrane-bound pump encoded by the MDR1 gene, and doxorubicin-resistant cell lines strongly induce its expression [123]. The cytotoxic effect of doxorubicin is based on its DNA intercalation and binding to topoisomerase, leading to apoptosis. The topoisomerase status may be a progonostic factor in survival and response to doxorubicin [6,124]. Alterations in p53 activity and apoptotic pathways also contribute to therapy resistance. The nature of the p53 aberrations is very heterogenic in osteosarcoma patients, providing controversial data regarding its contribution to therapy resistance. However, some individuals can benefit from p53 reactivation via small molecule RITA [125]. Osteosarcoma cells upregulate DNA repair pathways to fight direct and indirect DNA damage caused by cisplatin, ifosfamide, and doxorubicin. A pathway involved in the repair of DNA damage linked to chemotherapy resistance is nucleotide excision repair (NER). Upregulation of the involved protein ERCC1 and polymorphisms of ERCC2 are markers of poor prognosis [126].
6. Conclusions
Osteosarcomas exert high inter- and intratumor heterogeneity. A cellular background of the disease reveals that tumors consist of many cell subpopulations, including predominantly cells of mesenchymal origin, osteoclasts, and immune cells. Among those, there are populations of osteosarcoma cancer stem cells that are responsible for drug resistance, disease recurrence, and metastases. A genetic background reveals that osteosarcoma has very variable genetic alterations, but can be grouped by hereditary and sporadic subtypes. Since there are two peaks of incidence, the sporadic type is divided into pediatric- and adult-onset cases. Driver mutations generally include dysregulation of cell division, inactivation of p53 and pRb, and interference with DNA repair mechanisms. Dysregulation of signaling does not rely only on mutations, but also on ncRNA networks. ncRNA networks include the interactions between circRNAs, lncRNAs, and microRNAs that modulate the mRNA levels of oncogenes and tumor suppressors. Tumor cells also educate surrounding cells, changing local and distant environments by secretion of exosomes packed with proteins, as well as coding and non-coding RNA molecules. The proinflammatory microenvironment supports progression via cytokines and growth factors secreted from immune cells, osteoclasts, and tumor stroma.
Altogether, complexity and heterogeneity of the osteosarcoma system are the key reasons why targeted therapies have limited success and chemotherapy still dominates in OS treatment. Special focus should be put into combination therapies that target proliferating cells and cancer stem cells, change the microenvironment, improve the immune response, and fight drug resistance.
Funding
This research was funded by the Croatian Science Foundation, grant numbers IP-2018-01-7590 and IP-2020-02-9559.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mirabello, L.; Troisi, R.J.; Savage, S.A. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int. J. Cancer 2009, 125, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Creytens, D. Molecular Classification of Soft Tissue and Bone Tumors. Diagnostics 2021, 11, 2326. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.; Gorlick, R. Advancing therapy for osteosarcoma. Nat. Rev. Clin. Oncol. 2021, 18, 609–624. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, D.; Yang, Q.; Lv, X.; Huang, W.; Zhou, Z.; Wang, Y.; Zhang, Z.; Yuan, T.; Ding, X.; et al. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat. Commun. 2020, 11, 6322. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Dou, X.; Li, H.; Sun, Z.; Li, H.; Shen, Y.; Weng, W.; Min, J. Identification of Cell Subpopulations and Interactive Signaling Pathways From a Single-Cell RNA Sequencing Dataset in Osteosarcoma: A Comprehensive Bioinformatics Analysis. Front. Oncol. 2022, 12, 853979. [Google Scholar] [CrossRef]
- Lilienthal, I.; Herold, N. Targeting Molecular Mechanisms Underlying Treatment Efficacy and Resistance in Osteosarcoma: A Review of Current and Future Strategies. Int. J. Mol. Sci. 2020, 21, 6885. [Google Scholar] [CrossRef] [PubMed]
- Corre, I.; Verrecchia, F.; Crenn, V.; Redini, F.; Trichet, V. The Osteosarcoma Microenvironment: A Complex but Targetable Ecosystem. Cells 2020, 9, 976. [Google Scholar] [CrossRef] [PubMed]
- Verrecchia, F.; Rédini, F. Transforming Growth Factor-β Signaling Plays a Pivotal Role in the Interplay Between Osteosarcoma Cells and Their Microenvironment. Front. Oncol. 2018, 8, 133. [Google Scholar] [CrossRef]
- Zhou, W.-Y.; Zheng, H.; Du, X.-L.; Yang, J.-L. Characterization of FGFR signaling pathway as therapeutic targets for sarcoma patients. Cancer Biol. Med. 2016, 13, 260–268. [Google Scholar] [CrossRef]
- Kansara, M.; Teng, M.W.; Smyth, M.J.; Thomas, D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer 2014, 14, 722–735. [Google Scholar] [CrossRef]
- Mazzocca, A. The Systemic–Evolutionary Theory of the Origin of Cancer (SETOC): A New Interpretative Model of Cancer as a Complex Biological System. Int. J. Mol. Sci. 2019, 20, 4885. [Google Scholar] [CrossRef] [PubMed]
- Ryu, D.; Joung, J.-G.; Kim, N.K.D.; Kim, K.-T.; Park, W.-Y. Deciphering intratumor heterogeneity using cancer genome analysis. Hum. Genet. 2016, 135, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Abarrategi, A.; Tornin, J.; Martinez-Cruzado, L.; Hamilton, A.; Martinez-Campos, E.; Rodrigo, J.P.; González, M.V.; Baldini, N.; Garcia-Castro, J.; Rodriguez, R. Osteosarcoma: Cells-of-Origin, Cancer Stem Cells, and Targeted Therapies. Stem Cells Int. 2016, 2016, 3631764. [Google Scholar] [CrossRef]
- Halldorsson, A.; Brooks, S.; Montgomery, S.; Graham, S. Lung metastasis 21 years after initial diagnosis of osteosarcoma: A case report. J. Med. Case Rep. 2009, 3, 9298. [Google Scholar] [CrossRef]
- Valent, P.; Bonnet, D.; De Maria, R.; Lapidot, T.; Copland, M.; Melo, J.V.; Chomienne, C.; Ishikawa, F.; Schuringa, J.J.; Stassi, G.; et al. Cancer stem cell definitions and terminology: The devil is in the details. Nat. Rev. Cancer 2012, 12, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Hass, R.; Kasper, C.; Böhm, S.; Jacobs, R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Commun. Signal. 2011, 9, 12. [Google Scholar] [CrossRef]
- Brune, J.C.; Tormin, A.; Johansson, M.C.; Rissler, P.; Brosjö, O.; Löfvenberg, R.; von Steyern, F.V.; Mertens, F.; Rydholm, A.; Scheding, S. Mesenchymal stromal cells from primary osteosarcoma are non-malignant and strikingly similar to their bone marrow counterparts. Int. J. Cancer 2011, 129, 319–330. [Google Scholar] [CrossRef]
- Rubio, R.; Gutierrez-Aranda, I.; Sáez-Castillo, A.I.; Labarga, A.; Rosu-Myles, M.; Gonzalez-Garcia, S.; Toribio, M.L.; Menendez, P.; Rodriguez, R. The differentiation stage of p53-Rb-deficient bone marrow mesenchymal stem cells imposes the phenotype of in vivo sarcoma development. Oncogene 2013, 32, 4970–4980. [Google Scholar] [CrossRef]
- Wang, J.-Y.; Wu, P.-K.; Chen, P.C.-H.; Lee, C.-W.; Chen, W.-M.; Hung, S.-C. Generation of Osteosarcomas from a Combination of Rb Silencing and c-Myc Overexpression in Human Mesenchymal Stem Cells. STEM CELLS Transl. Med. 2016, 6, 512–526. [Google Scholar] [CrossRef]
- Quist, T.; Jin, H.; Zhu, J.-F.; Smith-Fry, K.; Capecchi, M.R.; Jones, K.B. The impact of osteoblastic differentiation on osteosarcomagenesis in the mouse. Oncogene 2015, 34, 4278–4284. [Google Scholar] [CrossRef]
- Le Nail, L.-R.; Brennan, M.; Rosset, P.; Deschaseaux, F.; Piloquet, P.; Pichon, O.; Le Caignec, C.; Crenn, V.; Layrolle, P.; Hérault, O.; et al. Comparison of Tumor- and Bone Marrow-Derived Mesenchymal Stromal/Stem Cells from Patients with High-Grade Osteosarcoma. Int. J. Mol. Sci. 2018, 19, 707. [Google Scholar] [CrossRef] [PubMed]
- Ridge, S.M.; Sullivan, F.J.; Glynn, S.A. Mesenchymal stem cells: Key players in cancer progression. Mol. Cancer 2017, 16, 31. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, G.; Chen, R.; Hua, Y.; Cai, Z. Mesenchymal stem cells in the osteosarcoma microenvironment: Their biological properties, influence on tumor growth, and therapeutic implications. Stem Cell Res. Ther. 2018, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Baglio, S.R.; Lagerweij, T.; Pérez-Lanzón, M.; Ho, X.D.; Léveillé, N.; Melo, S.A.; Cleton-Jansen, A.-M.; Jordanova, E.S.; Roncuzzi, L.; Greco, M.; et al. Blocking Tumor-Educated MSC Paracrine Activity Halts Osteosarcoma Progression. Clin. Cancer Res. 2017, 23, 3721–3733. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.L.; Liang, Y.; Xie, X.B.; Yin, J.Q.; Zou, C.Y.; Zhao, Z.Q.; Shen, J.N.; Wang, J. Enrichment of Osteosarcoma Stem Cells by Chemotherapy. Chin. J. Cancer 2011, 30, 426. [Google Scholar] [CrossRef]
- Kise, K.; Kinugasa-Katayama, Y.; Takakura, N. Tumor microenvironment for cancer stem cells. Adv. Drug Deliv. Rev. 2016, 99, 197–205. [Google Scholar] [CrossRef]
- Arnold, C.R.; Mangesius, J.; Skvortsova, I.-I.; Ganswindt, U. The Role of Cancer Stem Cells in Radiation Resistance. Front. Oncol. 2020, 10, 164. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.-G.; Lee, S.-H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef]
- Ayob, A.Z.; Ramasamy, T.S. Cancer stem cells as key drivers of tumour progression. J. Biomed. Sci. 2018, 25, 20. [Google Scholar] [CrossRef]
- Liu, J.; Mi, J.; Zhou, B.P. Metabolic rewiring in cancer-associated fibroblasts provides a niche for oncogenesis and metastatic dissemination. Mol. Cell. Oncol. 2016, 3, e1056331. [Google Scholar] [CrossRef]
- Geary, L.A.; Nash, K.A.; Adisetiyo, H.; Liang, M.; Liao, C.-P.; Jeong, J.H.; Zandi, E.; Roy-Burman, P. CAF-Secreted Annexin A1 Induces Prostate Cancer Cells to Gain Stem Cell–like Features. Mol. Cancer Res. 2014, 12, 607–621. [Google Scholar] [CrossRef]
- Bodo, M.; Lilli, C.; Bellucci, C.; Carinci, P.; Calvitti, M.; Pezzetti, F.; Stabellini, G.; Bellocchio, S.; Balducci, C.; Carinci, F.; et al. Basic Fibroblast Growth Factor Autocrine Loop Controls Human Osteosarcoma Phenotyping and Differentiation. Mol. Med. 2002, 8, 393–404. [Google Scholar] [CrossRef]
- Basu-Roy, U.; Basilico, C.; Mansukhani, A. Perspectives on cancer stem cells in osteosarcoma. Cancer Lett. 2013, 338, 158–167. [Google Scholar] [CrossRef]
- Bassi, G.; Panseri, S.; Dozio, S.M.; Sandri, M.; Campodoni, E.; Dapporto, M.; Sprio, S.; Tampieri, A.; Montesi, M. Scaffold-based 3D cellular models mimicking the heterogeneity of osteosarcoma stem cell niche. Sci. Rep. 2020, 10, 22294. [Google Scholar] [CrossRef]
- Wang, M.-L.; Xu, N.-Y.; Tang, R.-Z.; Liu, X.-Q. A 3D-printed scaffold-based osteosarcoma model allows to investigate tumor phenotypes and pathogenesis in an in vitro bone-mimicking niche. Mater. Today Bio 2022, 15, 100295. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, C.P.; Kukekov, V.G.; Reith, J.D.; Tchigrinova, O.; Suslov, O.N.; Scott, E.W.; Ghivizzani, S.C.; Ignatova, T.N.; Steindler, D.A. Stem-Like Cells in Bone Sarcomas: Implications for Tumorigenesis. Neoplasia 2005, 7, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Begicevic, R.-R.; Falasca, M. ABC Transporters in Cancer Stem Cells: Beyond Chemoresistance. Int. J. Mol. Sci. 2017, 18, 2362. [Google Scholar] [CrossRef]
- Tirino, V.; Desiderio, V.; Paino, F.; Papaccio, G.; De Rosa, M. Methods for Cancer Stem Cell Detection and Isolation. Methods Mol. Biol. 2012, 879, 513–529. [Google Scholar] [CrossRef]
- Murase, M.; Kano, M.; Tsukahara, T.; Takahashi, A.; Torigoe, T.; Kawaguchi, S.; Kimura, S.; Wada, T.; Uchihashi, Y.; Kondo, T.; et al. Side population cells have the characteristics of cancer stem-like cells/cancer-initiating cells in bone sarcomas. Br. J. Cancer 2009, 101, 1425–1432. [Google Scholar] [CrossRef]
- Yang, M.; Yan, M.; Zhang, R.; Li, J.; Luo, Z. Side population cells isolated from human osteosarcoma are enriched with tumor-initiating cells. Cancer Sci. 2011, 102, 1774–1781. [Google Scholar] [CrossRef]
- Martins-Neves, S.R.; Lopes, O.; Carmo, A.D.; Paiva, A.A.; Simões, P.C.P.S.S.; Abrunhosa, A.J.; Gomes, C.M.F. Therapeutic implications of an enriched cancer stem-like cell population in a human osteosarcoma cell line. BMC Cancer 2012, 12, 139. [Google Scholar] [CrossRef] [PubMed]
- Basu-Roy, U.; Seo, E.; Ramanathapuram, L.; Rapp, T.B.; Perry, J.A.; Orkin, S.H.; Mansukhani, A.; Basilico, C. Sox2 maintains self renewal of tumor-initiating cells in osteosarcomas. Oncogene 2012, 31, 2270–2282. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Wang, D.; Gu, D. Knockdown of Sox2 Inhibits OS Cells Invasion and Migration via Modulating Wnt/β-Catenin Signaling Pathway. Pathol. Oncol. Res. 2018, 24, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.N.; Lv, Y.F.; Guo, Q.N. Advances in Osteosarcoma Stem Cell Research and Opportunities for Novel Therapeutic Targets. Cancer Lett. 2016, 370, 268–274. [Google Scholar] [CrossRef]
- Honoki, K.; Fujii, H.; Tsujiuchi, T.; Kido, A.; Yoshitani, K.; Takakura, Y. Sphere-forming stem-like cell populations with drug resistance in human sarcoma cell lines. Int. J. Oncol. 2009, 34, 1381–1386. [Google Scholar] [CrossRef]
- Greco, N.; Schott, T.; Mu, X.; Rothenberg, A.; Voigt, C.; McGough, R.L., 3rd; Goodman, M.; Huard, J.; Weiss, K.R. ALDH Activity Correlates with Metastatic Potential in Primary Sarcomas of Bone. J. Cancer Ther. 2014, 5, 331–338. [Google Scholar] [CrossRef]
- Adhikari, A.S.; Agarwal, N.; Wood, B.M.; Porretta, C.; Ruiz, B.; Pochampally, R.R.; Iwakuma, T. CD117 and Stro-1 Identify Osteosarcoma Tumor-Initiating Cells Associated with Metastasis and Drug Resistance. Cancer Res 2010, 70, 4602–4612. [Google Scholar] [CrossRef]
- Morath, I.; Hartmann, T.; Orian-Rousseau, V. CD44: More than a mere stem cell marker. Int. J. Biochem. Cell Biol. 2016, 81, 166–173. [Google Scholar] [CrossRef]
- Zhu, H.; Mitsuhashi, N.; Klein, A.; Barsky, L.W.; Weinberg, K.; Barr, M.L.; Demetriou, A.; Wu, G.D. The Role of the Hyaluronan Receptor CD44 in Mesenchymal Stem Cell Migration in the Extracellular Matrix. Stem Cells 2006, 24, 928–935. [Google Scholar] [CrossRef]
- Li, J.; Zhong, X.-Y.; Li, Z.-Y.; Cai, J.-F.; Zou, L.; Li, J.-M.; Yang, T.; Liu, W. CD133 expression in osteosarcoma and derivation of CD133+ cells. Mol. Med. Rep. 2013, 7, 577–584. [Google Scholar] [CrossRef]
- He, A.; Qi, W.; Huang, Y.; Feng, T.; Chen, J.; Sun, Y.; Shen, Z.; Yao, Y. CD133 expression predicts lung metastasis and poor prognosis in osteosarcoma patients: A clinical and experimental study. Exp. Ther. Med. 2012, 4, 435–441. [Google Scholar] [CrossRef]
- Fonsatti, E.; Altomonte, M.; Nicotra, M.R.; Natali, P.G.; Maio, M. Endoglin (CD105): A powerful therapeutic target on tumor-associated angiogenetic blood vessels. Oncogene 2003, 22, 6557–6563. [Google Scholar] [CrossRef]
- Tian, J.; Li, X.; Si, M.; Liu, T.; Li, J. CD271+ Osteosarcoma Cells Display Stem-Like Properties. PLoS ONE 2014, 9, e98549. [Google Scholar] [CrossRef] [PubMed]
- Jubelin, C.; Muñoz-Garcia, J.; Cochonneau, D.; Moranton, E.; Heymann, M.-F.; Heymann, D. Biological evidence of cancer stem-like cells and recurrent disease in osteosarcoma. Cancer Drug Resist. 2022, 5, 184–198. [Google Scholar] [CrossRef] [PubMed]
- Kovac, M.; Blattmann, C.; Ribi, S.; Smida, J.; Mueller, N.S.; Engert, F.; Castro-Giner, F.; Weischenfeldt, J.; Kovacova, M.; Krieg, A.; et al. Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat. Commun. 2015, 6, 8940. [Google Scholar] [CrossRef]
- Mohaghegh, P. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum. Mol. Genet. 2001, 10, 741–746. [Google Scholar] [CrossRef]
- Brown, H.K.; Tellez-Gabriel, M.; Cartron, P.-F.; Vallette, F.M.; Heymann, M.-F.; Heymann, D. Characterization of circulating tumor cells as a reflection of the tumor heterogeneity: Myth or reality? Drug Discov. Today 2019, 24, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Czarnecka, A.M.; Synoradzki, K.; Firlej, W.; Bartnik, E.; Sobczuk, P.; Fiedorowicz, M.; Grieb, P.; Rutkowski, P. Molecular Biology of Osteosarcoma. Cancers 2020, 12, 2130. [Google Scholar] [CrossRef] [PubMed]
- Blandino, G.; Levine, A.J.; Oren, M. Mutant p53 gain of function: Differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene 1999, 18, 477–485. [Google Scholar] [CrossRef]
- Gokgoz, N.; Wunder, J.S.; Mousses, S.; Eskandarian, S.; Bell, R.S.; Andrulis, I.L. Comparison of P53 Mutations in Patients with Localized Osteosarcoma and Metastatic Osteosarcoma. Cancer 2001, 92, 2181–2189. [Google Scholar] [CrossRef]
- Ragland, B.D.; Bell, W.C.; Lopez, R.R.; Siegal, G.P. Cytogenetics and Molecular Biology of Osteosarcoma. Lab. Investig. 2002, 82, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Yeager, M.; Mai, P.L.; Gastier-Foster, J.M.; Gorlick, R.; Khanna, C.; Patiño-García, A.; Sierrasesúmaga, L.; Lecanda, F.; Andrulis, I.L.; et al. Germline TP53 Variants and Susceptibility to Osteosarcoma. J. Natl. Cancer Inst. 2015, 107, 101. [Google Scholar] [CrossRef]
- Bell, D.W.; Varley, J.M.; Szydlo, T.E.; Kang, D.H.; Wahrer, D.C.R.; Shannon, K.E.; Lubratovich, M.; Verselis, S.J.; Isselbacher, K.J.; Fraumeni, J.F.; et al. Heterozygous Germ Line hCHK2 Mutations in Li-Fraumeni Syndrome. Science 1999, 286, 2528–2531. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.L.; Boice, J.D.; Abramson, D.H.; Tarone, R.E.; Kleinerman, R.A.; Stovall, M.; Goldman, M.B.; Seddon, J.M.; Tarbell, N.; Fraumeni, J.F.; et al. Cancer Incidence After Retinoblastoma: Radiation Dose and Sarcoma Risk. JAMA 1997, 278, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Feugeas, O.; Guriec, N.; Babin-Boilletot, A.; Marcellin, L.; Simon, P.; Babin, S.; Thyss, A.; Hofman, P.; Terrier, P.; Kalifa, C.; et al. Loss of heterozygosity of the RB gene is a poor prognostic factor in patients with osteosarcoma. J. Clin. Oncol. 1996, 14, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.S.; Higgs, D.; Haddo, O.; Pringle, J.; Briggs, T.W.R. Osteosarcoma Associated With Diamond-Blackfan Anaemia: A Case of a Child Receiving Growth Hormone Therapy. Sarcoma 2004, 8, 47–49. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Healey, J.; Huvos, A.G.; Ladanyi, M. Amplification of the MYC Gene in Osteosarcoma Secondary to Paget’s Disease of Bone. Sarcoma 1997, 1, 159860. [Google Scholar] [CrossRef]
- Behjati, S.; Tarpey, P.S.; Haase, K.; Ye, H.; Young, M.D.; Alexandrov, L.B.; Farndon, S.J.; Collord, G.; Wedge, D.C.; Martincorena, I.; et al. Recurrent mutation of IGF signalling genes and distinct patterns of genomic rearrangement in osteosarcoma. Nat. Commun. 2017, 8, 15936. [Google Scholar] [CrossRef]
- Ribeiro, C.J.A.; Rodrigues, C.M.P.; Moreira, R.; Santos, M.M.M. Chemical Variations on the p53 Reactivation Theme. Pharmaceuticals 2016, 9, 25. [Google Scholar] [CrossRef]
- Cortés-Ciriano, I.; Lee, J.J.-K.; Xi, R.; Jain, D.; Jung, Y.L.; Yang, L.; Gordenin, D.; Klimczak, L.J.; Zhang, C.-Z.; Pellman, D.S.; et al. Comprehensive analysis of chromothripsis in 2658 human cancers using whole-genome sequencing. Nat. Genet. 2020, 52, 331–341. [Google Scholar] [CrossRef]
- Chen, X.; Bahrami, A.; Pappo, A.; Easton, J.; Dalton, J.; Hedlund, E.; Ellison, D.; Shurtleff, S.; Wu, G.; Wei, L.; et al. Recurrent Somatic Structural Variations Contribute to Tumorigenesis in Pediatric Osteosarcoma. Cell Rep. 2014, 7, 104–112. [Google Scholar] [CrossRef]
- Gianferante, D.M.; Mirabello, L.; Savage, S. Germline and somatic genetics of osteosarcoma—Connecting aetiology, biology and therapy. Nat. Rev. Endocrinol. 2017, 13, 480–491. [Google Scholar] [CrossRef] [PubMed]
- Gebert, L.F.R.; Macrae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Yang, Y.; Ma, Y.; Wang, F.; Xue, A.; Zhu, J.; Yang, H.; Chen, Q.; Chen, M.; Ye, L.; et al. Potential Regulatory Role of LncRNA-MiRNA-MRNA Axis in Osteosarcoma. Biomed. Pharmacother. 2020, 121, 109627. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Yu, L.; Xiong, M.; Dai, G. LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem. Biophys. Res. Commun. 2018, 495, 1822–1832. [Google Scholar] [CrossRef]
- Deng, Y.; Zhao, F.; Zhang, Z.; Sun, F.; Wang, M. Long Noncoding RNA SNHG7 Promotes the Tumor Growth and Epithelial-to-Mesenchymal Transition via Regulation of miR-34a Signals in Osteosarcoma. Cancer Biotherapy Radiopharm. 2018, 33, 365–372. [Google Scholar] [CrossRef]
- Xia, B.; Wang, L.; Feng, L.; Tian, B.; Tan, Y.; Du, B. Knockdown of Long Noncoding RNA CAT104 Inhibits the Proliferation, Migration, and Invasion of Human Osteosarcoma Cells by Regulating MicroRNA-381. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2018, 27, 89–98. [Google Scholar] [CrossRef]
- Wang, Y.; Kong, D. Knockdown of LncRNA MEG3 Inhibits Viability, Migration, and Invasion and Promotes Apoptosis by Sponging MiR-127 in Osteosarcoma Cell. J. Cell. Biochem. 2018, 119, 669–679. [Google Scholar] [CrossRef]
- Han, F.; Wang, C.; Wang, Y.; Zhang, L. Long noncoding RNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200s. Am. J. Cancer Res. 2017, 7, 770–783. [Google Scholar]
- Song, J.; Wu, X.; Liu, F.; Li, M.; Sun, Y.; Wang, Y.; Wang, C.; Zhu, K.; Jia, X.; Wang, B.; et al. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. Biochem. Biophys. Res. Commun. 2017, 490, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Luo, P.; Guo, W.; Shi, Y.; Xu, D.; Zheng, H.; Jia, L. LncRNA SNHG20 knockdown suppresses the osteosarcoma tumorigenesis through the mitochondrial apoptosis pathway by miR-139/RUNX2 axis. Biochem. Biophys. Res. Commun. 2018, 503, 1927–1933. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Chen, B.; Wu, B.; Guo, J.; Cao, Y. LncRNA TUG1 promotes cell proliferation and suppresses apoptosis in osteosarcoma by regulating miR-212-3p/FOXA1 axis. Biomed. Pharmacother. 2018, 97, 1645–1653. [Google Scholar] [CrossRef] [PubMed]
- Ba, Z.; Guangchen, N.; Hao, S.; Wang, X.; Cheng, Z.; Nie, G. Downregulation of lnc RNA CASC2 facilitates osteosarcoma growth and invasion through miR-181a. Cell Prolif. 2018, 51, e12409. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Liu, J. LncRNA-p21 inhibited the proliferation of osteosarcoma cells via the miR-130b/PTEN/AKT signaling pathway. Biomed. Pharmacother. 2018, 97, 911–918. [Google Scholar] [CrossRef]
- Yang, C.; Wang, G.; Yang, J.; Wang, L. Long noncoding RNA NBAT1 negatively modulates growth and metastasis of osteosarcoma cells through suppression of miR-21. Am. J. Cancer Res. 2017, 7, 2009–2019. [Google Scholar]
- Li, Z.; Li, X.; Xu, D.; Chen, X.; Li, S.; Zhang, L.; Chan, M.T.V.; Wu, W.K.K. An update on the roles of circular RNAs in osteosarcoma. Cell Prolif. 2021, 54, e12936. [Google Scholar] [CrossRef]
- Qiu, Y.; Pu, C.; Li, Y.; Qi, B. Construction of a circRNA-miRNA-mRNA network based on competitive endogenous RNA reveals the function of circRNAs in osteosarcoma. Cancer Cell Int. 2020, 20, 48. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, S.; Zhang, Q.; Dong, H.; Zhang, J. MicroRNA-206 Upregulation Relieves CircTCF25-induced Osteosarcoma Cell Proliferation and Migration. J. Cell. Physiol. 2020, 237, 4333. [Google Scholar] [CrossRef]
- Zhao, J.; Cheng, L. Long non-coding RNA CCAT1/miR-148a axis promotes osteosarcoma proliferation and migration through regulating PIK3IP1. Acta Biochim. Et Biophys. Sin. 2017, 49, 503–512. [Google Scholar] [CrossRef]
- Jiang, N.; Wang, X.; Xie, X.; Liao, Y.; Liu, N.; Liu, J.; Miao, N.; Shen, J.; Peng, T. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017, 405, 46–55. [Google Scholar] [CrossRef]
- Wang, Y.; Kong, D. LncRNA GAS5 Represses Osteosarcoma Cells Growth and Metastasis via Sponging MiR-203a. Cell. Physiol. Biochem. 2018, 45, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Niu, Y.; Li, D.; Liu, Z. lncRNA C2dat1 Promotes Cell Proliferation, Migration, and Invasion by Targeting miR-34a-5p in Osteosarcoma Cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2018, 26, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Liu, K.; Du, X. Long Non-Coding RNA TUG1 Promotes Proliferation and Inhibits Apoptosis of Osteosarcoma Cells by Sponging miR-132-3p and Upregulating SOX4 Expression. Yonsei Med. J. 2018, 59, 226–235. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, Q.; Wang, L.; Wang, S.; Sun, F.; Xu, D.; Jiang, J. Knockdown of the oncogene lncRNA NEAT1 restores the availability of miR-34c and improves the sensitivity to cisplatin in osteosarcoma. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Chen, F.; Zhao, J.; Li, B.; Liang, Y.; Pan, W.; Zhang, S.; Wang, X.; Zheng, D. Long Non-Coding RNA PVT1 Promotes Osteosarcoma Development by Acting as a Molecular Sponge to Regulate MiR-195. Oncotarget 2016, 7, 82620. [Google Scholar] [CrossRef]
- Guan, H.; Shang, G.; Cui, Y.; Liu, J.; Sun, X.; Cao, W.; Wang, Y.; Li, Y. Long noncoding RNA APTR contributes to osteosarcoma progression through repression of miR-132-3p and upregulation of yes-associated protein 1. J. Cell. Physiol. 2019, 234, 8998–9007. [Google Scholar] [CrossRef]
- Li, S.; Pei, Y.; Wang, W.; Liu, F.; Zheng, K.; Zhang, X. Circular RNA 0001785 regulates the pathogenesis of osteosarcoma as a ceRNA by sponging miR-1200 to upregulate HOXB2. Cell Cycle 2019, 18, 1281–1291. [Google Scholar] [CrossRef]
- Li, X.; Sun, X.-H.; Xu, H.-Y.; Pan, H.-S.; Liu, Y.; He, L. Circ_ORC2 enhances the regulatory effect of miR-19a on its target gene PTEN to affect osteosarcoma cell growth. Biochem. Biophys. Res. Commun. 2019, 514, 1172–1178. [Google Scholar] [CrossRef]
- Ren, C.; Liu, J.; Zheng, B.; Yan, P.; Sun, Y.; Yue, B. The circular RNA circ-ITCH acts as a tumour suppressor in osteosarcoma via regulating miR-22. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3359–3367. [Google Scholar] [CrossRef]
- Ji, X.; Shan, L.; Shen, P.; He, M. Circular RNA circ_001621 promotes osteosarcoma cells proliferation and migration by sponging miR-578 and regulating VEGF expression. Cell Death Dis. 2020, 11, 18. [Google Scholar] [CrossRef]
- Wu, Y.; Xie, Z.; Chen, J.; Chen, J.; Ni, W.; Ma, Y.; Huang, K.; Wang, G.; Wang, J.; Ma, J.; et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol. Cancer 2019, 18, 73. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.-X.; Guo, L.-X.; Pan, H.-S.; Liang, Y.-M.; Li, X. Circ_ANKIB1 stabilizes the regulation of miR-19b on SOCS3/STAT3 pathway to promote osteosarcoma cell growth and invasion. Hum. Cell 2019, 33, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, G.; Wu, Y.; Ma, J.; Wu, H.; Xie, Z.; Chen, S.; Yang, Y.; Wang, S.; Shen, P.; et al. CircMYO10 promotes osteosarcoma progression by regulating miR-370-3p/RUVBL1 axis to enhance the transcriptional activity of β-catenin/LEF1 complex via effects on chromatin remodeling. Mol. Cancer 2019, 18, 1. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Gao, H.; Lv, Z.; Liu, Y.; Zhao, S.; Gong, W.; Liu, W. Circular RNA PVT1 promotes metastasis via regulating of miR-526b/FOXC2 signals in OS cells. J. Cell. Mol. Med. 2020, 24, 5593–5604. [Google Scholar] [CrossRef]
- Pan, G.; Hu, T.; Chen, X.; Zhang, C. Upregulation Of circMMP9 Promotes Osteosarcoma Progression Via Targeting miR-1265/CHI3L1 Axis. Cancer Manag. Res. 2019, 11, 9225–9231. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, J.; Wu, B.; Wang, X.; Jiang, Y.; Zhu, D. Role of extracellular vesicles in osteosarcoma. Int. J. Med. Sci. 2022, 19, 1216–1226. [Google Scholar] [CrossRef]
- Sarhadi, V.K.; Daddali, R.; Seppänen-Kaijansinkko, R. Mesenchymal Stem Cells and Extracellular Vesicles in Osteosarcoma Pathogenesis and Therapy. Int. J. Mol. Sci. 2021, 22, 11035. [Google Scholar] [CrossRef]
- Wang, S.; Ma, F.; Feng, Y.; Liu, T.; He, S. Role of exosomal miR-21 in the tumor microenvironment and osteosarcoma tumorigenesis and progression (Review). Int. J. Oncol. 2020, 56, 1055–1063. [Google Scholar] [CrossRef]
- Han, F.; Pu, P.; Wang, C.; Ding, X.; Zhu, Z.; Xiang, W.; Wang, W. Osteosarcoma Cell-Derived Exosomal miR-1307 Promotes Tumorgenesis via Targeting AGAP1. BioMed Res. Int. 2021, 2021, 7358153. [Google Scholar] [CrossRef]
- Gong, L.; Bao, Q.; Hu, C.; Wang, J.; Zhou, Q.; Wei, L.; Tong, L.; Zhang, W.; Shen, Y. Exosomal miR-675 from metastatic osteosarcoma promotes cell migration and invasion by targeting CALN1. Biochem. Biophys. Res. Commun. 2018, 500, 170–176. [Google Scholar] [CrossRef]
- Raimondi, L.; De Luca, A.; Gallo, A.; Costa, V.; Russelli, G.; Cuscino, N.; Manno, M.; Raccosta, S.; Carina, V.; Bellavia, D.; et al. Osteosarcoma cell-derived exosomes affect tumor microenvironment by specific packaging of microRNAs. Carcinogenesis 2019, 41, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, W.; Zhang, Z.; Shi, D.; Wu, F.; Zhong, B.; Shao, Z. Prognostic Value of Programmed Cell Death 1 Ligand-1 (PD-L1) or PD-1 Expression in Patients with Osteosarcoma: A Meta-Analysis. J. Cancer 2018, 9, 2525–2531. [Google Scholar] [CrossRef] [PubMed]
- Torreggiani, E.; Roncuzzi, L.; Perut, F.; Zini, N.; Baldini, N. Multimodal transfer of MDR by exosomes in human osteosarcoma. Int. J. Oncol. 2016, 49, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Ciferri, M.C.; Quarto, R.; Tasso, R.; Bernardi, S. Extracellular Vesicles as Biomarkers and Therapeutic Tools: From Pre-Clinical to Clinical. Applications 2021, 10, 359. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, H.; Sun, X.; Wang, X.; Ren, T.; Huang, Y.; Zhang, R.; Zheng, B.; Guo, W. Exosomal PD-L1 and N-cadherin predict pulmonary metastasis progression for osteosarcoma patients. J. Nanobiotechnology 2020, 18, 151. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Fujiwara, T.; Uotani, K.; Morita, T.; Kiyono, M.; Yokoo, S.; Hasei, J.; Nakata, E.; Kunisada, T.; Ozaki, T. Clinical and Functional Significance of Intracellular and Extracellular microRNA-25-3p in Osteosarcoma. Acta Med. Okayama 2018, 72, 165–174. [Google Scholar]
- Jerez, S.; Araya, H.; Hevia, D.; Irarrázaval, C.E.; Thaler, R.; van Wijnen, A.J.; Galindo, M. Extracellular vesicles from osteosarcoma cell lines contain miRNAs associated with cell adhesion and apoptosis. Gene 2019, 710, 246–257. [Google Scholar] [CrossRef]
- Harrison, D.J.; Geller, D.S.; Gill, J.D.; Lewis, V.O.; Gorlick, R. Current and future therapeutic approaches for osteosarcoma. Expert Rev. Anticancer. Ther. 2018, 18, 39–50. [Google Scholar] [CrossRef]
- Chaiyawat, P.; Settakorn, J.; Sangsin, A.; Teeyakasem, P.; Klangjorhor, J.; Soongkhaw, A.; Pruksakorn, D. Exploring targeted therapy of osteosarcoma using proteomics data. OncoTargets Ther. 2017, 10, 565–577. [Google Scholar] [CrossRef]
- De Vita, A.; Vanni, S.; Miserocchi, G.; Fausti, V.; Pieri, F.; Spadazzi, C.; Cocchi, C.; Liverani, C.; Calabrese, C.; Casadei, R.; et al. A Rationale for the Activity of Bone Target Therapy and Tyrosine Kinase Inhibitor Combination in Giant Cell Tumor of Bone and Desmoplastic Fibroma: Translational Evidences. Biomedicines 2022, 10, 372. [Google Scholar] [CrossRef]
- Avnet, S.; Lemma, S.; Cortini, M.; Pellegrini, P.; Perut, F.; Zini, N.; Kusuzaki, K.; Chano, T.; Grisendi, G.; Dominici, M.; et al. Altered pH gradient at the plasma membrane of osteosarcoma cells is a key mechanism of drug resistance. Oncotarget 2016, 7, 63408–63423. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Healey, J.; Meyers, P.A.; Ladanyi, M.; Huvos, A.G.; Bertino, J.R.; Gorlick, R. Mechanisms of methotrexate resistance in osteosarcoma. Clin. Cancer Res. 1999, 5, 621–627. [Google Scholar] [PubMed]
- Rajkumar, T.; Yamuna, M. Multiple pathways are involved in drug resistance to doxorubicin in an osteosarcoma cell line. Anti-Cancer Drugs 2008, 19, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; Lasthaus, C.; Guerin, E.; Marcellin, L.; Pencreach, E.; Gaub, M.-P.; Guenot, D.; Entz-Werle, N. Role of Topoisomerases in Pediatric High Grade Osteosarcomas: TOP2A Gene Is One of the Unique Molecular Biomarkers of Chemoresponse. Cancers 2013, 5, 662–675. [Google Scholar] [CrossRef] [PubMed]
- Wiegering, A.; Matthes, N.; Mühling, B.; Koospal, M.; Quenzer, A.; Peter, S.; Germer, C.-T.; Linnebacher, M.; Otto, C. Reactivating p53 and Inducing Tumor Apoptosis (RITA) Enhances the Response of RITA-Sensitive Colorectal Cancer Cells to Chemotherapeutic Agents 5-Fluorouracil and Oxaliplatin. Neoplasia 2017, 19, 301–309. [Google Scholar] [CrossRef]
- Hattinger, C.M.; Michelacci, F.; Sella, F.; Magagnoli, G.; Benini, S.; Gambarotti, M.; Palmerini, E.; Picci, P.; Serra, M.; Ferrari, S. Excision repair cross-complementation group 1 protein expression predicts survival in patients with high-grade, non-metastatic osteosarcoma treated with neoadjuvant chemotherapy. Histopathology 2015, 67, 338–347. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).