Analysis of Non-Relapsed and Relapsed Adult Type Granulosa Cell Tumors Suggests Stable Transcriptomes during Tumor Progression
Abstract
1. Introduction
2. Materials and Methods
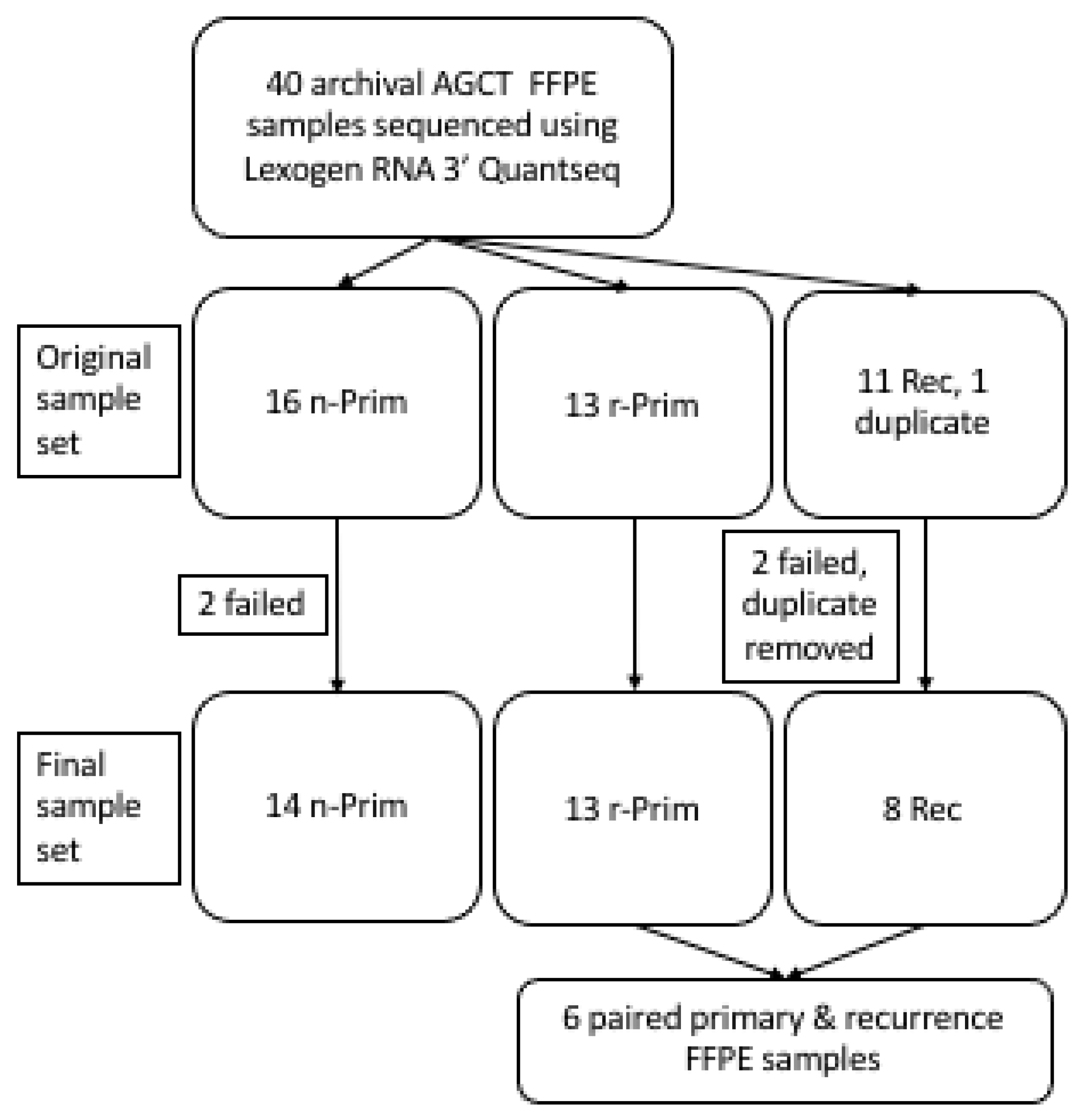
2.1. Patient Samples
2.2. RNA Extraction and RNA Quantity and Quality Assessment
2.3. Library Preparation and RNA Sequencing
2.4. Bioinformatics and Statistical Analysis
2.5. RNA In Situ Hybridization
3. Results
3.1. Transcriptional Profiling of Archival AGCT Samples
3.2. Primary AGCT Samples with or without Later Relapse Show Highly Similar Gene Expression Patterns
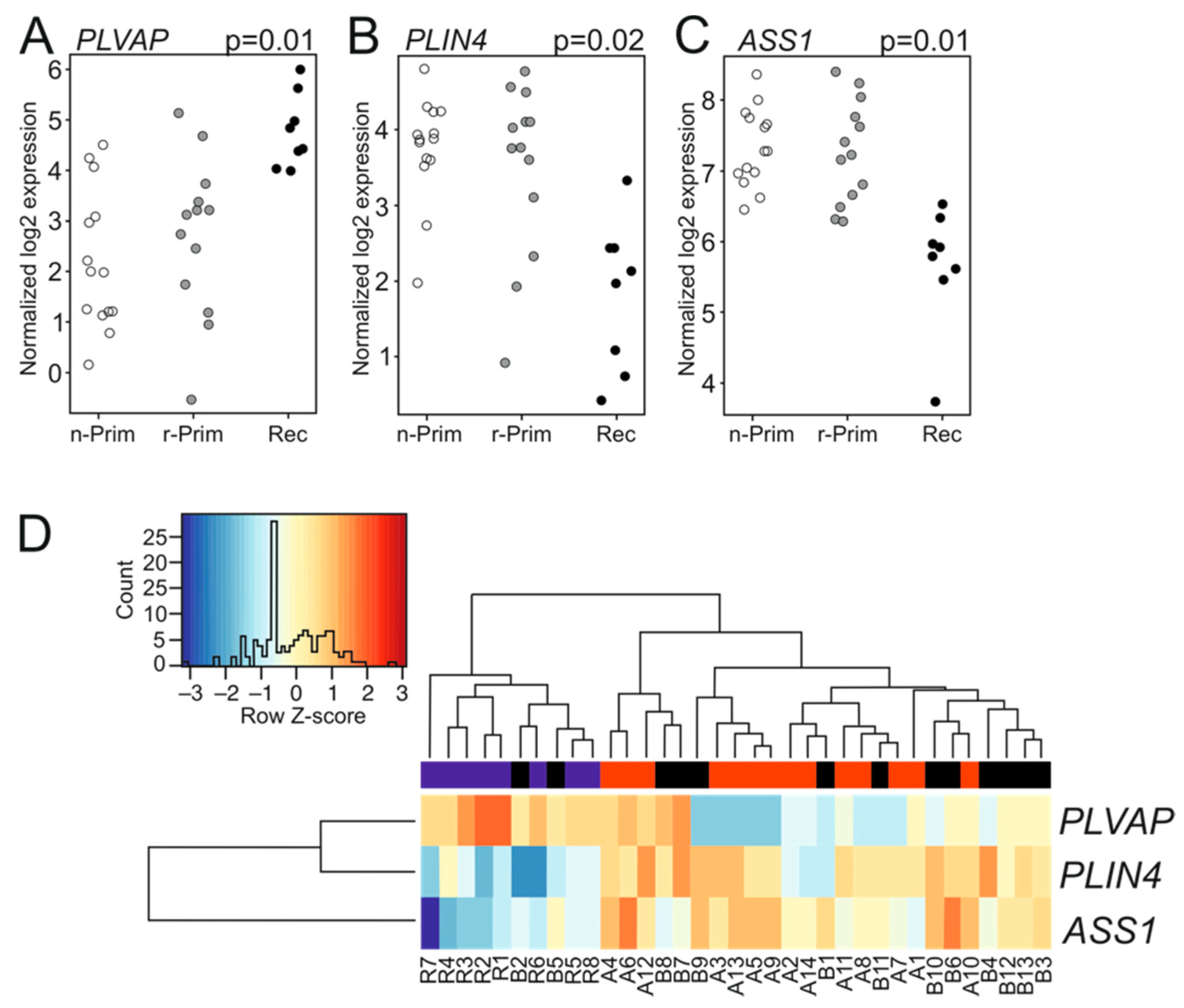
3.3. Relapsed AGCT Show Stable Transcriptomic Profiles as Compared to Primary Tumors
3.4. Gene Expression Patterns in AGCT Are Independent of TERT Mutation Status
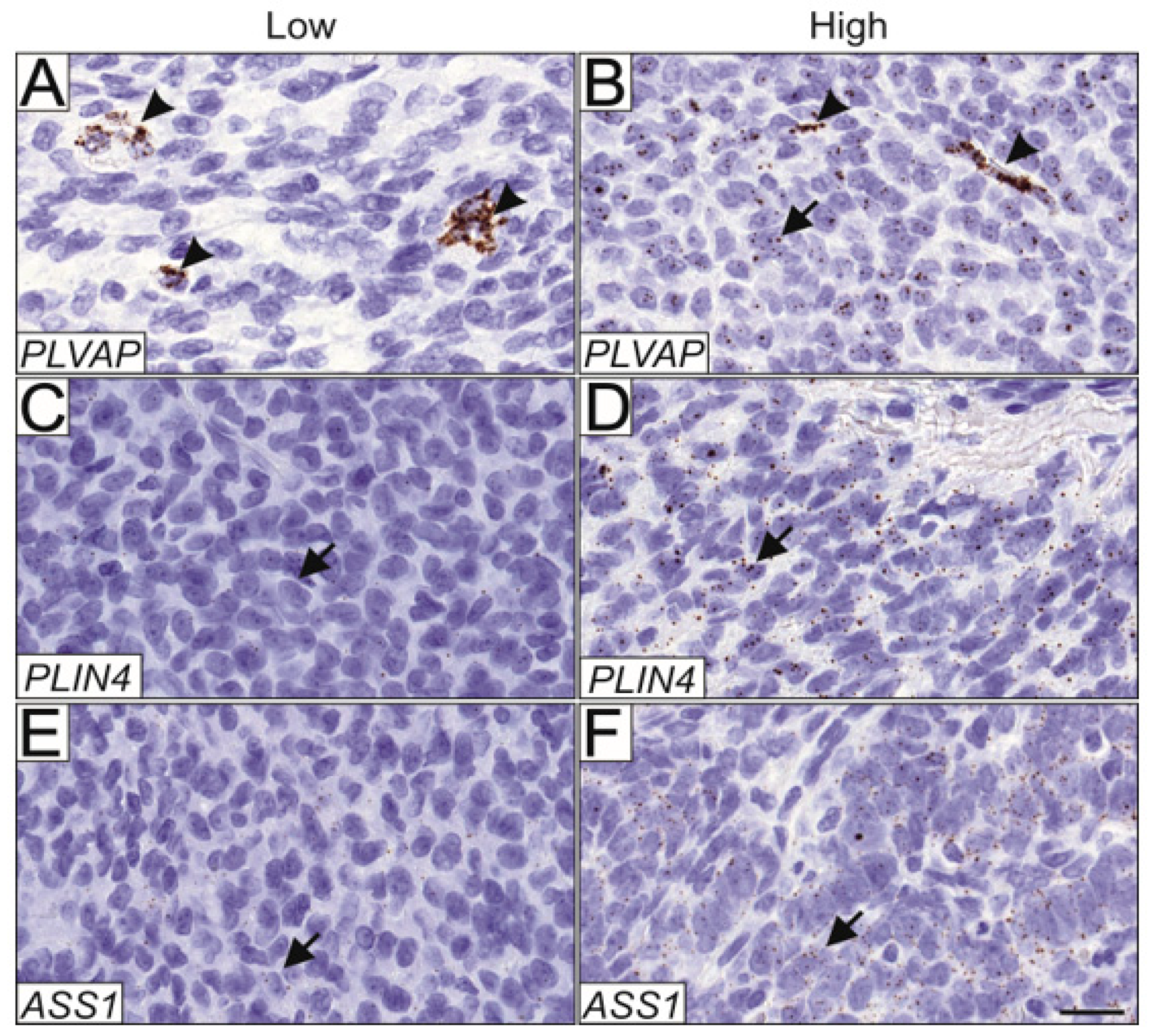
3.5. A TMA Validation Cohort Confirms PLVAP Upregulation Both in Tumor Cells and Endothelial Cells within Relapsed Samples
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jamieson, S.; Fuller, P.J. Molecular pathogenesis of granulosa cell tumors of the ovary. Endocr. Rev. 2012, 33, 109–144. [Google Scholar] [CrossRef] [PubMed]
- Bryk, S.; Farkkila, A.; Butzow, R.; Leminen, A.; Heikinheimo, M.; Anttonen, M.; Riska, A.; Unkila-Kallio, L. Clinical characteristics and survival of patients with an adult-type ovarian granulosa cell tumor: A 56-year single-center experience. Int. J. Gynecol. Cancer 2015, 25, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Boussios, S.; Zarkavelis, G.; Seraj, E.; Zerdes, I.; Tatsi, K.; Pentheroudakis, G. Non-epithelial Ovarian Cancer: Elucidating Uncommon Gynaecological Malignancies. Anticancer Res. 2016, 36, 5031–5042. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.P.; Kobel, M.; Senz, J.; Morin, R.D.; Clarke, B.A.; Wiengard, K.C.; Leung, G.; Zayed, A.; Mehl, E.; Kalloger, S.E.; et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N. Engl. J. Med. 2009, 360, 2719–2729. [Google Scholar] [CrossRef]
- McConechy, M.K.; Farkkila, A.; Horlings, H.M.; Talhouk, A.; Unkila-Kallio, L.; van Meurs, H.S.; Yang, W.; Rozenberg, N.; Andersson, N.; Zaby, K.; et al. Molecularly Defined Adult Granulosa Cell Tumor of the Ovary: The Clinical Phenotype. J. Natl. Cancer Inst. 2016, 108, djw134. [Google Scholar] [CrossRef] [PubMed]
- Färkkilä, A.; McConechy, M.; Yang, W.; Talhouk, A.; Ng, Y.; Lum, A.; Morin, R.D.; Bushell, K.; Riska, A.; McAlpine, J.; et al. FOXL2 402C>G Mutation Can Be Identified in the Circulating Tumor DNA of Patients with Adult-Type Granulosa Cell Tumor. J. Mol. Diagn. 2017, 19, 126–136. [Google Scholar] [CrossRef]
- Mangili, G.; Ottolina, J.; Gadducci, A.; Giorda, G.; Breda, E.; Savarese, A.; Candiani, M.; Frigerio, L.; Scarfone, G.; Pignata, S.; et al. Long-term follow-up is crucial after treatment for granulosa cell tumours of the ovary. Br. J. Cancer 2013, 109, 29–34. [Google Scholar] [CrossRef]
- Färkkilä, A.; Andersson, N.; Butzow, R.; Leminen, A.; Heikinheimo, M.; Anttonen, M.; Unkila-Kallio, L. HER2 and GATA4 are new prognostic factors for early-stage ovarian granulosa cell tumor-a long-term follow-up study. Cancer Med. 2014, 3, 526–536. [Google Scholar] [CrossRef]
- Evans, A.T.; Gaffey, T.A.; Malkasian, G.D.; Annegers, J.F. Clinicopathologic review of 118 granulosa and 82 theca cell tumors. Obstet. Gynecol. 1980, 55, 231–238. [Google Scholar]
- Bryk, S.; Farkkila, A.; Butzow, R.; Leminen, A.; Tapper, J.; Heikinheimo, M.; Unkila-Kallio, L.; Riska, A. Characteristics and outcome of recurrence in molecularly defined adult-type ovarian granulosa cell tumors. Gynecol. Oncol. 2016, 143, 571–577. [Google Scholar] [CrossRef]
- Pilsworth, J.A.; Cochrane, D.R.; Xia, Z.; Aubert, G.; Farkkila, A.E.M.; Horlings, H.M.; Yanagida, S.; Yang, W.; Lim, J.L.P.; Wang, Y.K.; et al. Correction: TERT promoter mutation in adult granulosa cell tumor of the ovary. Mod. Pathol. 2019, 32, 593. [Google Scholar] [CrossRef] [PubMed]
- Alexiadis, M.; Rowley, S.M.; Chu, S.; Leung, D.T.H.; Stewart, C.J.R.; Amarasinghe, K.C.; Campbell, I.; Fuller, P. Mutational Landscape of Ovarian Adult Granulosa Cell Tumors from Whole Exome and Targeted TERT Promoter Sequencing. Mol. Cancer Res. 2019, 17, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Hillman, R.T.; Celestino, J.; Terranova, C.; Beird, H.C.; Gumbs, C.; Little, L.; Nguyen, T.; Thornton, R.; Tippen, S.; Zhang, J.; et al. KMT2D/MLL2 inactivation is associated with recurrence in adult-type granulosa cell tumors of the ovary. Nat. Commun. 2018, 9, 2496. [Google Scholar] [CrossRef] [PubMed]
- Haltia, U.M.; Pihlajoki, M.; Andersson, N.; Makinen, L.; Tapper, J.; Cervera, A.; Horlings, H.M.; Turpeinen, U.; Anttonen, M.; Butzow, R.; et al. Functional profiling of FSH and estradiol in ovarian granulosa cell tumors. J. Endocr. Soc. 2020, 4, bvaa034. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.H.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Bossel Ben-Moshe, N.; Gilad, S.; Perry, G.; Benjamin, S.; Balint-Lahat, N.; Pavlovsky, A.; Halperin, S.; Markus, B.; Yosepovich, A.; Barshack, I.; et al. mRNA-seq whole transcriptome profiling of fresh frozen versus archived fixed tissues. BMC Genom. 2018, 19, 419. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, H.; Hou, Y.; Wei, T.; Liu, J. Plasmalemma vesicle-associated protein: A crucial component of vascular homeostasis. Exp. Ther. Med. 2016, 12, 1639–1644. [Google Scholar] [CrossRef]
- Sirois, I.; Aguilar-Mahecha, A.; Lafleur, J.; Fowler, E.; Vu, V.; Scriver, M.; Buchanan, M.; Chabot, C.; Ramanathan, A.; Balachandran, B.; et al. A Unique Morphological Phenotype in Chemoresistant Triple-Negative Breast Cancer Reveals Metabolic Reprogramming and PLIN4 Expression as a Molecular Vulnerability. Mol. Cancer Res. 2019, 17, 2492–2507. [Google Scholar] [CrossRef]
- Hsieh, K.; Lee, Y.K.; Londos, C.; Raaka, B.M.; Dalen, K.T.; Rimmel, A.R. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J. Cell Sci. 2012, 125, 4067–4076. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-Y.; Chi, H.-C.; Chi, L.-M.; Yang, H.-Y.; Tsai, M.-M.; Lee, K.-F.; Huang, H.-W.; Chou, L.-F.; Cheng, A.-J.; Yang, C.-W.; et al. Argininosuccinate synthetase 1 contributes to gastric cancer invasion and progression by modulating autophagy. FASEB J. 2018, 32, 2601–2614. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.L.; Kash, J.C.; Beres, S.B.; Sheng, Z.-M.; Musser, J.M.; Taubenberger, J.K. High-throughput RNA sequencing of a formalin-fixed, paraffin-embedded autopsy lung tissue sample from the 1918 influenza pandemic. J. Pathol. 2013, 229, 535–545. [Google Scholar] [CrossRef]
- Turnbull, A.K.; Selli, C.; Martinez-Perez, C.; Fernando, A.; Renshaw, L.; Keys, J.; Figureoa, J.D.; He, X.; Tanioka, M.; Munro, A.F.; et al. Unlocking the transcriptomic potential of formalin-fixed paraffin embedded clinical tissues: Comparison of gene expression profiling approaches. BMC Bioinform. 2020, 21, 30. [Google Scholar] [CrossRef]
- Mills, A.M.; Chinn, Z.; Rauch, L.A.; Dusenbery, A.C.; Whitehair, R.M.; Saks, E.; Duska, L.R. Emerging biomarkers in ovarian granulosa cell tumors. Int. J. Gynecol. Cancer 2019, 29, 560–565. [Google Scholar] [CrossRef] [PubMed]
- King, L.A.; Okagaki, T.; Gallup, D.G.; Twiggs, L.B.; Messing, M.J.; Carson, L.F. Mitotic count, nuclear atypia, and immunohistochemical determination of Ki-67, c-myc, p21-ras, c-erbB2, and p53 expression in granulosa cell tumors of the ovary: Mitotic count and Ki-67 are indicators of poor prognosis. Gynecol. Oncol. 1996, 61, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Caburet, S.; Anttonen, M.; Todeschini, A.-L.; Unkila-Kallio, L.; Mestivier, D.; Butzow, R.; Veitia, R.A. Combined comparative genomic hybridization and transcriptomic analyses of ovarian granulosa cell tumors point to novel candidate driver genes. BMC Cancer 2015, 15, 251. [Google Scholar] [CrossRef] [PubMed]
- Kraus, F.; Dremaux, J.; Altakfi, W.; Goux, M.; Pontois, L.; Sevestre, H.; Trudel, S. FOXL2 homozygous genotype and chromosome instability are associated with recurrence in adult granulosa cell tumors of the ovary. Oncotarget 2020, 11, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Benayoun, B.A.; Anttonen, M.; L’Hote, D.; Bailly-Bechet, M.; Andersson, N.; Heikinheimo, M.; Veitia, R.A. Adult ovarian granulosa cell tumor transcriptomics: Prevalence of FOXL2 target genes misregulation gives insights into the pathogenic mechanism of the p.Cys134Trp somatic mutation. Oncogene 2013, 32, 2739–2746. [Google Scholar] [CrossRef][Green Version]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, K.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in the Cancer Genome Atlas. Cell 2018, 173, 321.e10–337.e10. [Google Scholar] [CrossRef]
- Da Paula, A.C.; da Silva, E.M.; Segura, S.; Pareja, F.; Bi, R.; Selenica, P.; Kim, S.H.; Ferrando, L.; Vahdatinia, M.; Soslow, R.A.; et al. Genomic profiling of primary and recurrent adult granulosa cell tumors of the ovary. Mod. Pathol. 2020, 33, 1606–1617. [Google Scholar] [CrossRef]
- Herrnberger, L.; Ebner, K.; Junglas, B.; Tamm, E.R. The role of plasmalemma vesicle-associated protein (PLVAP) in endothelial cells of Schlemm’s canal and ocular capillaries. Exp. Eye Res. 2012, 105, 27–33. [Google Scholar] [CrossRef]
- Mehine, M.; Khamaiseh, S.; Ahvenainen, T.; Heikkinen, T.; Ayravainen, A.; Pakarinen, P.; Harkki, P.; Pasanen, A.; Butzow, R.; Vahteristo, P. 3′ RNA Sequencing Accurately Classifies Formalin-Fixed Paraffin-Embedded Uterine Leiomyomas. Cancers 2020, 12, 3839. [Google Scholar] [CrossRef] [PubMed]
- Roussel, E.; Kinget, L.; Verbiest, A.; Boeckx, B.; Zucman-Rossi, J.; Couchy, G.; Caruso, S.; Baldewijns, M.; Joniau, S.; Van Poppel, H.; et al. Molecular underpinnings of glandular tropism in metastatic clear cell renal cell carcinoma: Therapeutic implications. Acta Oncol. 2021, 60, 1499–1506. [Google Scholar] [CrossRef] [PubMed]
- Pilsworth, J.A.; Cochrane, D.H.; Neilson, S.J.; Moussavi, B.H.; Lai, D.; Munzur, A.D.; Senz, J.; Wang, Y.K.; Zareian, S.; Bashashati, A.; et al. Adult-type granulosa cell tumor of the ovary: A FOXL2-centric disease. J. Pathol. Clin. Res. 2021, 7, 234–252. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | Primary Tumor Only | Primary Tumor with Recurrence | Recurrent | Total |
|---|---|---|---|---|
| N (% or Range) | N (% or Range) | N (% or Range) | N (% or Range) | |
| Number of patients | 14 | 13 | 8 | 35 |
| Growth pattern | ||||
| Sarcomatoid | 5 (35.7) | 8 (61.5) | 4 (50) | 17 (48.6) |
| Differentiated | 9 (64.3) | 5 (38.5) | 4 (50) | 18 (51.4) |
| Menopause status | ||||
| Pre-menopausal | 5 (35.7) | 4 (30.8) | 1 (12.5) | 10 (28.6) |
| Post-menopausal | 9 (64.7) | 9 (69.2) | 7 (87.5) | 25 (71.4) |
| Age at diagnosis in years (average, range) | 52 (31–68) | 48 (29–66) | 57 (46–69) | 51 (29–69) |
| Tumor size | ||||
| <10 cm | 9 (64.7) | 6 (46.2) | 6 (75.0) | 21 (60.0) |
| >10 cm | 5 (35.7) | 7 (53.8) | 2 (25.0) | 14 (40.0) |
| Time to 1st recurrence in years (average, range) | - | 9.4 (1.8–18.9) | 8.1 (2.8–18.9) | 8.9 (1.8–18.9) |
| Follow-up time in years (average, range) | 20.2 (17.2–26.1) | 18.0 (6.9–33.9) | 16.4 (7.4–22.2) | 18.5 (6.9–33.9) |
| TERT promoter mutation status | ||||
| Positive | 3 (21.4) | 1 (7.7) | 1 (12.5) | 5 (14.3) |
| Wild type | 10 (71.4) | 7 (53.8) | 1 (12.5) | 18 (51.4) |
| N/A | 1 (7.1) | 5 (38.5) | 6 (75.0) | 12 (34.3) |
| Survival | ||||
| Alive | 13 (92.9) | 7 (53.8) | 5 (62.5) | 25 (71.4) |
| Died of AGCT | 1 (7.1) | 5 (38.5) | 3 (37.5) | 8 (22.9) |
| Died of other causes | 1 (7.1) | 1 (7.7) | - | 2 (5.7) |
| Primary tumor stage | ||||
| 1a | 6 (42.9) | 6 (46.2) | 2 (25.0) | 14 (40.0) |
| 1c | 5 (35.7) | 6 (46.2) | 5 (62.5) | 16 (45.7) |
| 2 | 2 (14.3) | 1 (7.7) | 1 (12.5) | 4 (11.4) |
| 3 | 1 (7.1) | - | - | |
| Sample archival age in years (average, range) | 22 (18–27) | 28 (13–43) | 15 (4–35) | 23 (4–43) |
| Marker | Score 0 n, (%) | Score 1 n, (%) | Score 2 n, (%) | Score 3 n, (%) | ||||
|---|---|---|---|---|---|---|---|---|
| PLVAP GCT cells (n = 168) | 125 | (74.4) | 41 | (24.4) | 2 | (1.2) | 0 | - |
| PLVAP endothelial cells (n = 168) | 82 | (48.8) | 30 | (17.9) | 40 | (23.8) | 16 | (9.5) |
| PLIN4 (n = 168) | 41 | (24.4) | 87 | (51.8) | 40 | (23.8) | 0 | - |
| ASS1 (n = 167) | 69 | (41.3) | 88 | (52.7) | 10 | (6.0) | 0 | - |
| PPIB (n = 168) | 11 | (6.5) | 63 | (37.5) | 61 | (36.6) | 33 | (19.6) |
| Marker | Positive n, (%) | Negative n, (%) | ||
|---|---|---|---|---|
| PLVAP AGCT cells (n = 157) | 43 | (27.4) | 114 | (72.6) |
| PLVAP endothelial cells (n = 156) | 84 | (53.8) | 72 | (46.2) |
| PLIN4 (n = 157) | 126 | (80.3) | 31 | (19.7) |
| ASS1 (n = 157) | 77 | (49.4) | 79 | (50.6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andersson, N.; Haltia, U.-M.; Färkkilä, A.; Wong, S.C.; Eloranta, K.; Wilson, D.B.; Unkila-Kallio, L.; Pihlajoki, M.; Kyrönlahti, A.; Heikinheimo, M. Analysis of Non-Relapsed and Relapsed Adult Type Granulosa Cell Tumors Suggests Stable Transcriptomes during Tumor Progression. Curr. Issues Mol. Biol. 2022, 44, 686-698. https://doi.org/10.3390/cimb44020048
Andersson N, Haltia U-M, Färkkilä A, Wong SC, Eloranta K, Wilson DB, Unkila-Kallio L, Pihlajoki M, Kyrönlahti A, Heikinheimo M. Analysis of Non-Relapsed and Relapsed Adult Type Granulosa Cell Tumors Suggests Stable Transcriptomes during Tumor Progression. Current Issues in Molecular Biology. 2022; 44(2):686-698. https://doi.org/10.3390/cimb44020048
Chicago/Turabian StyleAndersson, Noora, Ulla-Maija Haltia, Anniina Färkkilä, Swee Chong Wong, Katja Eloranta, David B. Wilson, Leila Unkila-Kallio, Marjut Pihlajoki, Antti Kyrönlahti, and Markku Heikinheimo. 2022. "Analysis of Non-Relapsed and Relapsed Adult Type Granulosa Cell Tumors Suggests Stable Transcriptomes during Tumor Progression" Current Issues in Molecular Biology 44, no. 2: 686-698. https://doi.org/10.3390/cimb44020048
APA StyleAndersson, N., Haltia, U.-M., Färkkilä, A., Wong, S. C., Eloranta, K., Wilson, D. B., Unkila-Kallio, L., Pihlajoki, M., Kyrönlahti, A., & Heikinheimo, M. (2022). Analysis of Non-Relapsed and Relapsed Adult Type Granulosa Cell Tumors Suggests Stable Transcriptomes during Tumor Progression. Current Issues in Molecular Biology, 44(2), 686-698. https://doi.org/10.3390/cimb44020048






