The Effects of Co-Culture of Embryonic Stem Cells with Neural Stem Cells on Differentiation
Abstract
1. Introduction
2. Materials and Methods
2.1. Types of Culture Medium
2.2. Undifferentiated ESC Culture
2.3. Neural Stem Cell Culture
2.4. Mouse Embryonic Fibroblast Culture
2.5. 2D Co-Culture of ESCs with NSC or MEF Feeders
2.6. 3D Co-Culture with NSCs or MEFs
2.7. 2D or 3D Culture of ESCs in CM
2.8. Reverse Transcription Polymerase Chain Reaction (qPCR)
2.9. FACS Analysis and Sorting
2.10. Equipment
2.11. Statistical Analysis
3. Results
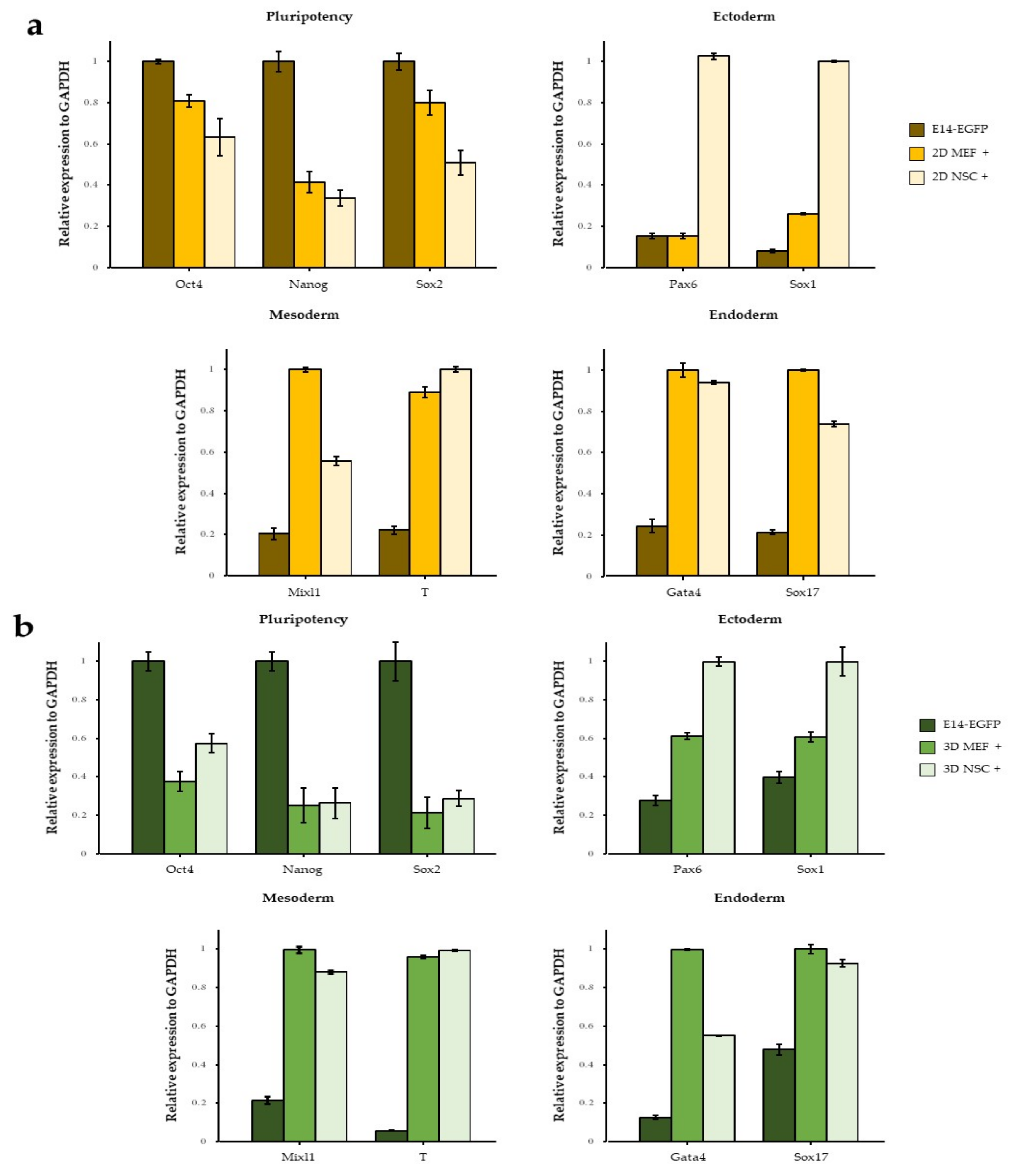
3.1. NSC Feeder Induced Differentiation of ESCs
3.2. NSC-CM Did Not Promote Differentiation of ESCs
3.3. The Effect of 3D Co-Culture on Differentiation of ESCs
3.4. Neuroectodermal Differentiation of ESCs Was Improved by NSC Co-Culture
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638. [Google Scholar] [CrossRef] [PubMed]
- Llames, S.; García-Pérez, E.; Meana, A.; Larcher, F.; Del Río, M. Feeder Layer Cell Actions and Applications. Tissue Eng. Part B Rev. 2015, 21, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Rangappa, S.; Entwistle, J.W.C.; Wechsler, A.S.; Kresh, J.Y. Cardiomyocyte-mediated contact programs human mesenchymal stem cells to express cardiogenic phenotype. J. Thorac. Cardiovasc. Surg. 2003, 126, 124–132. [Google Scholar] [CrossRef]
- Metallo, C.M.; Mohr, J.C.; Detzel, C.J.; de Pablo, J.J.; Van Wie, B.J.; Palecek, S.P. Engineering the Stem Cell Microenvironment. Biotechnol. Prog. 2007, 23, 18–23. [Google Scholar] [CrossRef]
- Nair, R.; Ngangan, A.V.; Kemp, M.L.; McDevitt, T.C. Gene expression signatures of extracellular matrix and growth factors during embryonic stem cell differentiation. PLoS ONE 2012, 7, e42580. [Google Scholar] [CrossRef][Green Version]
- Schugar, R.C.; Robbins, P.D.; Deasy, B.M. Small molecules in stem cell self-renewal and differentiation. Gene Ther. 2008, 15, 126–135. [Google Scholar] [CrossRef]
- Kubo, A.; Shinozaki, K.; Shannon, J.M.; Kouskoff, V.; Kennedy, M.; Woo, S.; Fehling, H.J.; Keller, G. Development of definitive endoderm from embryonic stem cells in culture. Development 2004, 131, 1651–1662. [Google Scholar] [CrossRef]
- Yasunaga, M.; Tada, S.; Torikai-Nishikawa, S.; Nakano, Y.; Okada, M.; Jakt, L.M.; Nishikawa, S.; Chiba, T.; Era, T.; Nishikawa, S. Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat. Biotechnol. 2005, 23, 1542–1550. [Google Scholar] [CrossRef]
- Ema, M.; Takahashi, S.; Rossant, J. Deletion of the selection cassette, but not cis-acting elements, in targeted Flk1-lacZ allele reveals Flk1 expression in multipotent mesodermal progenitors. Blood 2006, 107, 111–117. [Google Scholar] [CrossRef]
- Kataoka, H.; Takakura, N.; Nishikawa, S.; Tsuchida, K.; Kodama, H.; Kunisada, T.; Risau, W.; Kita, T.; Nishikawa, S.I. Expressions of PDGF receptor alpha, c-Kit and Flk1 genes clustering in mouse chromosome 5 define distinct subsets of nascent mesodermal cells. Dev. Growth Differ. 1997, 39, 729–740. [Google Scholar] [CrossRef]
- Nostro, M.C.; Cheng, X.; Keller, G.M.; Gadue, P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell 2008, 2, 60–71. [Google Scholar] [CrossRef]
- Era, T.; Izumi, N.; Hayashi, M.; Tada, S.; Nishikawa, S.; Nishikawa, S. Multiple mesoderm subsets give rise to endothelial cells, whereas hematopoietic cells are differentiated only from a restricted subset in embryonic stem cell differentiation culture. Stem Cells 2008, 26, 401–411. [Google Scholar] [CrossRef]
- Ying, Q.L.; Stavridis, M.; Griffiths, D.; Li, M.; Smith, A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003, 21, 183–186. [Google Scholar] [CrossRef]
- Aubert, J.; Dunstan, H.; Chambers, I.; Smith, A. Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat. Biotechnol. 2002, 20, 1240–1245. [Google Scholar] [CrossRef]
- Schlessinger, D.I.; Patino, S.C.; Syed, S.Y.B.; Sonthalia, S. Embryology, Epidermis; StatPearls Publishing: Tampa, FL, USA, 2021. [Google Scholar]
- Ou, D.B.; Zeng, D.; Jin, Y.; Liu, X.T.; Teng, J.W.; Guo, W.G.; Wang, H.T.; Su, F.F.; He, Y.; Zheng, Q.S. The long-term differentiation of embryonic stem cells into cardiomyocytes: An indirect co-culture model. PLoS ONE 2013, 8, e55233. [Google Scholar] [CrossRef]
- Bahmani, L.; Taha, M.F.; Javeri, A. Coculture with embryonic stem cells improves neural differentiation of adipose tissue-derived stem cells. Neuroscience 2014, 272, 229–239. [Google Scholar] [CrossRef]
- Bidarra, S.J.; Barrias, C.C.; Barbosa, M.A.; Soares, R.; Amédée, J.; Granja, P.L. Phenotypic and proliferative modulation of human mesenchymal stem cells via crosstalk with endothelial cells. Stem Cell Res. 2011, 7, 186–197. [Google Scholar] [CrossRef]
- Van der Meer, A.D.; Orlova, V.V.; ten Dijke, P.; van den Berg, A.; Mummery, C.L. Three-dimensional co-cultures of human endothelial cells and embryonic stem cell-derived pericytes inside a microfluidic device. Lab A Chip 2013, 13, 3562–3568. [Google Scholar] [CrossRef]
- Lacham-Kaplan, O.; Chy, H.; Trounson, A. Testicular Cell Conditioned Medium Supports Differentiation of Embryonic Stem Cells into Ovarian Structures Containing Oocytes. Stem Cells 2005, 24, 266–273. [Google Scholar] [CrossRef]
- Tsai, Z.-Y.; Singh, S.; Yu, S.-L.; Chou, C.-H.; Li, S.S.-L. A feeder-free culture using autogeneic conditioned medium for undifferentiated growth of human embryonic stem cells: Comparative expression profiles of mRNAs, microRNAs and proteins among different feeders and conditioned media. BMC Cell Biol. 2010, 11, 76. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.-Q.; Lin, G.; Yuan, D.; Wang, J.; Liu, T.-C.; Lu, G.-X. Proliferative feeder cells support prolonged expansion of human embryonic stem cells. Cell Biol. Int. 2005, 29, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Greber, B.; Lehrach, H.; Adjaye, J. Fibroblast growth factor 2 modulates transforming growth factor β signaling in mouse embryonic fibroblasts and human ESCs (hESCs) to support hESC self-renewal. Stem Cells 2007, 25, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.J.; Muotri, A.; Gage, F.; Varki, A. Human embryonic stem cells express an immunogenic nonhuman sialic acid. Nat. Med. 2005, 11, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.-S.; Fujishiro, M.; Nishioka, C.; Arai, T.; Takahashi, E.; Gong, J.-S.; Akaike, T.; Ito, Y. Feeder cells support the culture of induced pluripotent stem cells even after chemical fixation. PLoS ONE 2012, 7, e32707. [Google Scholar] [CrossRef]
- Takahashi, K.; Narita, M.; Yokura, M.; Ichisaka, T.; Yamanaka, S. Human induced pluripotent stem cells on autologous feeders. PLoS ONE 2009, 4, e8067. [Google Scholar] [CrossRef]
- Pan, C.; Hicks, A.; Guan, X.; Chen, H.; Bishop, C.E. SNL fibroblast feeder layers support derivation and maintenance of human induced pluripotent stem cells. J. Genet. Genom. 2010, 37, 241–248. [Google Scholar] [CrossRef]
- Khoo, M.L.M.; McQuade, L.R.; Smith, M.S.R.; Lees, J.G.; Sidhu, K.S.; Tuch, B.E. Growth and Differentiation of Embryoid Bodies Derived from Human Embryonic Stem Cells: Effect of Glucose and Basic Fibroblast Growth Factor1. Biol. Reprod. 2005, 73, 1147–1156. [Google Scholar] [CrossRef]
- Nieto, A.; Cabrera, C.; Catalina, P.; Cobo, F.; Barnie, A.; Cortés, J.; Del Jesus, A.B.; Montes, R.; Concha, A. Effect of mitomycin-C on human foreskin fibroblasts used as feeders in human embryonic stem cells: Immunocytochemistry MIB1 score and DNA ploidy and apoptosis evaluated by flow cytometry. Cell Biol. Int. 2007, 31, 269–278. [Google Scholar] [CrossRef]
- Eiselleova, L.; Peterkova, I.; Neradil, J.; Slaninova, I.; Hampl, A.; Dvorak, P. Comparative study of mouse and human feeder cells for human embryonic stem cells. Int. J. Dev. Biol. 2004, 52, 353–363. [Google Scholar] [CrossRef]
- Hovatta, O.; Mikkola, M.; Gertow, K.; Strömberg, A.M.; Inzunza, J.; Hreinsson, J.; Rozell, B.r.; Blennow, E.; Andäng, M.; Ährlund-Richter, L. A culture system using human foreskin fibroblasts as feeder cells allows production of human embryonic stem cells. Hum. Reprod. 2003, 18, 1404–1409. [Google Scholar] [CrossRef]
- Anchan, R.M.; Quaas, P.; Gerami-Naini, B.; Bartake, H.; Griffin, A.; Zhou, Y.; Day, D.; Eaton, J.L.; George, L.L.; Naber, C. Amniocytes can serve a dual function as a source of iPS cells and feeder layers. Hum. Mol. Genet. 2011, 20, 962–974. [Google Scholar] [CrossRef]
- Choi, H.W.; Kim, J.S.; Choi, S.; Jang, H.J.; Kim, M.J.; Choi, Y.; Schöler, H.R.; Chung, H.M.; Do, J.T. Neural stem cells achieve and maintain pluripotency without feeder cells. PLoS ONE 2011, 6, e21367. [Google Scholar] [CrossRef]
- Williams, R.L.; Hilton, D.J.; Pease, S.; Willson, T.A.; Stewart, C.L.; Gearing, D.P.; Wagner, E.F.; Metcalf, D.; Nicola, N.A.; Gough, N.M. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 1988, 336, 684–687. [Google Scholar] [CrossRef]
- Walther, C.; Gruss, P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development 1991, 113, 1435–1449. [Google Scholar] [CrossRef]
- Stoykova, A.; Gruss, P. Roles of Pax-genes in developing and adult brain as suggested by expression patterns. J. Neurosci. 1994, 14, 1395–1412. [Google Scholar] [CrossRef]
- Grindley, J.C.; Davidson, D.R.; Hill, R.E. The role of Pax-6 in eye and nasal development. Development 1995, 121, 1433–1442. [Google Scholar] [CrossRef]
- Grindley, J.C.; Hargett, L.K.; Hill, R.E.; Ross, A.; Hogan, B.L. Disruption of PAX6 function in mice homozygous for the Pax6Sey-1Neu mutation produces abnormalities in the early development and regionalization of the diencephalon. Mech. Dev. 1997, 64, 111–126. [Google Scholar] [CrossRef]
- Ericson, J.; Rashbass, P.; Schedl, A.; Brenner-Morton, S.; Kawakami, A.; van Heyningen, V.; Jessell, T.M.; Briscoe, J. Pax6 Controls Progenitor Cell Identity and Neuronal Fate in Response to Graded Shh Signaling. Cell 1997, 90, 169–180. [Google Scholar] [CrossRef]
- Kan, L.; Jalali, A.; Zhao, L.R.; Zhou, X.; McGuire, T.; Kazanis, I.; Episkopou, V.; Bassuk, A.G.; Kessler, J.A. Dual function of Sox1 in telencephalic progenitor cells. Dev. Biol. 2007, 310, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Pevny, L.H.; Sockanathan, S.; Placzek, M.; Lovell-Badge, R. A role for SOX1 in neural determination. Development 1998, 125, 1967–1978. [Google Scholar] [CrossRef] [PubMed]
- Malas, S.; Postlethwaite, M.; Ekonomou, A.; Whalley, B.; Nishiguchi, S.; Wood, H.; Meldrum, B.; Constanti, A.; Episkopou, V. Sox1-deficient mice suffer from epilepsy associated with abnormal ventral forebrain development and olfactory cortex hyperexcitability. Neuroscience 2003, 119, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Yue, F.; Johkura, K.; Tomotsune, D.; Shirasawa, S.; Yokoyama, T.; Nagai, M.; Sasaki, K. Bone marrow stromal cells as an inducer for cardiomyocyte differentiation from mouse embryonic stem cells. Ann. Anat. Anat. Anz. 2010, 192, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Söhl, G.; Willecke, K. Gap junctions and the connexin protein family. Cardiovasc. Res. 2004, 62, 228–232. [Google Scholar] [CrossRef]
- Clevers, H. Modeling development and disease with organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef]
- Dutta, D.; Heo, I.; Clevers, H. Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol. Med. 2017, 23, 393–410. [Google Scholar] [CrossRef]
- Song, L.; Tsai, A.-C.; Yuan, X.; Bejoy, J.; Sart, S.; Ma, T.; Li, Y. Neural differentiation of spheroids derived from human induced pluripotent stem cells–mesenchymal stem cells coculture. Tissue Eng. Part A 2018, 24, 915–929. [Google Scholar] [CrossRef]
- Baghdadi, M.B.; Kim, T.-H. Analysis of mouse intestinal organoid culture with conditioned media isolated from mucosal enteric glial cells. STAR Protoc. 2022, 3, 101351. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]




| Gene | Sequence 5′-3′ | |
|---|---|---|
| Gapdh | F | ATGAATACGGCTACAGCAACAGG |
| R | CTCTTGCTCAGTGCCTTGCTG | |
| Oct4 | F | CCAATCAGCTTGGGCTAGAG |
| R | CTGGGAAAGGTGTCCCTGTA | |
| Nanog | F | CTTTCACCTATTAAGGTGCTTGC |
| R | TGGCATCGGTTCATCATGGTA | |
| Sox2 | F | CATGAGAGCAAGTACTGGCAAG |
| R | CCAACGATATCAACCTGCATGG | |
| Pax6 | F | ACCAGTGTCTACCAGCCAATCC |
| R | GCACGAGTATGAGGAGGTCTGA | |
| Sox1 | F | GCCGAGTGGAAGGTCATGTC |
| R | TTGAGCAGCGTCTTGGTCTTG | |
| Mixl1 | F | TCCTCCATTGCCCTGCTCCT |
| R | ACGCCTCCTCCAGTCATGCT | |
| T | F | ATCAGAGTCCTTTGCTAGGTAG |
| R | GTTACAATCTTCTGGCTATGC | |
| Gata4 | F | CAGCAGCAGCAGTCAAGAGATG |
| R | ACCAGGCTGTTCCAAGAGTCC | |
| Sox17 | F | TTCTGTACACTTTAATGAGGCTGTTC |
| R | TTGTGGGAAGTGGGATCAAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.R.; Jang, S.W.; Han, J.H.; Na, G.R.; Jang, H.; Choi, H.W. The Effects of Co-Culture of Embryonic Stem Cells with Neural Stem Cells on Differentiation. Curr. Issues Mol. Biol. 2022, 44, 6104-6116. https://doi.org/10.3390/cimb44120416
Kim YR, Jang SW, Han JH, Na GR, Jang H, Choi HW. The Effects of Co-Culture of Embryonic Stem Cells with Neural Stem Cells on Differentiation. Current Issues in Molecular Biology. 2022; 44(12):6104-6116. https://doi.org/10.3390/cimb44120416
Chicago/Turabian StyleKim, Ye Rim, Si Won Jang, Jae Ho Han, Ga Rim Na, Hoon Jang, and Hyun Woo Choi. 2022. "The Effects of Co-Culture of Embryonic Stem Cells with Neural Stem Cells on Differentiation" Current Issues in Molecular Biology 44, no. 12: 6104-6116. https://doi.org/10.3390/cimb44120416
APA StyleKim, Y. R., Jang, S. W., Han, J. H., Na, G. R., Jang, H., & Choi, H. W. (2022). The Effects of Co-Culture of Embryonic Stem Cells with Neural Stem Cells on Differentiation. Current Issues in Molecular Biology, 44(12), 6104-6116. https://doi.org/10.3390/cimb44120416