Modelling 3D Tumour Microenvironment In Vivo: A Tool to Predict Cancer Fate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Rearing
2.2. Cell Culture Maintenance
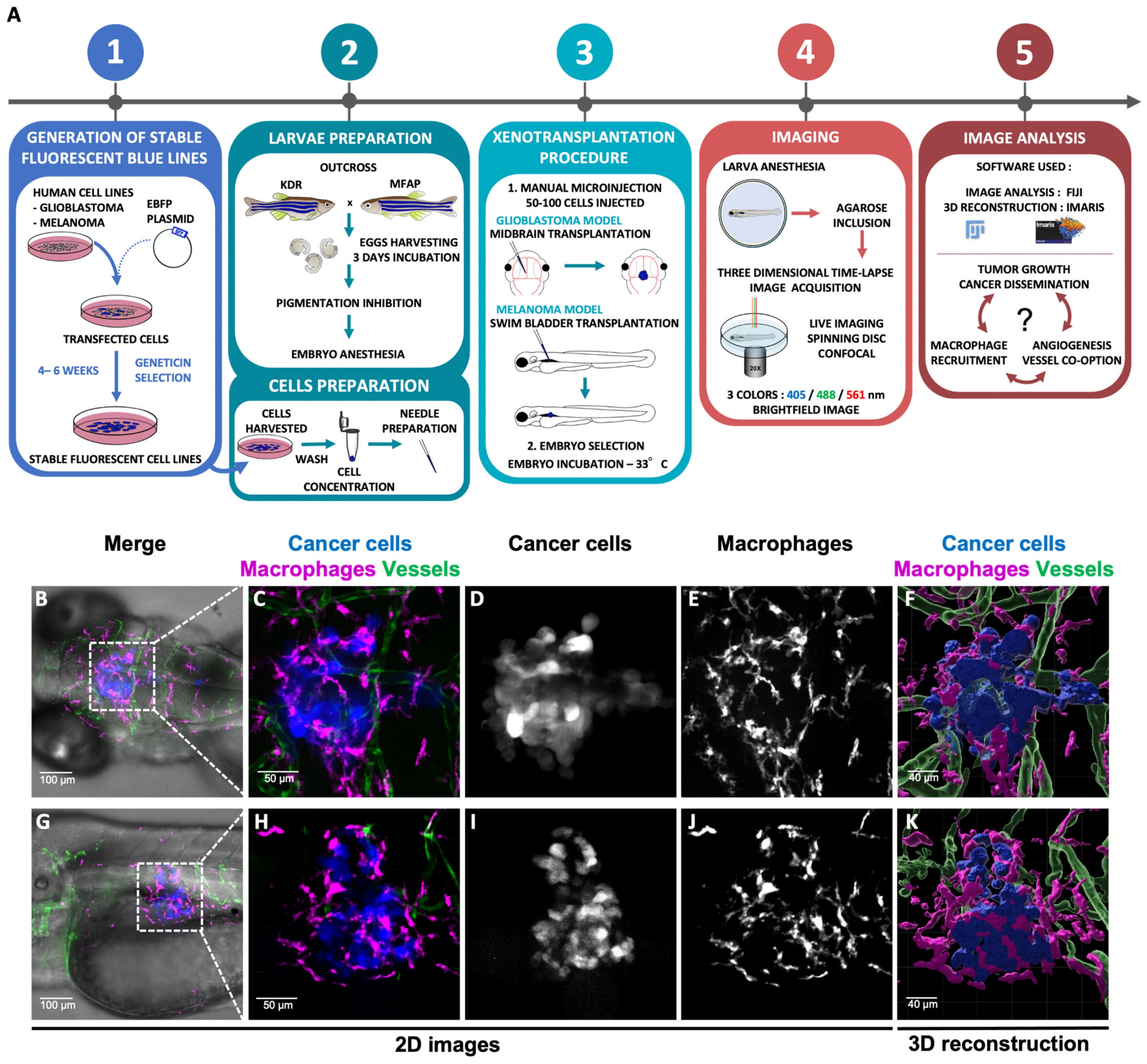
2.3. Generation of Stable Fluorescent Human Cancer Cell Lines
2.4. Embryo and Cell Preparation Prior to Xenotransplantation
2.5. Xenotransplantation of Human Cancer Cell Lines
2.6. The 3D and 4D Live-Imaging Acquisition
2.7. Image Analysis
3. Results
3.1. Cell Lines Engineering, Transplanted into Zebrafish Embryos and Live Imaging
3.2. Live Imaging of Interactions between Cancer Cells and Their Microenvironment
3.3. Advantage of 3D Reconstruction for Monitoring the Fate of Tumour Vasculature
3.4. Importance of Macrophages in Tumour Angiogenesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 1–19. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, S.; Dudley, A.C. Models and molecular mechanisms of blood vessel co-option by cancer cells. Angiogenesis 2020, 23, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Nishida, N.; Yano, H.; Nishida, T.; Kamura, T.; Kojiro, M. Angiogenesis in cancer. Vasc. Health Risk Manag. 2006, 2, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Song, Z.; Zhang, T.; He, X.; Huang, K.; Zhang, Q.; Shen, J.; Pan, J. Identification of Immune Cell Infiltration and Immune-Related Genes in the Tumor Microenvironment of Glioblastomas. Front. Immunol. 2020, 11, 2708. [Google Scholar] [CrossRef] [PubMed]
- Ladányi, A. Prognostic and predictive significance of immune cells infiltrating cutaneous melanoma. Pigment Cell Melanoma Res. 2015, 28, 490–500. [Google Scholar] [CrossRef] [PubMed]
- Sabdyusheva Litschauer, I.; Becker, K.; Saghafi, S.; Ballke, S.; Bollwein, C.; Foroughipour, M.; Gaugeler, J.; Foroughipour, M.; Schavelova, V.; Laszlo, V.; et al. 3D histopathology of human tumours by fast clearing and ultramicroscopy. Sci. Rep. 2020, 10, 17619. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.M.; Jung, D.; Kiemen, A.; Gaida, M.M.; Yoshizawa, T.; Braxton, A.M.; Noe, M.; Lionheart, G.; Oshima, K.; Thompson, E.D.; et al. Three-dimensional visualization of cleared human pancreas cancer reveals that sustained epithelial-to-mesenchymal transition is not required for venous invasion. Mod. Pathol. 2020, 33, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and animal models: Are 3D cultures the ideal tool to study cancer-microenvironment interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef] [PubMed]
- Lieschke, G.J.; Currie, P.D. Animal models of human disease: Zebrafish swim into view. Nat. Rev. Genet. 2007, 8, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Glasgow, E.; Agarwal, S. Zebrafish Xenografts for Drug Discovery and Personalized Medicine. Trends Cancer 2020, 6, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Novoa, B.; Figueras, A. Zebrafish: Model for the study of inflammation and the innate immune response to infectious diseases. Adv. Exp. Med. Biol. 2012, 946, 253–275. [Google Scholar] [PubMed]
- Hamilton, L.; Astell, K.R.; Velikova, G.; Sieger, D. A Zebrafish Live Imaging Model Reveals Differential Responses of Microglia Toward Glioblastoma Cells In Vivo. Zebrafish 2016, 13, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cao, Z.; Zhang, X.M.; Nakamura, M.; Sun, M.; Hartman, J.; Harris, R.A.; Sun, Y.; Cao, Y. Novel mechanism of macrophage-mediated metastasis revealed in a zebrafish model of tumor development. Cancer Res. 2015, 75, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Lal, S.; La Du, J.; Tanguay, R.L.; Greenwood, J.A. Calpain 2 is required for the invasion of glioblastoma cells in the zebrafish brain microenvironment. J. Neurosci. Res. 2012, 90, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, X.; Zhao, Y.; Li, Z.; Lin, S.; Wei, Y.; Yang, H. A Novel Xenograft Model in Zebrafish for High-Resolution Investigating Dynamics of Neovascularization in Tumors. PLoS ONE 2011, 6, e21768. [Google Scholar] [CrossRef] [PubMed]
- Mealiea, D.; Boudreau, E.; De Silva, N.; Okamoto, L.; Ho, T.; Fish, J.E.; McCart, J.A. Modeling oncolytic virus dynamics in the tumor microenvironment using zebrafish. Cancer Gene Ther. 2021, 28, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Roh-Johnson, M.; Shah, A.N.; Stonick, J.A.; Poudel, K.R.; Kargl, J.; Yang, G.H.; di Martino, J.; Hernandez, R.E.; Gast, C.E.; Zarour, L.R.; et al. Macrophage-Dependent Cytoplasmic Transfer during Melanoma Invasion In Vivo. Dev. Cell 2017, 43, 549–562.e6. [Google Scholar] [CrossRef] [PubMed]
- Britto, D.D.; Wyroba, B.; Chen, W.; Lockwood, R.A.; Tran, K.B.; Shepherd, P.R.; Hall, C.J.; Crosier, K.E.; Crosier, P.S.; Astin, J.W. Macrophages enhance Vegfa-driven angiogenesis in an embryonic zebrafish tumour xenograft model. DMM Dis. Model. Mech. 2018, 11, dmm035998. [Google Scholar] [CrossRef] [PubMed]
- Shurin, M.R.; Shurin, G.V.; Zlotnikov, S.B.; Bunimovich, Y.L. The Neuroimmune Axis in the Tumor Microenvironment. J. Immunol. 2020, 204, 280–285. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marines, J.; Lorenzini, F.; Kissa, K.; Fontenille, L. Modelling 3D Tumour Microenvironment In Vivo: A Tool to Predict Cancer Fate. Curr. Issues Mol. Biol. 2023, 45, 9076-9083. https://doi.org/10.3390/cimb45110569
Marines J, Lorenzini F, Kissa K, Fontenille L. Modelling 3D Tumour Microenvironment In Vivo: A Tool to Predict Cancer Fate. Current Issues in Molecular Biology. 2023; 45(11):9076-9083. https://doi.org/10.3390/cimb45110569
Chicago/Turabian StyleMarines, Johanna, Francesca Lorenzini, Karima Kissa, and Laura Fontenille. 2023. "Modelling 3D Tumour Microenvironment In Vivo: A Tool to Predict Cancer Fate" Current Issues in Molecular Biology 45, no. 11: 9076-9083. https://doi.org/10.3390/cimb45110569
APA StyleMarines, J., Lorenzini, F., Kissa, K., & Fontenille, L. (2023). Modelling 3D Tumour Microenvironment In Vivo: A Tool to Predict Cancer Fate. Current Issues in Molecular Biology, 45(11), 9076-9083. https://doi.org/10.3390/cimb45110569






