Punicalagin Targets Atherosclerosis: Gene Expression Profiling of THP-1 Macrophages Treated with Punicalagin and Molecular Docking
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. THP1 Cell Culture
2.3. Total Cellular RNA Preparation from the Control and Treated THP1 Cells
2.4. Preparation of the Sense Strand DNA for Microarray Gene Expression Analysis
2.5. Sense Strand DNA Labelling and Hybridization Gene 1.0 S.T Arrays
2.6. Microarray Data Normalization and Analysis
2.7. Molecular Pathway Analysis
2.8. Real-Time PCR Validation
2.9. LXR/RXR Activation and Network Interaction
2.10. Molecular Docking
3. Results
3.1. Normalization and Visualization
3.2. Identification of Canonical Pathways and Associated Disease Functions
3.3. Validation of Microarray Data
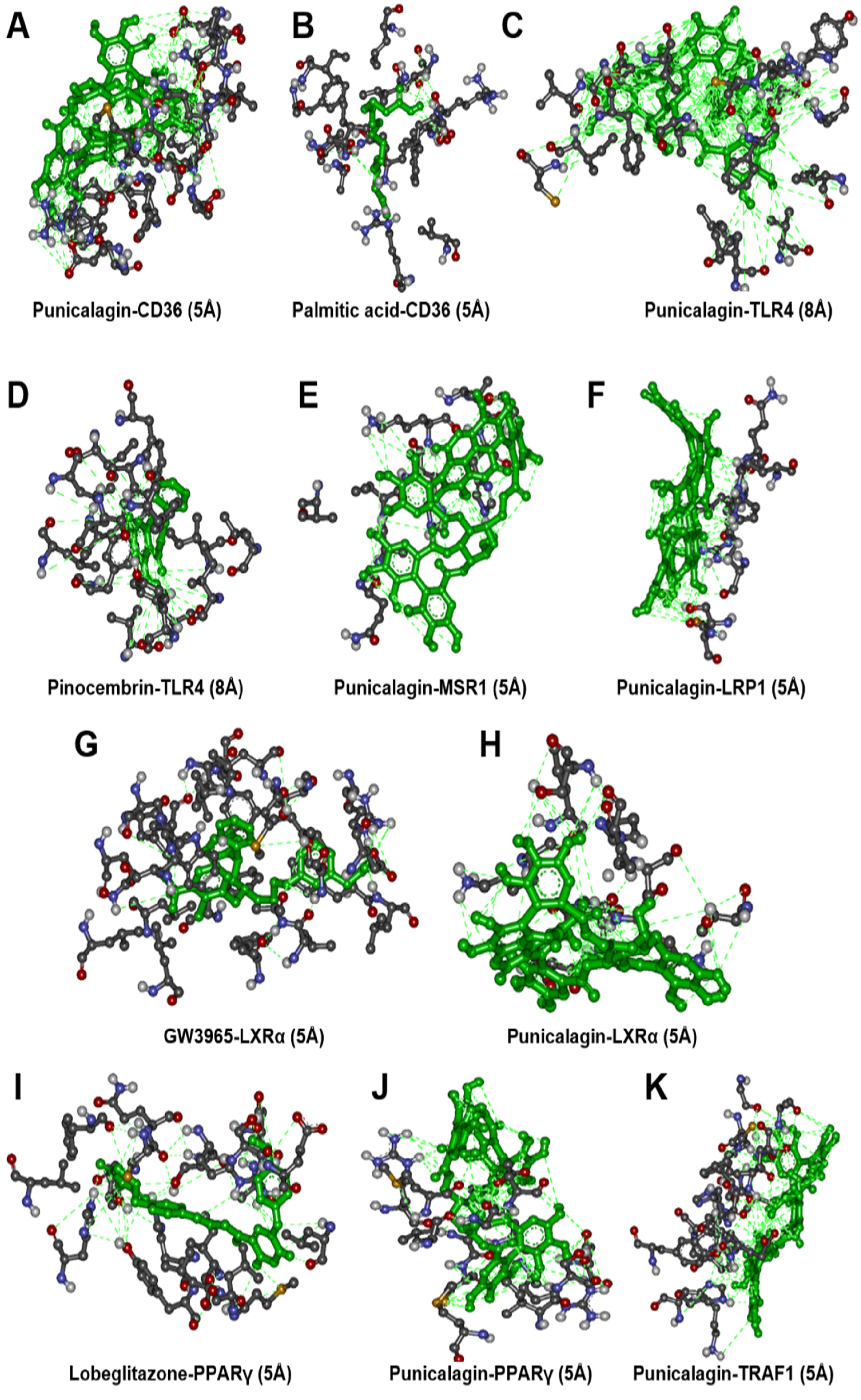
3.4. Molecular Docking Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cardiovascular Diseases (CVDs). Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 15 September 2021).
- WHO Global Action Plan for the Prevention and Control of Noncommunicable Diseases 2013–2020; World Health Organization: Geneva, Switzerland, 2013; p. 102.
- AJ, L. Atherosclerosis. Nature 2000, 407, 233–241. [Google Scholar] [CrossRef]
- Frostegård, J. Immunity, atherosclerosis and cardiovascular disease. BMC Med. 2013, 11, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frostegard, J.; Ulfgren, A.-K.; Nyberg, P.; Hedin, U.; Swedenborg, J.; Andersson, U.; Hansson, G.K. Cytokine expression in advanced human atherosclerotic plaques: Dominance of pro-inflammatory (Th1) and macrophage-stimulating cytokines. Atherosclerosis 1999, 145, 33–43. [Google Scholar] [CrossRef]
- Cheng, Y.-C.; Sheen, J.-M.; Hu, W.L.; Hung, Y.-C. Polyphenols and Oxidative Stress in Atherosclerosis-Related Ischemic Heart Disease and Stroke. Oxid. Med. Cell. Longev. 2017, 2017, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Tresserra-Rimbau, A.; Rimm, E.B.; Medina-Remón, A.; Martínez-González, M.A.; de la Torre, R.; Corella, D.; Salas-Salvadó, J.; Gómez-Gracia, E.; Lapetra, J.; Arós, F.; et al. Inverse association between habitual polyphenol intake and incidence of cardiovascular events in the PREDIMED study. Nutr. Metab. Cardiovasc. Dis. 2014, 24, 639–647. [Google Scholar] [CrossRef]
- Tangney, C.; Rasmussen, H.E. Polyphenols, Inflammation, and Cardiovascular Disease. Curr. Atheroscler. Rep. 2013, 15, 324. [Google Scholar] [CrossRef]
- Heber, D. Pomegranate Ellagitannins; CRC Press: Boca Raton, FL, USA, 2011; Volume 2, pp. 201–209. [Google Scholar]
- Gil, M.I.; Tomas-Barberan, F.A.; Hess-Pierce, B.; Holcroft, D.M.; Kader, A.A. Antioxidant activity of pomegranate juice and its relationship with phenolic composition and processing. J. Agric. Food Chem. 2000, 48, 4581–4589. [Google Scholar] [CrossRef]
- Aviram, M.; Rosenblat, M. Pomegranate Protection against Cardiovascular Diseases. Evid. Based. Complement. Alternat. Med. 2012, 2012, 382763. [Google Scholar] [CrossRef]
- Aviram, M.; Dornfeld, L.; Rosenblat, M.; Volkova, N.; Kaplan, M.; Coleman, R.; Hayek, T.; Presser, D.; Fuhrman, B. Pomegranate juice consumption reduces oxidative stress, atherogenic modifications to LDL, and platelet aggregation: Studies in humans and in atherosclerotic apolipoprotein E-deficient mice. Am. J. Clin. Nutr. 2000, 71, 1062–1076. [Google Scholar] [CrossRef]
- De Nigris, F.; Williams-Ignarro, S.; Lerman, L.O.; Crimi, E.; Botti, C.; Mansueto, G.; D’Armiento, F.P.; De Rosa, G.; Sica, V.; Ignarro, L.J.; et al. Beneficial Effects of Pomegranate Juice on Oxidation-Sensitive Genes and Endothelial Nitric Oxide Synthase Activity at Sites of Perturbed Shear Stress. Proc. Natl. Acad. Sci. USA 2005, 102, 4896–4901. [Google Scholar] [CrossRef] [Green Version]
- Aviram, M.; Volkova, N.; Coleman, R.; Dreher, M.; Reddy, M.K.; Ferreira, D.; Rosenblat, M. Pomegranate phenolics from the peels, arils, and flowers are antiatherogenic: Studies in vivo in atherosclerotic apolipoprotein E-deficient (E0) mice and in vitro in cultured macrophages and lipoproteins. Proc. J. Agric. Food Chem. 2008, 56, 1148–1157. [Google Scholar] [CrossRef] [PubMed]
- Atrahimovich, D.; Samson, A.O.; Khattib, A.; Vaya, J.; Khatib, S. Punicalagin Decreases Serum Glucose Levels and Increases PON1 Activity and HDL Anti-Inflammatory Values in Balb/c Mice Fed a High-Fat Diet. Oxid. Med. Cell. Longev. 2018, 2018, 2673076. [Google Scholar] [CrossRef] [PubMed]
- De Nigris, F.; Williams-Ignarro, S.; Sica, V.; Lerman, L.O.; D’Armiento, F.P.; Byrns, R.E.; Casamassimi, A.; Carpentiero, D.; Schiano, C.; Sumi, D.; et al. Effects of a Pomegranate Fruit Extract rich in punicalagin on oxidation-sensitive genes and eNOS activity at sites of perturbed shear stress and atherogenesis. Cardiovasc. Res. 2007, 73, 414–423. [Google Scholar] [CrossRef] [Green Version]
- Pomegranate Juice Flavonoids Inhibit Low-Density Lipoprotein Oxidation and Cardiovascular Diseases: Studies in Atherosclerotic Mice and in Humans—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/12224378/ (accessed on 15 September 2021).
- Atrahimovich, D.; Khatib, S.; Sela, S.; Vaya, J.; Samson, A.O. Punicalagin Induces Serum Low-Density Lipoprotein Influx to Macrophages. Oxid. Med. Cell. Longev. 2016, 2016, 7124251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seeram, N.P.; Henning, S.M.; Zhang, Y.; Suchard, M.; Li, Z.; Heber, D. Pomegranate juice ellagitannin metabolites are present in human plasma and some persist in urine for up to 48 hours. J. Nutr. 2006, 136, 2481–2485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Chen, J.; Ren, G.; Zhang, Y.; Tan, X.; Yang, L. Punicalagin prevents inflammation in lps-induced raw264.7 macrophages by inhibiting foxo3a/autophagy signaling pathway. Nutrients 2019, 11, 2794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- BenSaad, L.A.; Kim, K.H.; Quah, C.C.; Kim, W.R.; Shahimi, M. Anti-inflammatory potential of ellagic acid, gallic acid and punicalagin A&B isolated from Punica granatum. BMC Complement. Altern. Med. 2017, 17, 47. [Google Scholar] [CrossRef] [Green Version]
- Michael, D.R.; Salter, R.C.; Ramji, D.P. TGF-β inhibits the uptake of modified low density lipoprotein by human macrophages through a Smad-dependent pathway: A dominant role for Smad-2. Biochim. Biophys. Acta-Mol. Basis Dis. 2012, 1822, 1608–1616. [Google Scholar] [CrossRef] [Green Version]
- Almowallad, S.; Huwait, E.; Al-Massabi, R.; Saddeek, S.; Gauthaman, K.; Prola, A. Punicalagin regulates key processes associated with atherosclerosis in thp-1 cellular model. Pharmaceuticals 2020, 13, 372. [Google Scholar] [CrossRef]
- Xu, W.; Yu, L.; Zhou, W.; Luo, M. Resistin increases lipid accumulation and CD36 expression in human macrophages. Biochem. Biophys. Res. Commun. 2006, 351, 376–382. [Google Scholar] [CrossRef]
- Saenz, J.; Santa-María, C.; Reyes-Quiroz, M.E.; Geniz, I.; Jiménez, J.; Sobrino, F.; Alba, G. Grapefruit Flavonoid Naringenin Regulates the Expression of LXRα in THP-1 Macrophages by Modulating AMP-Activated Protein Kinase. Mol. Pharm. 2018, 15, 1735–1745. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, H.; Williams, J.O.; Ferekidis, N.; Ismail, A.; Chan, Y.H.; Michael, D.R.; Guschina, I.A.; Tyrrell, V.J.; O’Donnell, V.B.; Harwood, J.L.; et al. Dihomo-γ-linolenic acid inhibits several key cellular processes associated with atherosclerosis. Biochim. Biophys. Acta-Mol. Basis Dis. 2019, 1865, 2538–2550. [Google Scholar] [CrossRef] [PubMed]
- Pepino, M.Y.; Kuda, O.; Samovski, D.; Abumrad, N.A. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu. Rev. Nutr. 2014, 34, 281–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, L.; Duan, Y.; Ma, F.; Lou, L. Punicalagin inhibits the viability, migration, invasion, and EMT by regulating GOLPH3 in breast cancer cells. J. Recept. Signal Transduct. 2020, 40, 173–180. [Google Scholar] [CrossRef]
- Tang, J.M.; Min, J.; Li, B.S.; Hong, S.S.; Liu, C.; Hu, M.; Li, Y.; Yang, J.; Hong, L. Therapeutic Effects of Punicalagin Against Ovarian Carcinoma Cells in Association with β-Catenin Signaling Inhibition. Int. J. Gynecol. Cancer 2016, 26, 1557–1563. [Google Scholar] [CrossRef]
- Malik, A.; Mukhtar, H. Prostate cancer prevention through pomegranate fruit. Cell Cycle 2006, 5, 371–373. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.J.; Kong, F.Q.; Jie, J.; Li, Q.; Liu, H.; Di Xu, A.; Yang, Y.Q.; Jiang, B.; Wang, D.D.; Zhou, Z.Q.; et al. Macrophage MSR1 promotes BMSC osteogenic differentiation and M2-like polarization by activating PI3K/AKT/GSK3β/β-catenin pathway. Theranostics 2020, 10, 17–35. [Google Scholar] [CrossRef]
- Alvarez-Cubero, M.J.; Saiz, M.; Martinez-Gonzalez, L.J.; Alvarez, J.C.; Lorente, J.A.; Cozar, J.M. Genetic analysis of the principal genes related to prostate cancer: A review. Urol. Oncol. Semin. Orig. Investig. 2013, 31, 1419–1429. [Google Scholar] [CrossRef]
- Quirós-Fernández, R.; López-Plaza, B.; Bermejo, L.M.; Palma-Milla, S.; Gómez-Candela, C. Supplementation with Hydroxytyrosol and Punicalagin Improves Early Atherosclerosis Markers Involved in the Asymptomatic Phase of Atherosclerosis in the Adult Population: A Randomized, Placebo-Controlled, Crossover Trial. Nutrients 2019, 11, 640. [Google Scholar] [CrossRef] [Green Version]
- Metwally, H.; Tanaka, T.; Li, S.; Parajuli, G.; Kang, S.; Hanieh, H.; Hashimoto, S.; Chalise, J.P.; Gemechu, Y.; Standley, D.M.; et al. Noncanonical STAT1 Phosphorylation Expands Its Transcriptional Activity into Promoting LPS-Induced IL-6 and IL-12p40 Production. Sci. Signal. 2020, 13, eaay0574. [Google Scholar] [CrossRef]
- Lalani, A.I.; Zhu, S.; Gokhale, S.; Jin, J.; Xie, P. TRAF Molecules in Inflammation and Inflammatory Diseases. Curr. Pharmacol. Rep. 2018, 4, 64–90. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, M.K.; Choudhury, S.; Borah, A. An in silico investigation on the inhibitory potential of the constituents of Pomegranate juice on antioxidant defense mechanism: Relevance to neurodegenerative diseases. IBRO Rep. 2019, 6, 153–159. [Google Scholar] [CrossRef] [PubMed]
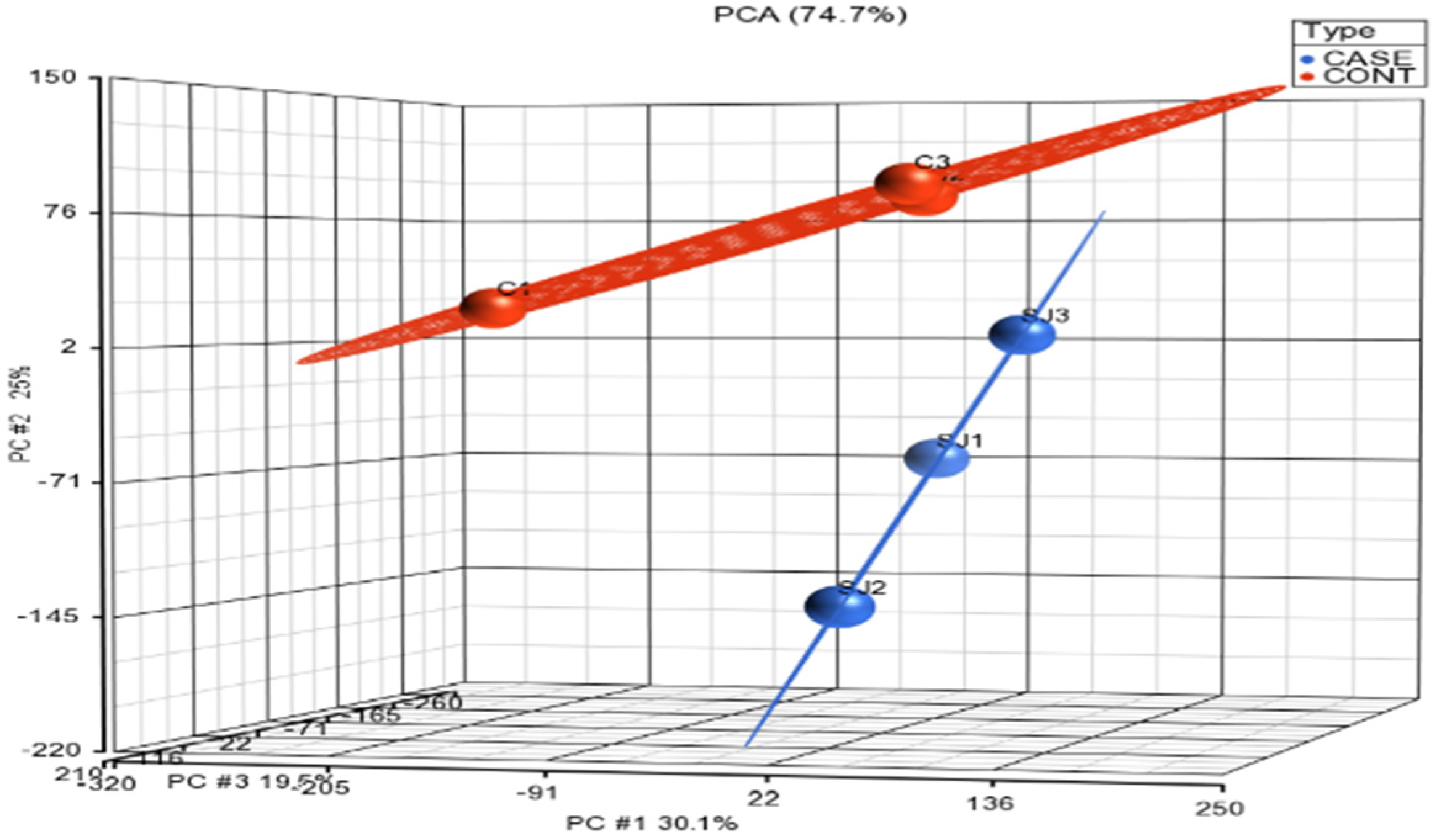
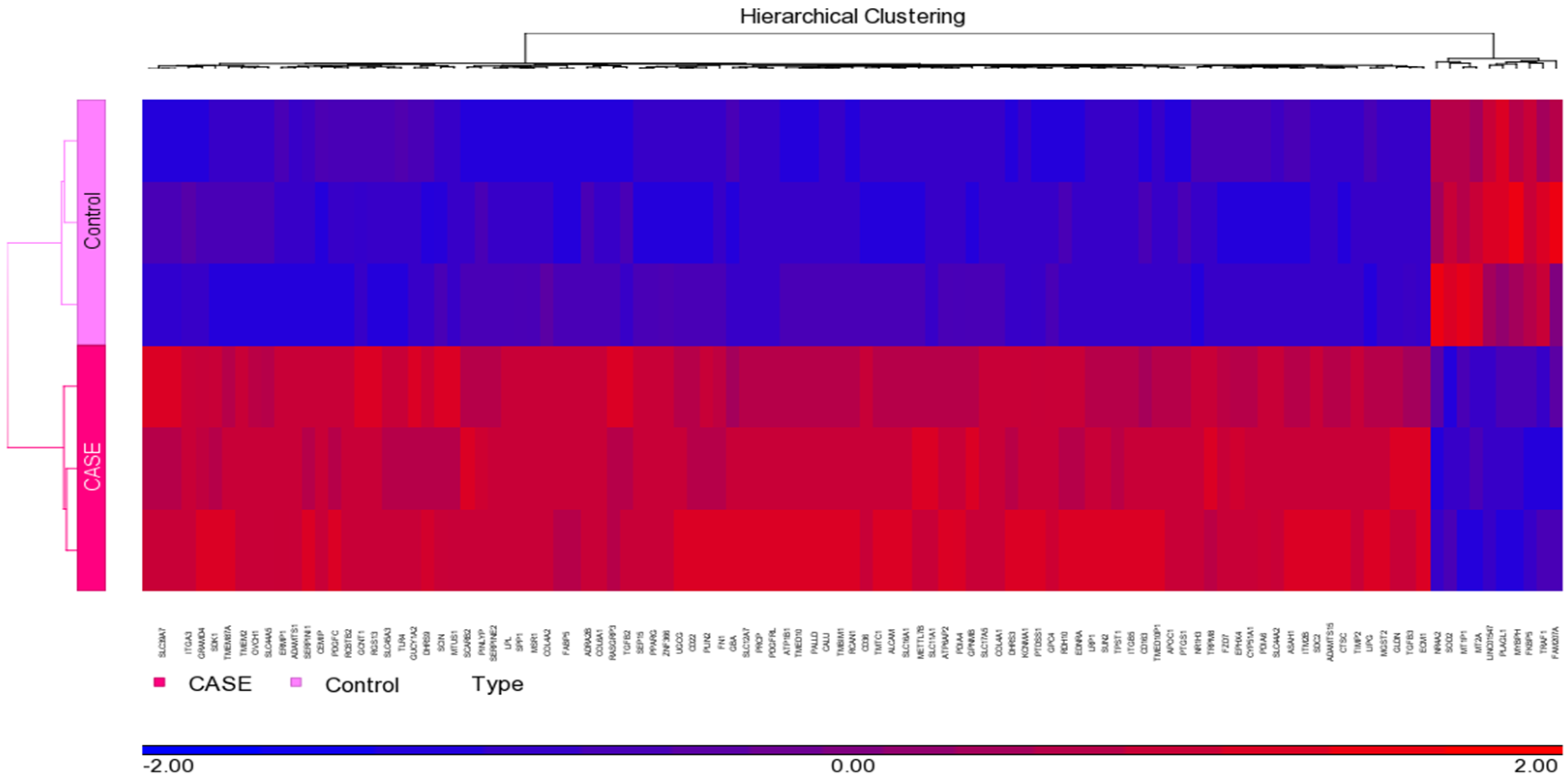

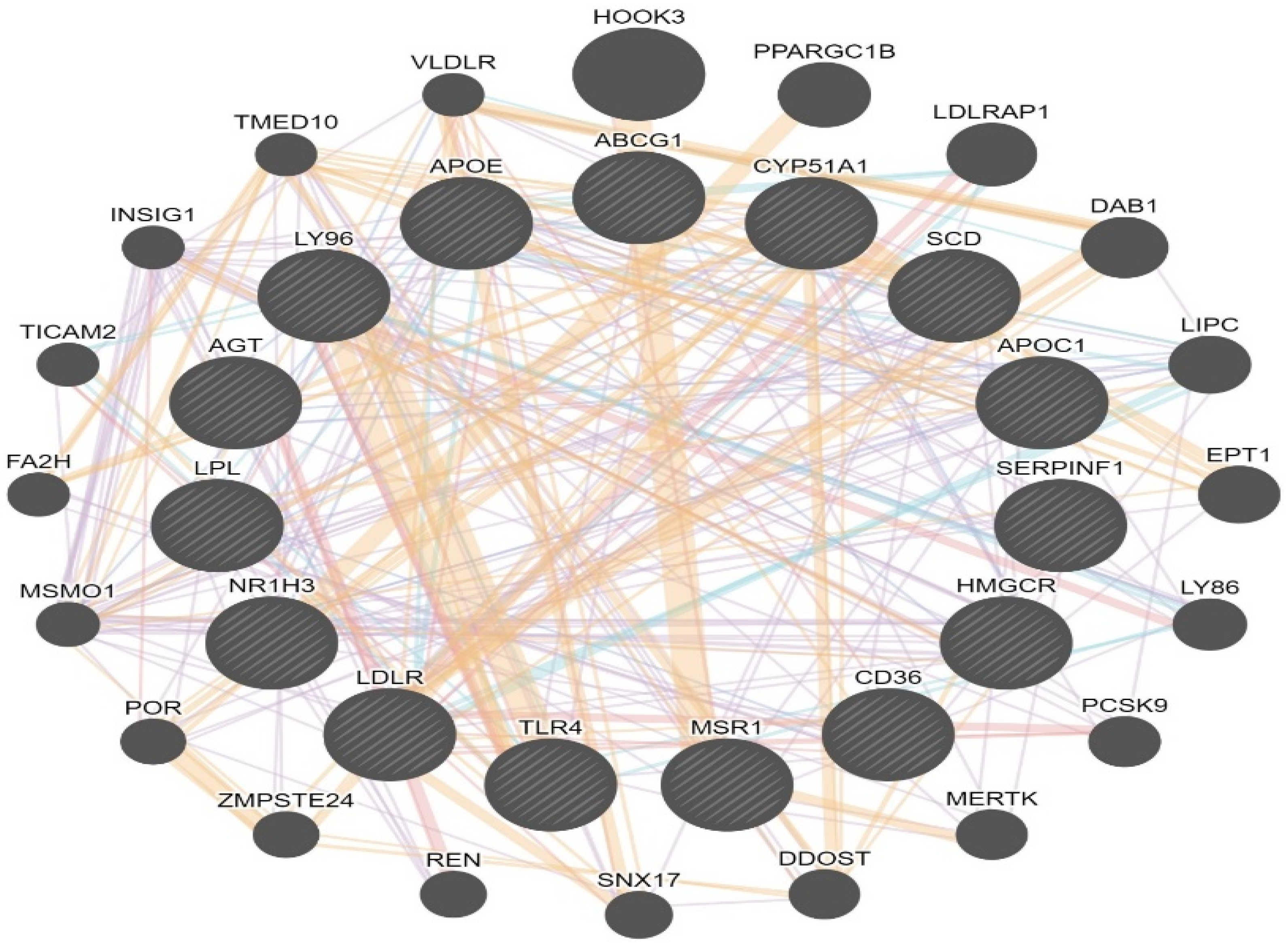

| Gene | Forward Primer | Reverse Primer | Ref. |
|---|---|---|---|
| MSR1 | ATTGCCCTTTACCTCCTCGT | TCATTTCCTTTCCCGTGAG | [24] |
| CD36 | AGATGCAGCCTCATTTCCAC | GCCTTGGATGGAAGAACAAA | [24] |
| NR1H3 | AAGCCCTGCATGCCTACGT | TGCAGACGCAGTGCAAACA | [25] |
| GAPDH | CTTTTGCGTCGCCAGCCGAG | GCCCAATACGACCAAATCCGTTGACT | [26] |
| Gene Symbol | p-Value | Fold-Change (Treated vs. Control) |
|---|---|---|
| FABP4 | 0.00261999 | 39.7893 |
| CD36 | 0.000175484 | 19.5949 |
| MSR1 | 0.000696357 | 16.8431 |
| CEMIP | 0.000847506 | 10.5071 |
| LPL | 0.000899872 | 10.0902 |
| TDO2 | 0.0102073 | 8.73151 |
| GPC4 | 0.0001794 | 8.39639 |
| SLC8A1 | 0.00194115 | 7.82049 |
| SERPINE2 | 0.00115135 | 7.67443 |
| FCER2 | 0.00844659 | 7.00205 |
| Gene Symbol | p-Value | Fold-Change (Treated vs. Control) |
|---|---|---|
| MT2A | 0.00236401 | −10.6361 |
| MT1P1 | 0.00268895 | −4.41548 |
| FAM207A | 0.0407155 | −2.72934 |
| PLAGL1 | 0.00595623 | −2.62005 |
| SOD2 | 0.00148297 | −2.60906 |
| MYBPH | 0.00864804 | −2.58598 |
| FKBP5 | 0.000695019 | −2.4766 |
| TRAF1 | 0.00389735 | −2.40964 |
| NR4A2 | 0.0183868 | −2.37157 |
| SNORD1A | 0.0350301 | −2.36584 |
| Ingenuity Canonical Pathways | −log (p-Value) | z-Score | Molecules |
|---|---|---|---|
| Cholesterol Biosynthesis I | 11.3 | 2.828 | CYP51A1, DHCR24, DHCR7, EBP, HSD17B7, LBR, MSMO1, SC5D |
| Cholesterol Biosynthesis II (via 24,25-dihydrolanosterol) | 11.3 | 2.828 | CYP51A1, DHCR24, DHCR7, EBP, HSD17B7, LBR, MSMO1, SC5D |
| Cholesterol Biosynthesis III (via Desmosterol) | 11.3 | 2.828 | CYP51A1, DHCR24, DHCR7, EBP, HSD17B7, LBR, MSMO1, SC5D |
| Superpathway of Cholesterol Biosynthesis | 9.31 | 3 | CYP51A1, DHCR24, DHCR7, EBP, HMGCR, HSD17B7, LBR, MSMO1, SC5D |
| LXR/RXR Activation | 8.97 | 1.265 | ABCG1, AGT, APOC1, APOE, CD36, CYP51A1, HMGCR, LDLR, LPL, LY96, MSR1, NR1H3, SCD, SERPINF1, TLR4 |
| Hepatic Fibrosis/Hepatic Stellate Cell Activation | 7.22 | 0 | AGT, COL4A1, COL4A2, COL6A1, EDNRA, FN1, IFNGR1, IGFBP3, LY96, MET, MMP1, PDGFC, TGFB2, TGFB3, TIMP2, TLR4 |
| Inhibition of Matrix Metalloproteases | 6.79 | 0 | LRP1, MMP1, MMP12, MMP8, RECK, SDC2, THBS2, TIMP2 |
| Zymosterol Biosynthesis | 6.02 | 2 | CYP51A1, HSD17B7, LBR, MSMO1 |
| Phagosome Formation | 5.24 | 0 | C3AR1, C5AR1, FCER1G, FCER2, FCGR1A, FN1, ITGA3, ITGA4, MSR1, TLR1, TLR4 |
| Caveolar-mediated Endocytosis Signaling | 4.65 | 0 | CAV1, HLA-A, INSR, ITGA3, ITGA4, ITGA6, ITGAL, ITGB5 |
| Receptor Symbol | PDB ID | Binding Affinity w/Punicalagin | Reference Ligand | Binding Affinity w/Ligand | Common H-Bonding |
|---|---|---|---|---|---|
| CD36 | 5LGD | −9.3 (Kcal/mol) | Palmitic acid | −6.8 (Kcal/mol) | 0 |
| TLR4 | 2Z65 | −9.0 (Kcal/mol) | Pinocembrin | −8.2(Kcal/mol) | 7 |
| MSR1 | Modelled | −7.4 (Kcal/mol) | - | − | - |
| LRP1 | 1CR8 | −6.4 (Kcal/mol) | - | − | - |
| NR1H3 | 3IPQ | −7.1 (Kcal/mol) | GW3965 | −13.9 (Kcal/mol) | 0 |
| TRAF1 | 5EIT | −9.0 (Kcal/mol) | - | − | - |
| PPAR-γ | 4EMA | −8.7 (Kcal/mol) | Lobeglitazone | −8.6 (Kcal/mol) | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huwait, E.; Almowallad, S.; Al-Massabi, R.; Saddeek, S.; Gauthaman, K.; Prola, A. Punicalagin Targets Atherosclerosis: Gene Expression Profiling of THP-1 Macrophages Treated with Punicalagin and Molecular Docking. Curr. Issues Mol. Biol. 2022, 44, 2153-2166. https://doi.org/10.3390/cimb44050145
Huwait E, Almowallad S, Al-Massabi R, Saddeek S, Gauthaman K, Prola A. Punicalagin Targets Atherosclerosis: Gene Expression Profiling of THP-1 Macrophages Treated with Punicalagin and Molecular Docking. Current Issues in Molecular Biology. 2022; 44(5):2153-2166. https://doi.org/10.3390/cimb44050145
Chicago/Turabian StyleHuwait, Etimad, Sanaa Almowallad, Rehab Al-Massabi, Salma Saddeek, Kalamegam Gauthaman, and Alexandre Prola. 2022. "Punicalagin Targets Atherosclerosis: Gene Expression Profiling of THP-1 Macrophages Treated with Punicalagin and Molecular Docking" Current Issues in Molecular Biology 44, no. 5: 2153-2166. https://doi.org/10.3390/cimb44050145
APA StyleHuwait, E., Almowallad, S., Al-Massabi, R., Saddeek, S., Gauthaman, K., & Prola, A. (2022). Punicalagin Targets Atherosclerosis: Gene Expression Profiling of THP-1 Macrophages Treated with Punicalagin and Molecular Docking. Current Issues in Molecular Biology, 44(5), 2153-2166. https://doi.org/10.3390/cimb44050145







