Investigating the Link between Alpha-1 Antitrypsin and Human Neutrophil Elastase in Bronchoalveolar Lavage Fluid of COVID-19 Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Patients
2.3. BALf Collection and Processing
2.4. Neutrophil Isolation and NETosis
2.5. BCA Protein Assay
2.6. 1D-PAGE
2.7. Western Blotting
2.8. In-Situ Digestion
2.9. Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS)
2.10. Preparation of Fluorescent Alpha 1-Antitrypsin
2.11. Measurement of Elastase Activity Using a Nanodrop System
2.12. ELISA Assays
2.13. Interaction of AAT and Histones
2.14. Confocal Microscopy
3. Results
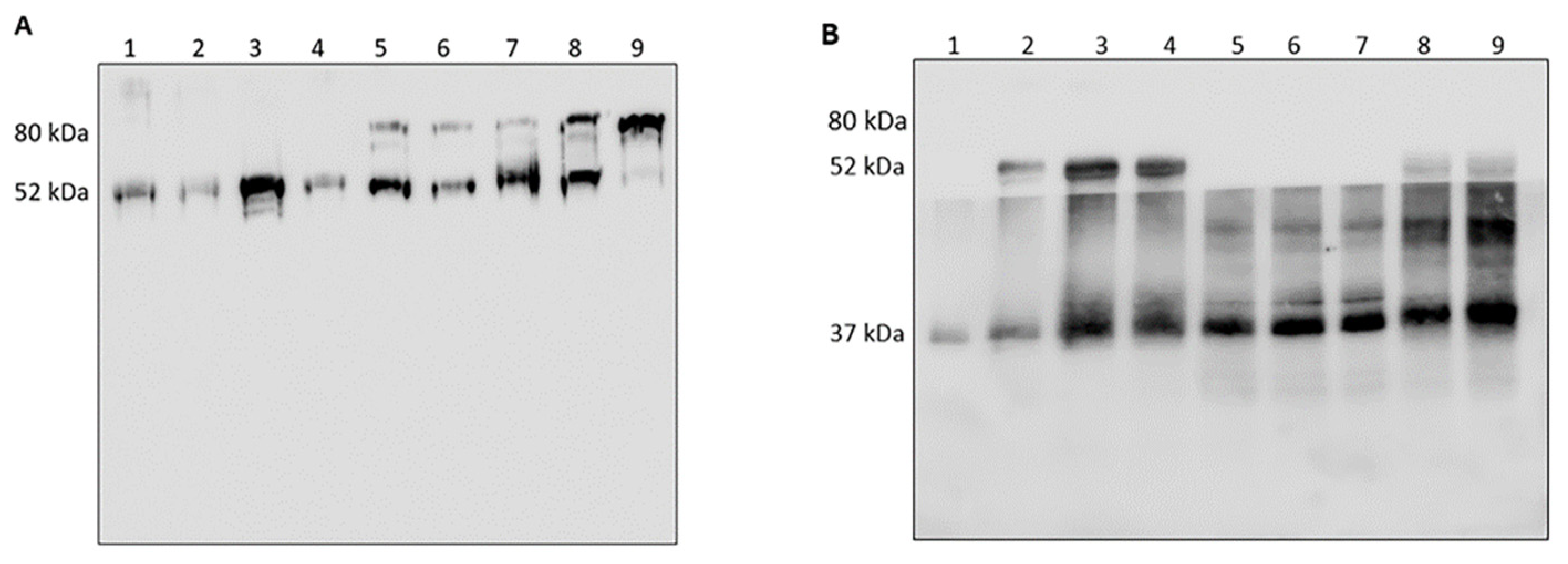
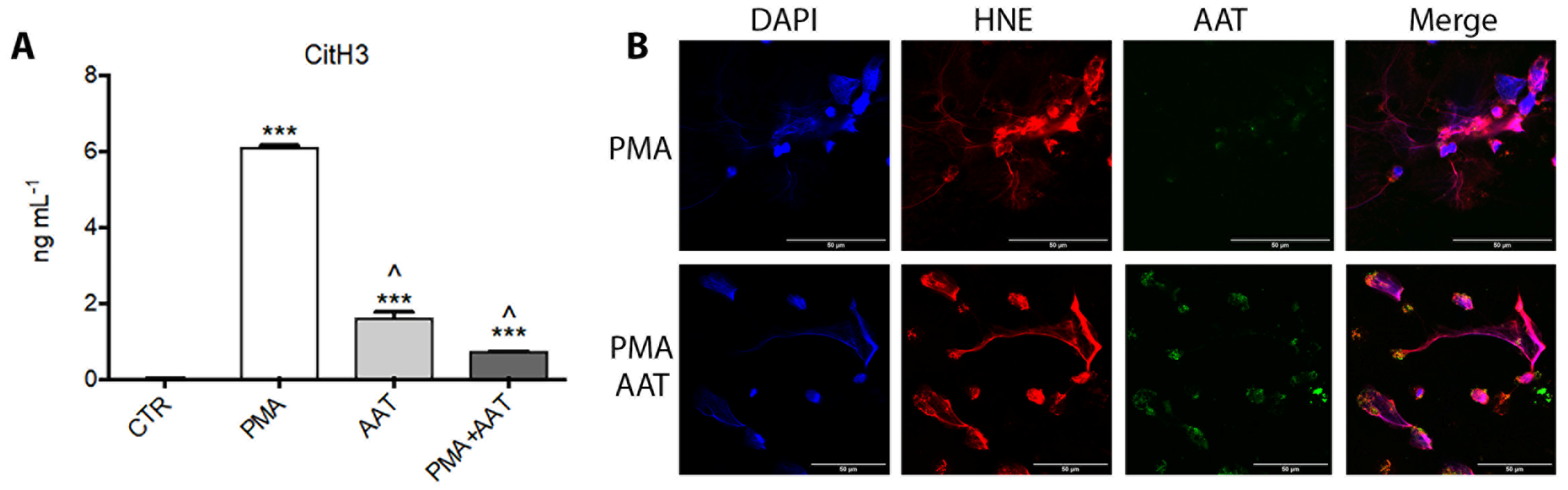
3.1. Analysis of HNE/AAT Balance in BALf of COVID-19 Patients
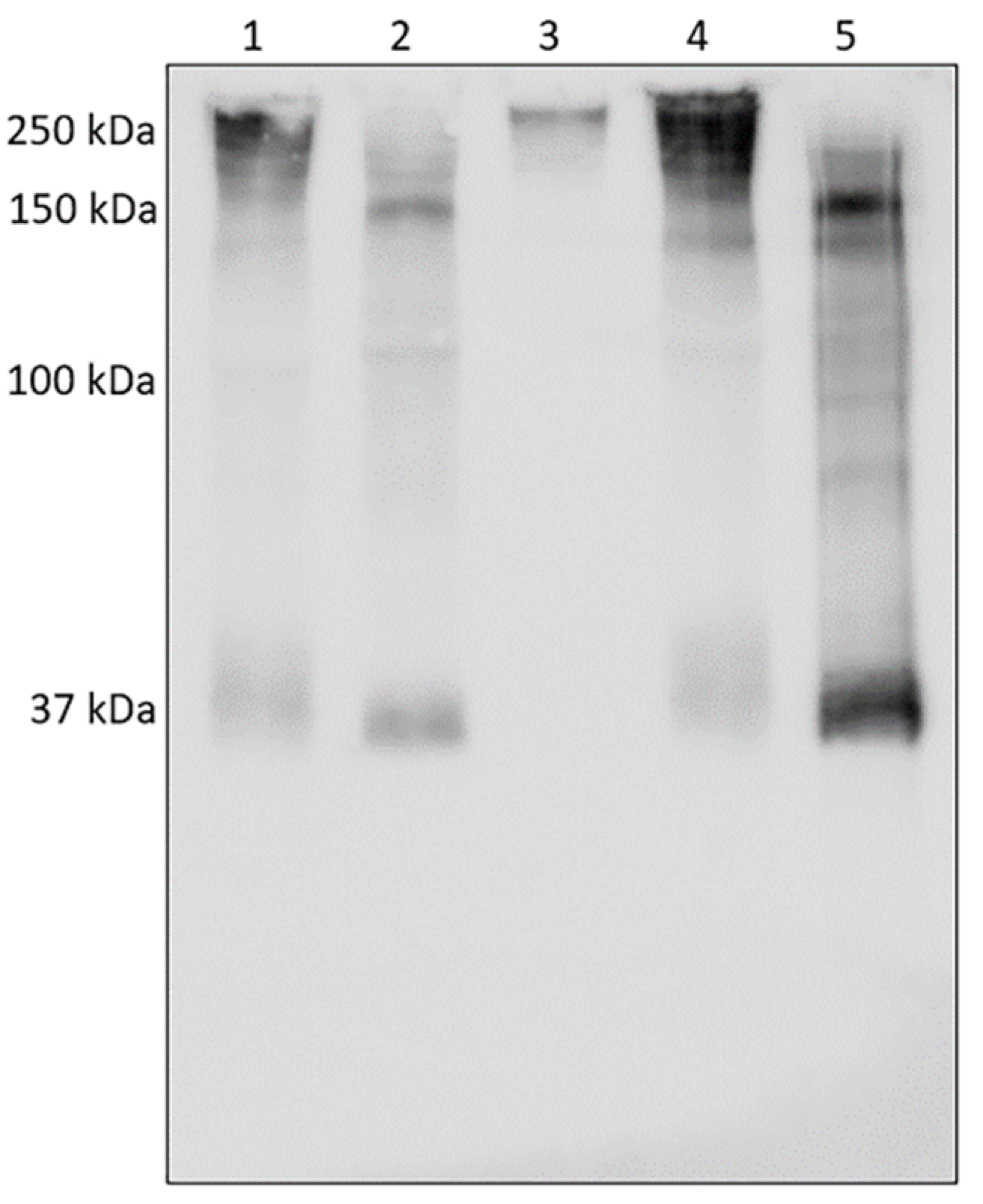
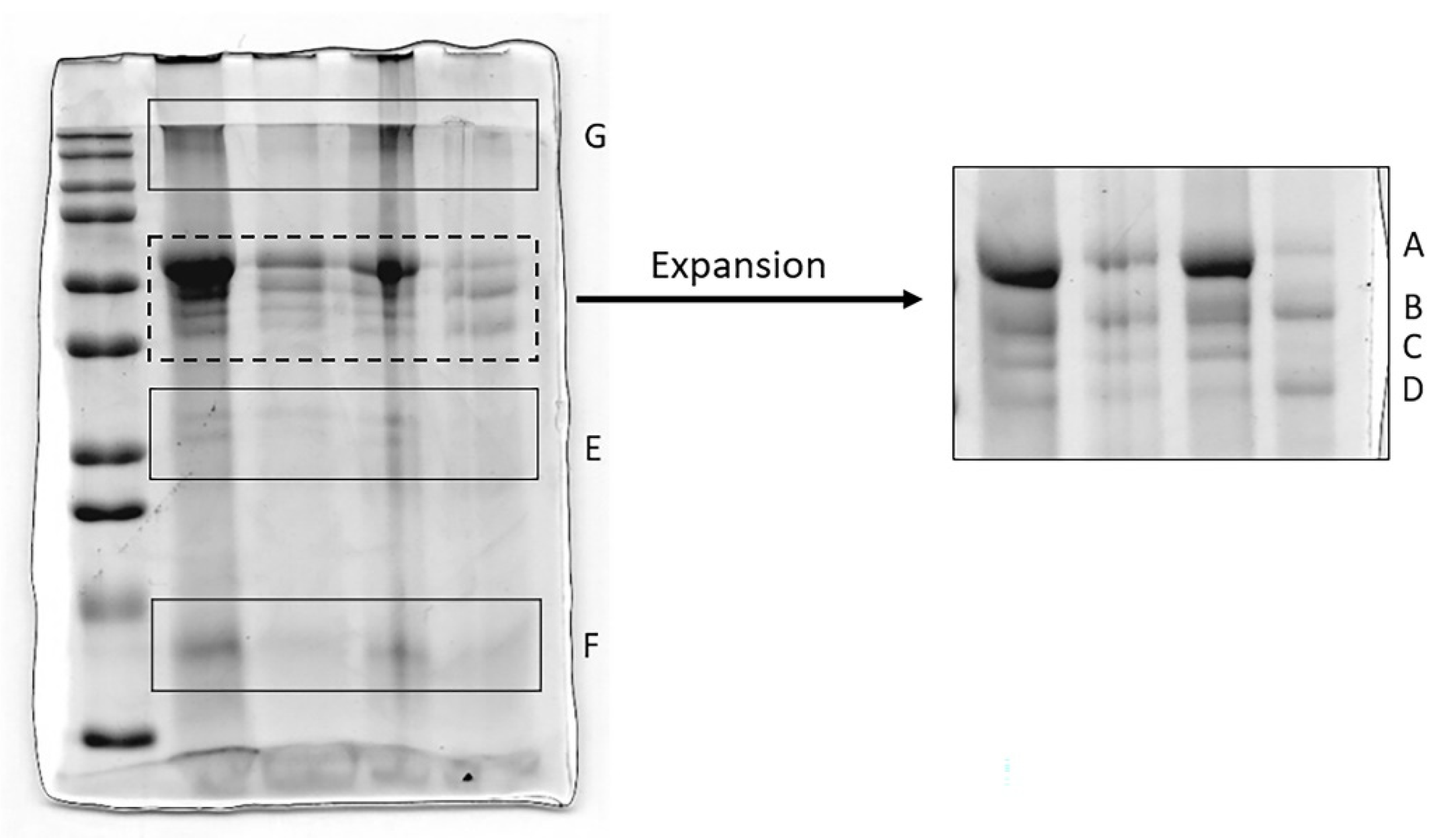
3.2. Analysis of HNE and Identification of Complexes with Other Proteins
3.3. Analysis of AAT
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Bronchoalveolar lavage | BAL |
| Human neutrophil elastase | HNE |
| Alpha1-antitrypsin | AAT |
| Reactive oxygen species | ROS |
| Neutrophil extracellular traps | NETs |
| Bronchiolitis obliterans syndrome | BOS |
| Acute respiratory distress syndrome | ARDS |
| Human lactoferrin | LF |
| Hemopexin | HPX |
| Haptoglobin | Hp |
| Fibrinogen alpha chain | FGA protein |
| Phorbol myristate acetale | PMA |
| Trichloroacetic acid | TCA |
| Sodium dodecyl sulphate | SDS |
| Bromophenol blue | BPB |
| Acetonitrile | ACN |
| Formic acid | FA |
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. China Novel Coronavirus Investigating and Research Team. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Thakur, V.; Ratho, R.K.; Kumar, P.; Bhatia, S.K.; Bora, I.; Mohi, G.K.; Saxena, S.K.; Devi, M.; Yadav, D.; Mehariya, S. Multi-Organ Involvement in COVID-19: Beyond Pulmonary Manifestations. J. Clin. Med. 2021, 10, 446. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, B.; Mao, J. The pathogenesis and treatment of the “Cytokine Storm” in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef]
- Pandolfi, L.; Fossali, T.; Frangipane, V.; Bozzini, S.; Morosini, M.; D’Amato, M.; Lettieri, S.; Urtis, M.; Di Toro, A.; Saracino, L.; et al. Broncho-alveolar inflammation in COVID-19 patients: A correlation with clinical outcome. BMC Pulm. Med. 2020, 20, 301. [Google Scholar] [CrossRef]
- McElvaney, O.J.; McEvoy, N.L.; McElvaney, O.F.; Carroll, T.P.; Murphy, M.P.; Dunlea, D.M.; Ní Choileáin, O.; Clarke, J.; O’Connor, E.; Hogan, G.; et al. Characterization of the Inflammatory Response to Severe COVID-19 Illness. Am. J. Respir. Crit. Care Med. 2020, 202, 812–821. [Google Scholar] [CrossRef]
- Magro, C.; Mulvey, J.J.; Berlin, D.; Nuovo, G.; Salvatore, S.; Harp, J.; Baxter-Stoltzfus, A.; Laurence, J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: A report of five cases. Transl. Res. 2020, 220, 1–13. [Google Scholar] [CrossRef]
- Kolaczkowska, E.; Kubes, P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 2013, 13, 159–175. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Wiegman, C.H.; Li, F.; Ryffel, B.; Togbe, D.; Chung, K.F. Oxidative Stress in Ozone-Induced Chronic Lung Inflammation and Emphysema: A Facet of Chronic Obstructive Pulmonary Disease. Front. Immunol. 2020, 11, 1957. [Google Scholar] [CrossRef]
- Cheresh, P.; Kim, S.J.; Tulasiram, S.; Kamp, D.W. Oxidative stress and pulmonary fibrosis. Biochim. Biophys. Acta 2013, 1832, 1028–1040. [Google Scholar] [CrossRef]
- Polverino, E.; Rosales-Mayor, E.; Dale, G.E.; Dembowsky, K.; Torres, A. The Role of Neutrophil Elastase Inhibitors in Lung Diseases. Chest 2017, 152, 249–262. [Google Scholar] [CrossRef]
- Cagnone, M.; Piloni, D.; Ferrarotti, I.; Di Venere, M.; Viglio, S.; Magni, S.; Bardoni, A.; Salvini, R.; Fumagalli, M.; Iadarola, P.; et al. A Pilot Study to Investigate the Balance between Proteases and α1-Antitrypsin in Bronchoalveolar Lavage Fluid of Lung Transplant Recipients. High Throughput 2019, 8, 5. [Google Scholar] [CrossRef]
- Seren, S.; Derian, L.; Keleş, I.; Guillon, A.; Lesner, A.; Gonzalez, L.; Baranek, T.; Si-Tahar, M.; Marchand-Adam, S.; Jenne, D.E.; et al. Proteinase release from activated neutrophils in mechanically ventilated patients with non-COVID-19 and COVID-19 pneumonia. Eur. Respir. J. 2021, 57, 2003755. [Google Scholar] [CrossRef]
- Guéant, J.L.; Guéant-Rodriguez, R.M.; Fromonot, J.; Oussalah, A.; Louis, H.; Chery, C.; Gette, M.; Gleye, S.; Callet, J.; Raso, J.; et al. Elastase and exacerbation of neutrophil innate immunity are involved in multi-visceral manifestations of COVID-19. Allergy 2021, 76, 1846–1858. [Google Scholar] [CrossRef]
- Akgun, E.; Tuzuner, M.B.; Sahin, B.; Kilercik, M.; Kulah, C.; Cakiroglu, H.N.; Serteser, M.; Unsal, I.; Baykal, A.T. Proteins associated with neutrophil degranulation are upregulated in nasopharyngeal swabs from SARS-CoV-2 patients. PLoS ONE 2020, 15, e0240012. [Google Scholar] [CrossRef]
- Bhattacharyya, C.; Das, C.; Ghosh, A.; Singh, A.K.; Mukherjee, S.; Majumder, P.P.; Basu, A.; Biswas, N.K. SARS-CoV-2 mutation 614G creates an elastase cleavage site enhancing its spread in high AAT-deficient regions. Infect. Genet. Evol. 2021, 90, 104760. [Google Scholar] [CrossRef]
- Vianello, A.; Braccioni, F. Geographical Overlap between Alpha-1 Antitrypsin Deficiency and COVID-19 Infection in Italy: Casual or Causal? Arch. Bronconeumol. 2020, 56, 609–610. [Google Scholar] [CrossRef]
- Oguntuyo, K.Y.; Stevens, C.S.; Siddiquey, M.N.; Schilke, R.M.; Woolard, M.D.; Zhang, H.; Acklin, J.A.; Ikegame, S.; Hung, C.T.; Lim, J.K.; et al. In plain sight: The role of alpha-1-antitrypsin in COVID-19 pathogenesis and therapeutics. bioRxiv 2020, 15, 2020.08.14.248880. [Google Scholar]
- Ritzmann, F.; Chitirala, P.; Krüger, N.; Hoffmann, M.; Zuo, W.; Lammert, F.; Smola, S.; Tov, N.; Alagem, N.; Lepper, P.M.; et al. AAT-in-COVID-19 Study Group. Therapeutic Application of Alpha-1 Antitrypsin in COVID-19. Am. J. Respir. Crit. Care Med. 2021, 204, 224–227. [Google Scholar]
- McElvaney, O.J.; Hobbs, B.D.; Qiao, D.; McElvaney, O.F.; Moll, M.; McEvoy, N.L.; Clarke, J.; O’Connor, E.; Walsh, S.; Cho, M.H.; et al. A linear prognostic score based on the ratio of interleukin-6 to interleukin-10 predicts outcomes in COVID-19. EBioMedicine 2020, 61, 103026. [Google Scholar] [CrossRef]
- Fuchs, T.A.; Abed, U.; Goosmann, C.; Hurwitz, R.; Schulze, I.; Wahn, V.; Weinrauch, Y.; Brinkmann, V.; Zychlinsky, A. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007, 176, 231–241. [Google Scholar] [CrossRef]
- Pilsczek, F.H.; Salina, D.; Poon, K.K.; Fahey, C.; Yipp, B.G.; Sibley, C.D.; Robbins, S.M.; Green, F.H.; Surette, M.G.; Sugai, M.; et al. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J. Immunol. 2010, 185, 7413–7425. [Google Scholar] [CrossRef]
- Vorobjeva, N.V.; Chernyak, B.V. NETosis: Molecular Mechanisms, Role in Physiology and Pathology. Biochemistry 2020, 85, 1178–1190. [Google Scholar] [CrossRef]
- Feng, X.; Li, S.; Sun, Q.; Zhu, J.; Chen, B.; Xiong, M.; Cao, G. Immune-Inflammatory Parameters in COVID-19 Cases: A Systematic Review and Meta-Analysis. Front. Med. 2020, 7, 301. [Google Scholar] [CrossRef]
- Jimeno, S.; Ventura, P.S.; Castellano, J.M.; García-Adasme, S.I.; Miranda, M.; Touza, P.; Lllana, I.; López-Escobar, A. Prognostic implications of neutrophil-lymphocyte ratio in COVID-19. Eur. J. Clin. Investig. 2021, 51, e13404. [Google Scholar] [CrossRef]
- Pandolfi, L.; Bozzini, S.; Frangipane, V.; Percivalle, E.; De Luigi, A.; Violatto, M.B.; Lopez, G.; Gabanti, E.; Carsana, L.; D’Amato, M.; et al. Neutrophil Extracellular Traps Induce the Epithelial-Mesenchymal Transition: Implications in Post-COVID-19 Fibrosis. Front. Immunol. 2021, 12, 663303. [Google Scholar] [CrossRef]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Weber, A.; Barnes, B.J.; Egeblad, M.; et al. Neutrophil extracellular traps in COVID-19. JCI Insight 2020, 5, e138999. [Google Scholar] [CrossRef]
- Saffarzadeh, M.; Juenemann, C.; Queisser, M.A.; Lochnit, G.; Barreto, G.; Galuska, S.P.; Lohmeyer, J.; Preissner, K.T. Neutrophil extracellular traps directly induce epithelial and endothelial cell death: A predominant role of histones. PLoS ONE 2012, 7, e32366. [Google Scholar] [CrossRef]
- Ackermann, M.; Anders, H.J.; Bilyy, R.; Bowlin, G.L.; Daniel, C.; De Lorenzo, R.; Egeblad, M.; Henneck, T.; Hidalgo, A.; Hoffmann, M.; et al. Patients with COVID-19: In the dark-NETs of neutrophils. Cell Death Differ. 2021, 28, 3125–3139. [Google Scholar] [CrossRef]
- Middleton, E.A.; He, X.Y.; Denorme, F.; Campbell, R.A.; Ng, D.; Salvatore, S.P.; Mostyka, M.; Baxter-Stoltzfus, A.; Borczuk, A.C.; Loda, M.; et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood 2020, 136, 1169–1179. [Google Scholar] [CrossRef]
- Radermecker, C.; Detrembleur, N.; Guiot, J.; Cavalier, E.; Henket, M.; d’Emal, C.; Vanwinge, C.; Cataldo, D.; Oury, C.; Delvenne, P.; et al. Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID-19. J. Exp. Med. 2020, 217, e20201012. [Google Scholar] [CrossRef] [PubMed]
- Arcanjo, A.; Logullo, J.; Menezes, C.C.B.; de Souza Carvalho Giangiarulo, T.C.; Dos Reis, M.C.; de Castro, G.M.M.; da Silva Fontes, Y.; Todeschini, A.R.; Freire-de-Lima, L.; Decoté-Ricardo, D.; et al. The emerging role of neutrophil extracellular traps in severe acute respiratory syndrome coronavirus 2 (COVID-19). Sci. Rep. 2020, 10, 19630. [Google Scholar] [CrossRef] [PubMed]
- Ducastel, M.; Chenevier-Gobeaux, C.; Ballaa, Y.; Meritet, J.F.; Brack, M.; Chapuis, N.; Pene, F.; Carlier, N.; Szwebel, T.A.; Roche, N.; et al. Oxidative Stress and Inflammatory Biomarkers for the Prediction of Severity and ICU Admission in Unselected Patients Hospitalized with COVID-19. Int. J. Mol. Sci. 2021, 22, 7462. [Google Scholar] [CrossRef] [PubMed]
- Frenzel, E.; Korenbaum, E.; Hegermann, J.; Ochs, M.; Koepke, J.; Koczulla, A.R.; Welte, T.; Köhnlein, T.; Janciauskiene, S. Does augmentation with alpha1-antitrypsin affect neutrophil extracellular traps formation? Int. J. Biol. Sci. 2012, 8, 1023–1025. [Google Scholar] [CrossRef][Green Version]
- Majewski, P.; Majchrzak-Gorecka, M.; Grygier, B.; Skrzeczynska-Moncznik, J.; Osiecka, O.; Cichy, J. Inhibitors of Serine Proteases in Regulating the Production and Function of Neutrophil Extracellular Traps. Front. Immunol. 2016, 7, 261. [Google Scholar] [CrossRef]
- Campbell, R.A.; Campbell, H.D.; Bircher, J.S.; de Araujo, C.V.; Denorme, F.; Crandell, J.L.; Rustad, J.L.; Monts, J.; Cody, M.J.; Kosaka, Y.; et al. Placental HTRA1 cleaves α1-antitrypsin to generate a NET-inhibitory peptide. Blood 2021, 138, 977–988. [Google Scholar] [CrossRef]
- Zabieglo, K.; Majewski, P.; Majchrzak-Gorecka, M.; Wlodarczyk, A.; Grygier, B.; Zegar, A.; Kapinska-Mrowiecka, M.; Naskalska, A.; Pyrc, K.; Dubin, A.; et al. The inhibitory effect of secretory leukocyte protease inhibitor (SLPI) on formation of neutrophil extracellular traps. J. Leukoc. Biol. 2015, 98, 99–106. [Google Scholar] [CrossRef]
- da Cruz, D.B.; Helms, J.; Aquino, L.R.; Stiel, L.; Cougourdan, L.; Broussard, C.; Chafey, P.; Riès-Kautt, M.; Meziani, F.; Toti, F.; et al. DNA-bound elastase of neutrophil extracellular traps degrades plasminogen, reduces plasmin formation, and decreases fibrinolysis: Proof of concept in septic shock plasma. FASEB J. 2019, 33, 14270–14280. [Google Scholar] [CrossRef]
- Dubois, A.V.; Gauthier, A.; Bréa, D.; Varaigne, F.; Diot, P.; Gauthier, F.; Attucci, S. Influence of DNA on the activities and inhibition of neutrophil serine proteases in cystic fibrosis sputum. Am. J. Respir. Cell Mol. Biol. 2012, 47, 80–86. [Google Scholar] [CrossRef]
- Gogol, M.; Ostrowska, D.; Klaga, K.; Bochenska, O.; Wolak, N.; Aoki, W.; Ueda, M.; Kozik, A.; Rapala-Kozik, M. Inactivation of α1-proteinase inhibitor by Candida albicans aspartic proteases favors the epithelial and endothelial cell colonization in the presence of neutrophil extracellular traps. Acta Biochim. Pol. 2016, 63, 167–175. [Google Scholar] [CrossRef]
- Ciaramelli, C.; Fumagalli, M.; Viglio, S.; Bardoni, A.M.; Piloni, D.; Meloni, F.; Iadarola, P.; Airoldi, C. 1H NMR To Evaluate the Metabolome of Bronchoalveolar Lavage Fluid (BALf) in Bronchiolitis Obliterans Syndrome (BOS): Toward the Development of a New Approach for Biomarker Identification. J. Proteome Res. 2017, 16, 1669–1682. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Yvon, M.; Chabanet, C.; Pélissier, J.P. Solubility of peptides in trichloroacetic acid (TCA) solutions. Hypothesis on the precipitation mechanism. Int. J. Pept. Protein Res. 1989, 34, 166–176. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Candiano, G.; Bruschi, M.; Musante, L.; Santucci, L.; Ghiggeri, G.M.; Carnemolla, B.; Orecchia, P.; Zardi, L.; Righetti, P.G. Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 2004, 25, 13. [Google Scholar] [CrossRef] [PubMed]
- Di Venere, M.; Fumagalli, M.; Cafiso, A.; De Marco, L.; Epis, S.; Plantard, O.; Bardoni, A.; Salvini, R.; Viglio, S.; Bazzocchi, C.; et al. Ixodes ricinus and Its Endosymbiont Midichloria mitochondrii: A Comparative Proteomic Analysis of Salivary Glands and Ovaries. PLoS ONE 2015, 10, e0138842. [Google Scholar] [CrossRef]
- Cochrane, C.G.; Spragg, R.; Revak, S.D. Pathogenesis of the adult respiratory distress syndrome. Evidence of oxidant activity in bronchoalveolar lavage fluid. J. Clin. Investig. 1983, 71, 754–761. [Google Scholar] [CrossRef]
- Luisetti, M.; Piccioni, P.D.; Donnetta, A.M.; Bulgheroni, A.; Peona, V. Protease-antiprotease imbalance: Local evaluation with bronchoalveolar lavage. Respiration 1992, 59, 24–27. [Google Scholar] [CrossRef]
- Wewers, M.D.; Herzyk, D.J.; Gadek, J.E. Alveolar fluid neutrophil elastase activity in the adult respiratory distress syndrome is complexed to alpha-2-macroglobulin. J. Clin. Investig. 1988, 82, 1260–1267. [Google Scholar] [CrossRef]
- Nakazawa, D.; Kumar, S.V.; Marschner, J.; Desai, J.; Holderied, A.; Rath, L.; Kraft, F.; Lei, Y.; Fukasawa, Y.; Moeckel, G.W.; et al. Histones and Neutrophil Extracellular Traps Enhance Tubular Necrosis and Remote Organ Injury in Ischemic AKI. J. Am. Soc. Nephrol. 2017, 28, 1753–1768. [Google Scholar] [CrossRef]
- Taggart, C.; Cervantes-Laurean, D.; Kim, G.; McElvaney, N.G.; Wehr, N.; Moss, J.; Levine, R.L. Oxidation of either methionine 351 or methionine 358 in alpha 1-antitrypsin causes loss of anti-neutrophil elastase activity. J. Biol. Chem. 2000, 275, 27258–27265. [Google Scholar] [CrossRef]
- Kell, D.B.; Heyden, E.L.; Pretorius, E. The Biology of Lactoferrin, an Iron-Binding Protein that Can Help Defend against Viruses and Bacteria. Front. Immunol. 2020, 11, 1221. [Google Scholar] [CrossRef] [PubMed]
- Kühnle, A.; Lütteke, T.; Bornhöfft, K.F.; Galuska, S.P. Polysialic Acid Modulates the Binding of External Lactoferrin in Neutrophil Extracellular Traps. Biology 2019, 8, 20. [Google Scholar] [CrossRef]
- Redwan, E.M.; Uversky, V.N.; El-Fakharany, E.M.; Al-Mehdar, H. Potential lactoferrin activity against pathogenic viruses. Comptes Rendus Biol. 2014, 337, 581–595. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. Cold Spring Harbor Perspect. Biol. 2011, 3, a004952. [Google Scholar] [CrossRef] [PubMed]
- Tolosano, E.; Altruda, F. Hemopexin: Structure, function, and regulation. DNA Cell Biol. 2002, 21, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Kwak, Y.H.; Sammy, F.; He, P.; Thundivalappil, S.; Sun, G.; Chao, W.; Warren, H.S. Synergistic inflammation is induced by blood degradation products with microbial Toll-like receptor agonists and is blocked by hemopexin. J. Infect. Dis. 2010, 202, 624–632. [Google Scholar] [CrossRef]
- Chintagari, N.R.; Jana, S.; Alayash, A.I. Oxidized Ferric and Ferryl Forms of Hemoglobin Trigger Mitochondrial Dysfunction and Injury in Alveolar Type I Cells. Am. J. Respir. Cell Mol. Biol. 2016, 55, 288–298. [Google Scholar] [CrossRef]
- Sirivongrangson, P.; Kulvichit, W.; Payungporn, S.; Pisitkun, T.; Chindamporn, A.; Peerapornratana, S.; Pisitkun, P.; Chitcharoen, S.; Sawaswong, V.; Worasilchai, N.; et al. Endotoxemia and circulating bacteriome in severe COVID-19 patients. Intensive Care Med. Exp. 2020, 8, 72. [Google Scholar] [CrossRef]
- di Masi, A.; De Simone, G.; Ciaccio, C.; D’Orso, S.; Coletta, M.; Ascenzi, P. Haptoglobin: From hemoglobin scavenging to human health. Mol. Asp. Med. 2020, 73, 100851. [Google Scholar] [CrossRef]
- Naryzny, S.N.; Legina, O.K. Haptoglobin as a Biomarker. Biochem. Mosc. Suppl. B Biomed. Chem. 2021, 15, 184–198. [Google Scholar] [CrossRef] [PubMed]
- Schaer, D.J.; Vinchi, F.; Ingoglia, G.; Tolosano, E.; Buehler, P.W. Haptoglobin, hemopexin, and related defense pathways-basic science, clinical perspectives, and drug development. Front. Physiol. 2014, 5, 415. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wang, J.Y. Role of Polymeric Immunoglobulin Receptor in IgA and IgM Transcytosis. Int. J. Mol. Sci. 2021, 22, 2284. [Google Scholar] [CrossRef] [PubMed]
- Turula, H.; Wobus, C.E. The Role of the Polymeric Immunoglobulin Receptor and Secretory Immunoglobulins during Mucosal Infection and Immunity. Viruses 2018, 10, 237. [Google Scholar] [CrossRef]
- Pilette, C.; Ouadrhiri, Y.; Dimanche, F.; Vaerman, J.P.; Sibille, Y. Secretory component is cleaved by neutrophil serine proteinases but its epithelial production is increased by neutrophils through NF-kappa B- and p38 mitogen-activated protein kinase-dependent mechanisms. Am. J. Respir. Cell Mol. Biol. 2003, 28, 485–498. [Google Scholar] [CrossRef]
- Healy, L.D.; Puy, C.; Itakura, A.; Chu, T.; Robinson, D.K.; Bylund, A.; Phillips, K.G.; Gardiner, E.E.; McCarty, O.J. Colocalization of neutrophils, extracellular DNA and coagulation factors during NETosis: Development and utility of an immunofluorescence-based microscopy platform. J. Immunol. Methods 2016, 435, 77–84. [Google Scholar] [CrossRef]
- Fisher, J.; Mohanty, T.; Karlsson, C.A.Q.; Khademi, S.M.H.; Malmström, E.; Frigyesi, A.; Nordenfelt, P.; Malmstrom, J.; Linder, A. Proteome Profiling of Recombinant DNase Therapy in Reducing NETs and Aiding Recovery in COVID-19 Patients. Mol. Cell Proteom. 2021, 20, 100113. [Google Scholar] [CrossRef]
| Numbering | Age | Gender | Macrophages (%) | Lymphocytes (%) | Neutrophils (%) | Eosinophils (%) | Days after Intubation | P/F ** |
|---|---|---|---|---|---|---|---|---|
| COVID-01 | 67 | M | n.d. | n.d. | n.d. | n.d. | 1 | 123 |
| COVID-02 | 54 | M | 9 | 7 | 81 | 2 | 2 | 107 |
| COVID-03 | 41 | M | 41 | 0 | 60 | 0 | 3 | 92 |
| COVID-04 | 69 | M | n.d. | n.d. | n.d. | n.d. | 1 | 107 |
| COVID-05 | 60 | M | 15 | 0 | 85 | 0 | 4 | 68 |
| COVID-06 | 50 | F | 2 | 6 | 92 | 0 | 3 | 180 |
| COVID-07 | 47 | F | 23 | 12 | 65 | 0 | 3 | 98 |
| COVID-08 | 66 | M | n.d. | n.d. | n.d. | n.d. | 2 | 111 |
| COVID-09 | 78 | F | 33 | 24 | 43 | 0 | 2 | 209 |
| COVID-10 | 51 | F | 34 | 8 | 58 | 0 | 3 | 142 |
| COVID-11 | 60 | M | 64 | 25 | 11 | 0 | 3 | 123 |
| COVID-12 | 66 | F | 45 | 29 | 26 | 0 | 4 | 71 |
| COVID-13 | 60 | M | 81 | 9 | 10 | 0 | 2 | 73 |
| COVID-14 | 53 | M | 56 | 13 | 31 | 0 | 2 | 200 |
| COVID-15 | 43 | M | 17 | 10 | 74 | 0 | 3 | 113 |
| COVID-16 | 60 | M | n.d. | n.d. | n.d. | n.d. | 1 | 145 |
| COVID-17 | 61 | M | 33 | 12 | 61 | 0 | 1 | 192 |
| COVID-18 | 72 | F | 10 | 14 | 77 | 0 | 1 | 300 |
| COVID-19 | 56 | F | n.d. | n.d. | n.d. | n.d. | 1 | 132 |
| COVID-20 | 60 | M | 18 | 13 | 81 | 0 | 2 | 145 |
| COVID-21 | 28 | M | n.d. | n.d. | n.d. | n.d. | 1 | 188 |
| COVID-22 | 65 | M | 14 | 10 | 76 | 0 | 3 | 121 |
| COVID-23 | 55 | M | 68 | 10 | 22 | 0 | 1 | 185 |
| COVID-24 | 63 | M | 35 | 14 | 51 | 0 | 1 | 176 |
| COVID-25 | 80 | M | 8 | 1 | 91 | 0 | 2 | 148 |
| COVID-26 | 64 | M | 8 | 2 | 90 | 0 | 2 | 200 |
| COVID-27 | 67 | M | n.d. | n.d. | n.d. | n.d. | 2 | 179 |
| COVID-28 | 29 | M | 18 | 4 | 78 | 0 | 1 | 210 |
| COVID-29 | 74 | M | 19 | 6 | 75 | 0 | 4 | 300 |
| COVID-30 | 70 | M | 13 | 5 | 82 | 0 | 3 | 192 |
| COVID-31 | 63 | M | n.d. | n.d. | n.d. | n.d. | 2 | 175 |
| COVID-32 | 69 | M | n.d. | n.d. | n.d. | n.d. | 2 | 187 |
| COVID-33 | 68 | F | 12 | 11 | 77 | 0 | 1 | 158 |
| Numbering | BCA (mg/mL) | Elastase (ng/mL) | Elastase Specific Activity (mU/mg) | NETs (Cit H3 ng/mL) |
|---|---|---|---|---|
| COVID-01 | 1.24 | 40.70 | 2.79 | 2.98 |
| COVID-02 | 0.72 | 60.69 | 1.22 | 0.96 |
| COVID-03 | 0.95 | 68.36 | 1.07 | n.d. |
| COVID-04 | 2.64 | 7.92 | 1.34 | n.d. |
| COVID-05 | 1.44 | 92.92 | 1.66 | 1.52 |
| COVID-06 | 3.16 | 32.99 | 3.69 | n.d. |
| COVID-07 | 0.39 | 29.44 | 1.10 | 0.32 |
| COVID-08 | 0.52 | 11.82 | 0.04 | 0.13 |
| COVID-09 | 1.29 | 82.29 | 0.74 | 0.05 |
| COVID-10 | 1.67 | 29.19 | 0.41 | 0.10 |
| COVID-11 | 0.38 | 2.65 | 3.68 | 0.06 |
| COVID-12 | 0.36 | 6.63 | 0.64 | 0.06 |
| COVID-13 | 0.38 | 2.99 | 0.32 | n.d. |
| COVID-14 | 0.42 | 22.64 | 0.29 | 0.17 |
| COVID-15 | 0.41 | 25.81 | 0.19 | 0.29 |
| COVID-16 | 1.55 | 80.01 | 0.27 | 2.64 |
| COVID-17 | 0.81 | 36.94 | 0.81 | 0.18 |
| COVID-18 | 1.02 | 47.54 | 0.43 | 0.09 |
| COVID-19 | 1.03 | 12.80 | 1.45 | 0.09 |
| COVID-20 | 0.33 | 2.41 | 1.24 | 0.27 |
| COVID-21 | 1.25 | 37.63 | 1.97 | 2.89 |
| COVID-22 | 0.78 | 2.62 | 0.17 | 0.38 |
| COVID-23 | 0.43 | 2.79 | 0.56 | n.d. |
| COVID-24 | 1.69 | 13.50 | 0.80 | 1.06 |
| COVID-25 | 0.99 | 13.60 | 2.04 | 0.16 |
| COVID-26 | 0.54 | 2.02 | 1.37 | n.d. |
| COVID-27 | 0.97 | 12.52 | 1.77 | 0.17 |
| COVID-28 | 0.36 | 4.15 | 0.97 | 0.13 |
| COVID-29 | 0.58 | 15.24 | 1.53 | 3.26 |
| COVID-30 | 0.57 | 14.23 | 0.91 | 0.07 |
| COVID-31 | 0.57 | 1.88 | 0.74 | 0.71 |
| COVID-32 | 0.73 | 10.78 | 0.46 | 0.54 |
| COVID-33 | 0.55 | 8.51 | 1.62 | 0.33 |
| Accession Number | Gene Name | Protein Name | Score (%) | Coverage (%) | Peptide Count |
|---|---|---|---|---|---|
| tr|Q19KS2|Q19KS2_HUMAN | LTF | Lactoferrin | 92 | 3.68 | 2 |
| sp|P61626|LYSC_HUMAN | LYZ | Lysozyme C | 99 | 23.65 | 5 |
| sp|P02790|HEMO_HUMAN | HPX | Hemopexin | 99 | 8.44 | 4 |
| tr|Q8WW76|Q8WW76_HUMAN | FGA | Fibrinogen alpha chain | 99 | 20.18 | 6 |
| sp|P01833|PIGR_HUMAN | PIGR | Polymeric immunoglobulin receptor | 99 | 4.32 | 4 |
| tr|Q6NSB4|Q6NSB4_HUMAN | HP | haptoglobin (HP protein) | 91 | 16.01 | 3 |
| sp|P01024|CO3_HUMAN | C3 | Complement C3 | 99 | 5.17 | 10 |
| tr|H9KV48|H9KV48_HUMAN | SERPING1 | Plasma protease C1 inhibitor | 90 | 5.18 | 4 |
| sp|P62807|H2B1C_HUMAN | H2BC4 | Histone H2B | 99 | 35.71 | 7 |
| tr|B2R5B3|B2R5B3_HUMAN | H2AFB1 | Histone H2A | 99 | 16.92 | 4 |
| sp|P62805|H4_HUMAN | H4C1 | Histone H4 | 99 | 45.63 | 7 |
| sp|P01009|A1AT_HUMAN | SERPINA1 | Alpha-1-antitrypsin | 99 | 22.73 | 13 |
| sp|P08246|ELNE_HUMAN | ELANE | Human Neutrophil Elastase | 69 | 12.36 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Amato, M.; Vertui, V.; Pandolfi, L.; Bozzini, S.; Fossali, T.; Colombo, R.; Aliberti, A.; Fumagalli, M.; Iadarola, P.; Didò, C.; et al. Investigating the Link between Alpha-1 Antitrypsin and Human Neutrophil Elastase in Bronchoalveolar Lavage Fluid of COVID-19 Patients. Curr. Issues Mol. Biol. 2022, 44, 2122-2138. https://doi.org/10.3390/cimb44050143
D’Amato M, Vertui V, Pandolfi L, Bozzini S, Fossali T, Colombo R, Aliberti A, Fumagalli M, Iadarola P, Didò C, et al. Investigating the Link between Alpha-1 Antitrypsin and Human Neutrophil Elastase in Bronchoalveolar Lavage Fluid of COVID-19 Patients. Current Issues in Molecular Biology. 2022; 44(5):2122-2138. https://doi.org/10.3390/cimb44050143
Chicago/Turabian StyleD’Amato, Maura, Valentina Vertui, Laura Pandolfi, Sara Bozzini, Tommaso Fossali, Riccardo Colombo, Anna Aliberti, Marco Fumagalli, Paolo Iadarola, Camilla Didò, and et al. 2022. "Investigating the Link between Alpha-1 Antitrypsin and Human Neutrophil Elastase in Bronchoalveolar Lavage Fluid of COVID-19 Patients" Current Issues in Molecular Biology 44, no. 5: 2122-2138. https://doi.org/10.3390/cimb44050143
APA StyleD’Amato, M., Vertui, V., Pandolfi, L., Bozzini, S., Fossali, T., Colombo, R., Aliberti, A., Fumagalli, M., Iadarola, P., Didò, C., Viglio, S., & Meloni, F. (2022). Investigating the Link between Alpha-1 Antitrypsin and Human Neutrophil Elastase in Bronchoalveolar Lavage Fluid of COVID-19 Patients. Current Issues in Molecular Biology, 44(5), 2122-2138. https://doi.org/10.3390/cimb44050143








