Heat-Induced Proteotoxic Stress Response in Placenta-Derived Stem Cells (PDSCs) Is Mediated through HSPA1A and HSPA1B with a Potential Higher Role for HSPA1B
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Culture of Placenta-Derived Stem Cells
2.2. Heat Stress Experimentation
2.3. RNA Isolation and cDNA Synthesis
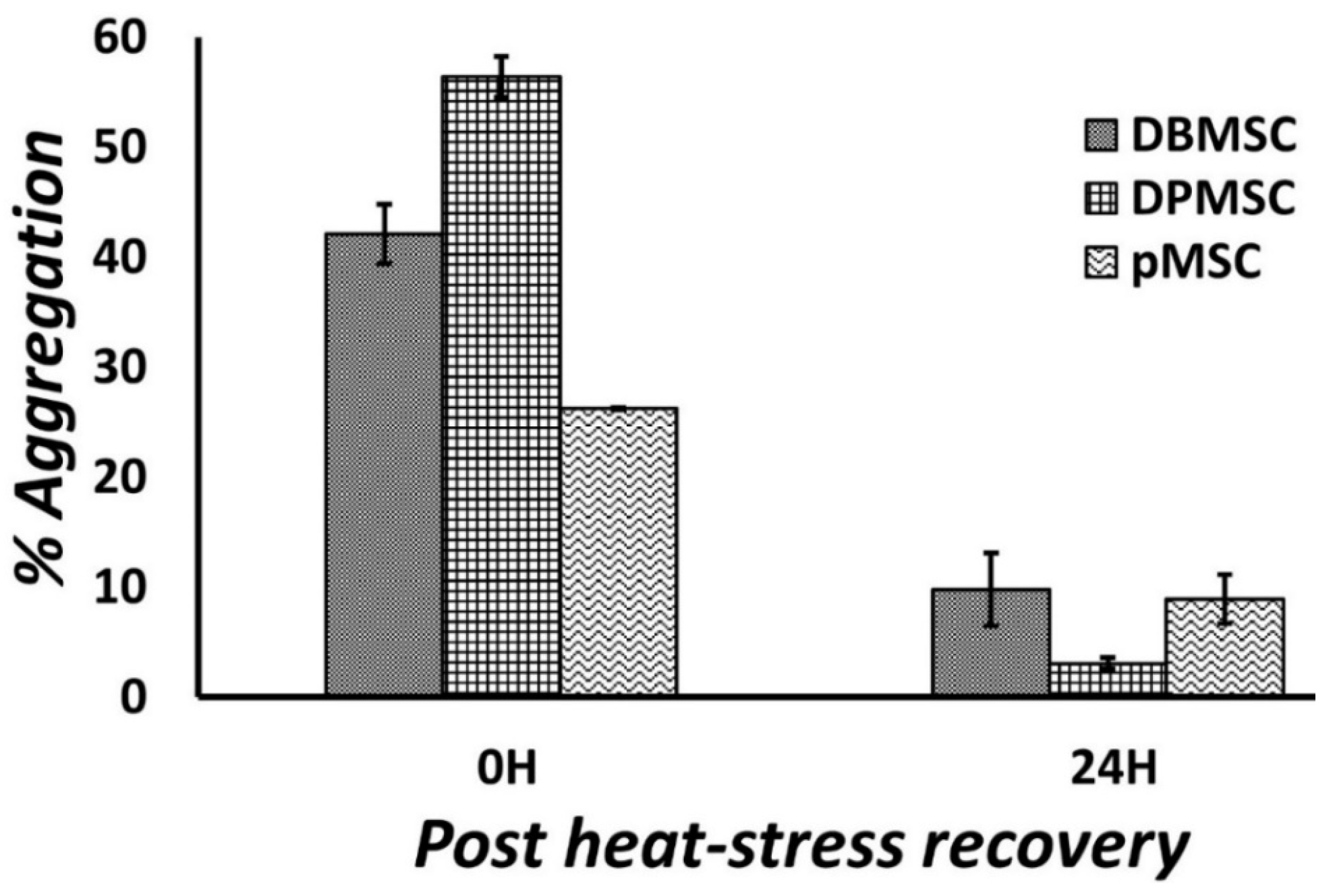
2.4. Protein Aggregation Assay
2.5. Cell Proliferation Assays
2.6. Gene Arrays and RT-PCR
2.7. Protein Extraction and Concentration Determination
2.8. Immunoblotting
2.9. Expression Data Analysis
2.10. Expression Kinetics Analysis
3. Results
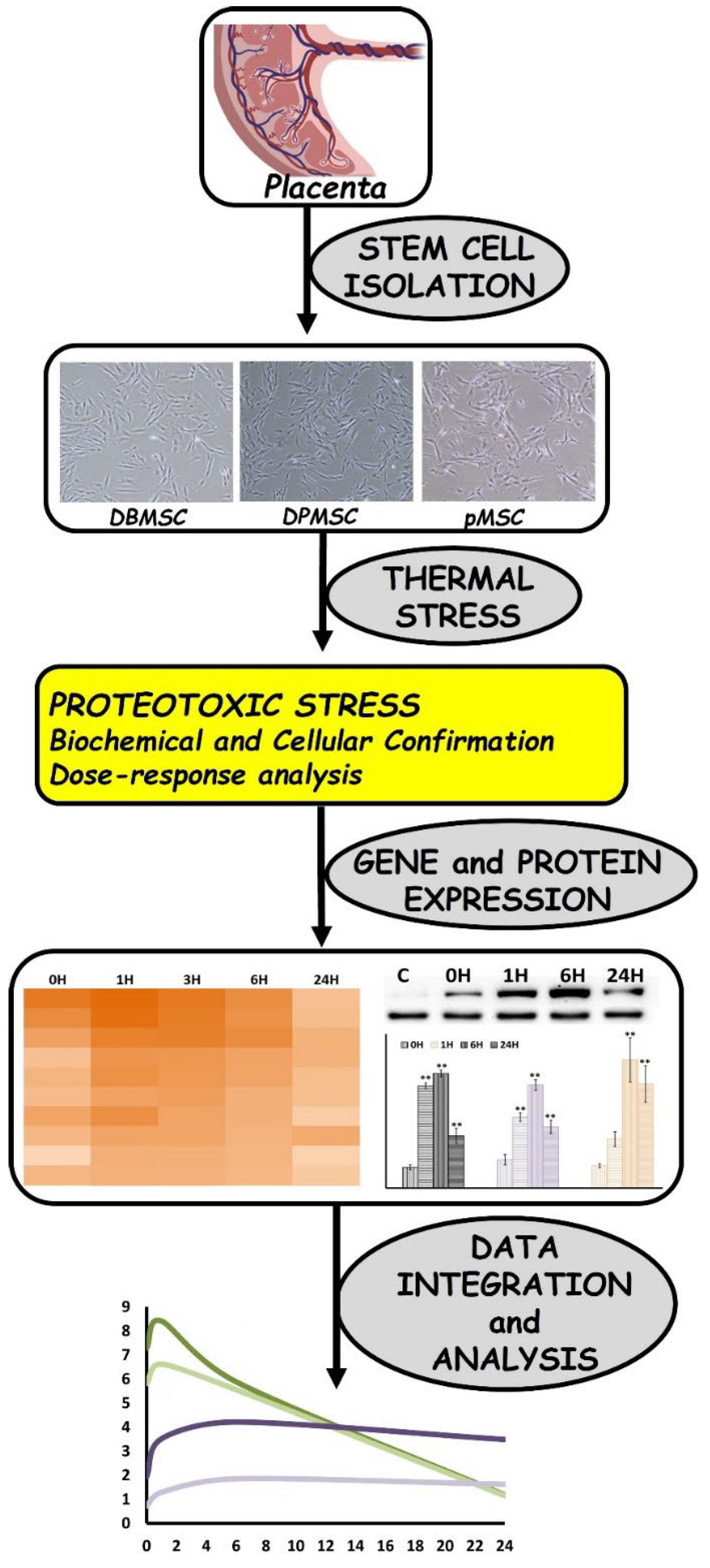
3.1. Cellular Models of Proteotoxic Stress
3.2. Dose-Responsive Characteristic of Heat-Stress and Stress Response
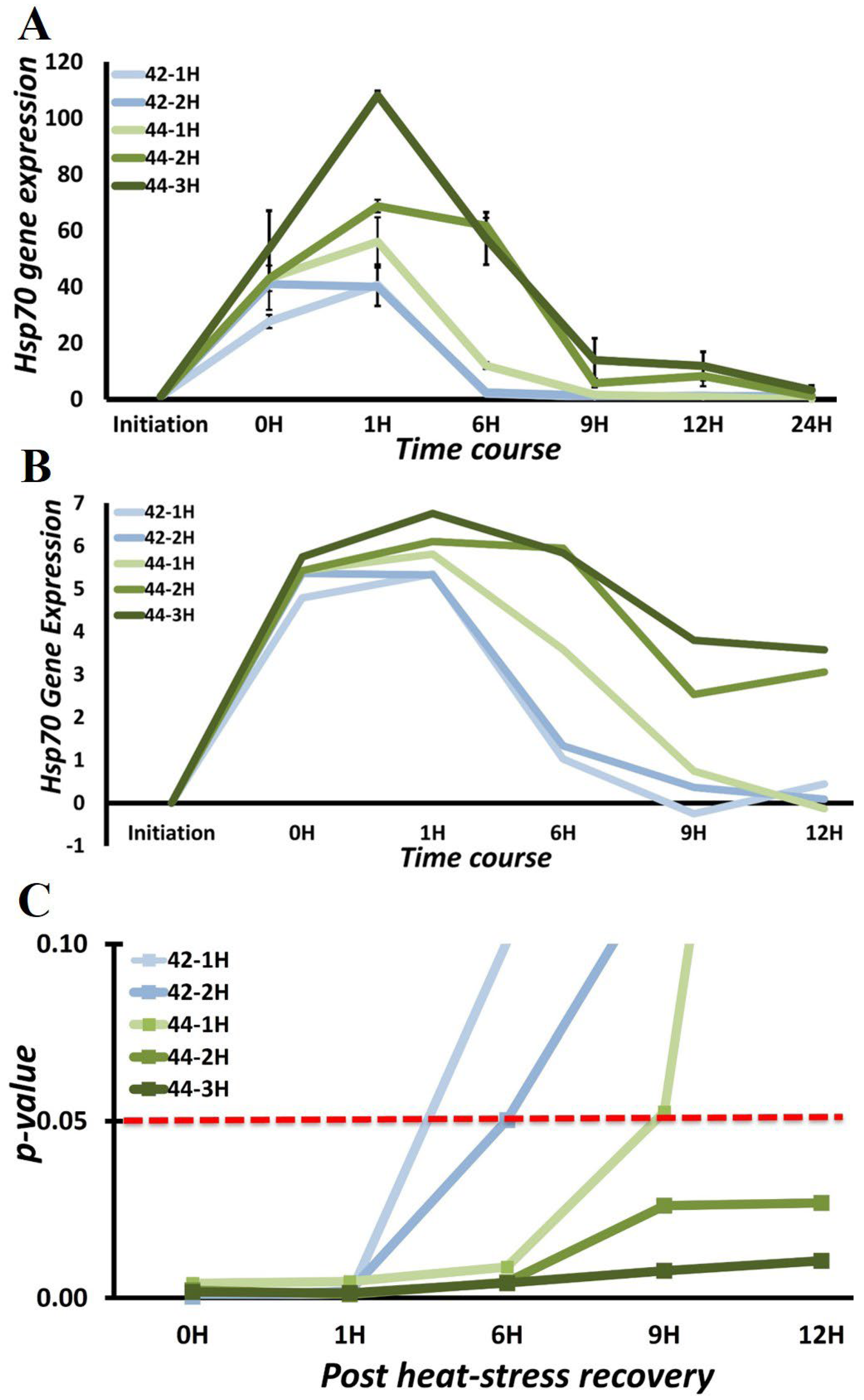
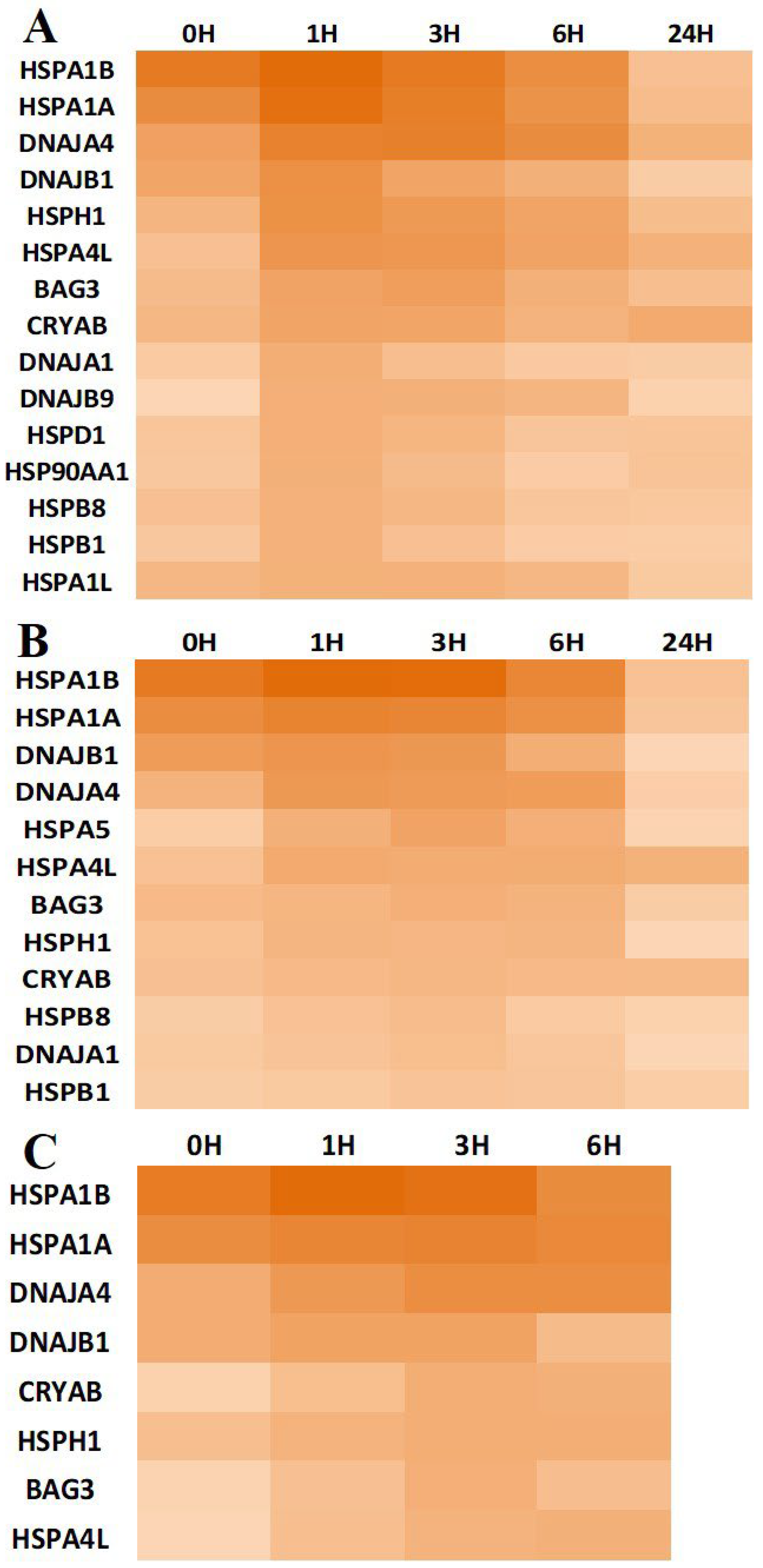
3.3. Chaperone Gene Expression during Heat-Shock Response
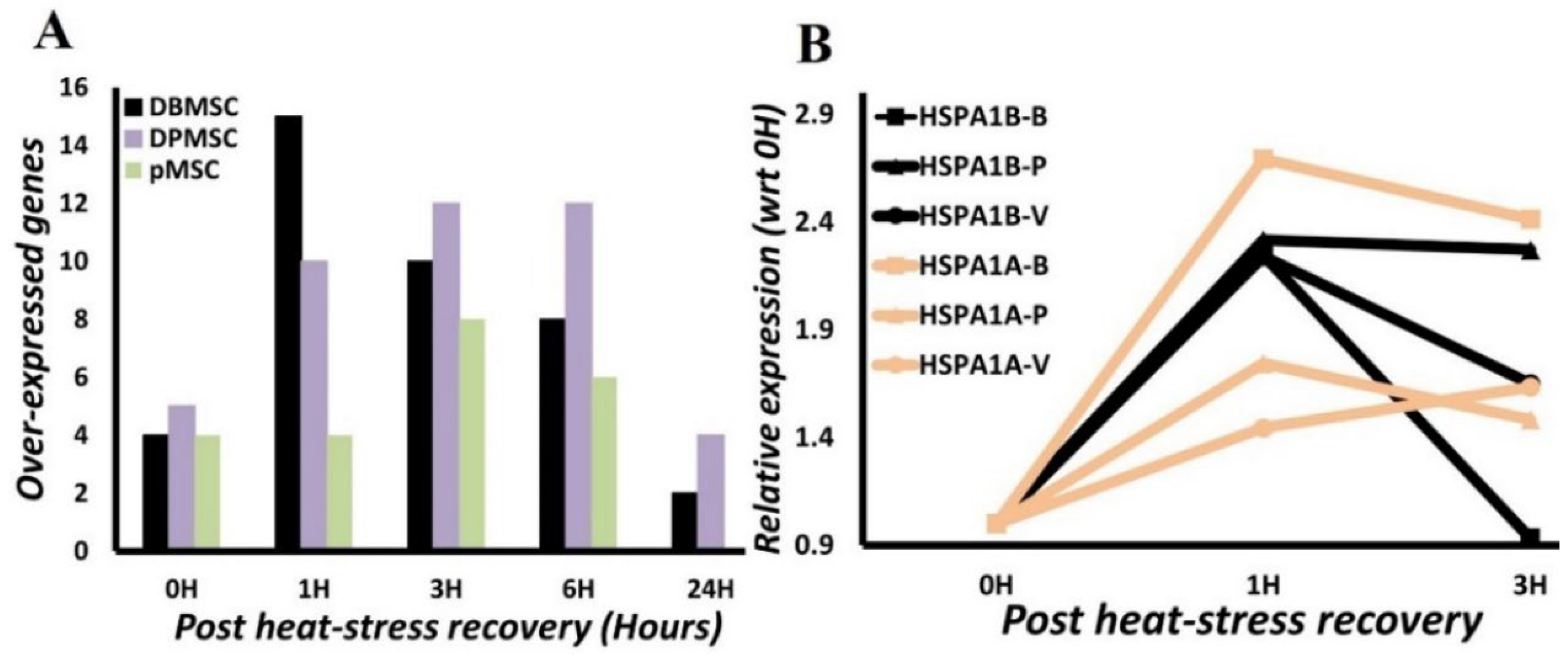
3.4. HSPA1A and HSPA1B Are Primary Determinants of Heat-Induced Proteotoxic Stress Response
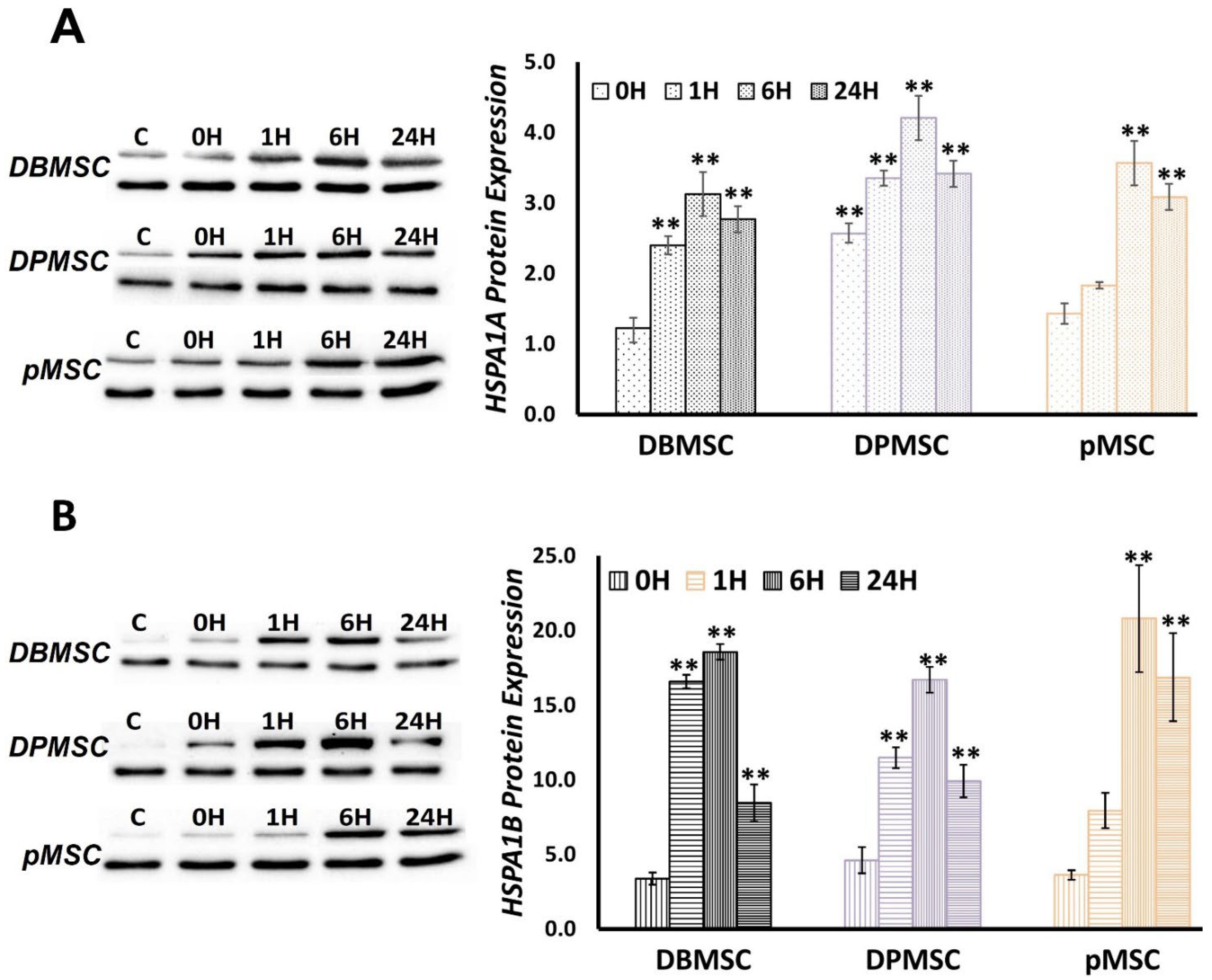
3.5. HSPA1A and HSPA1B Protein Expression follows a Temporal Pattern
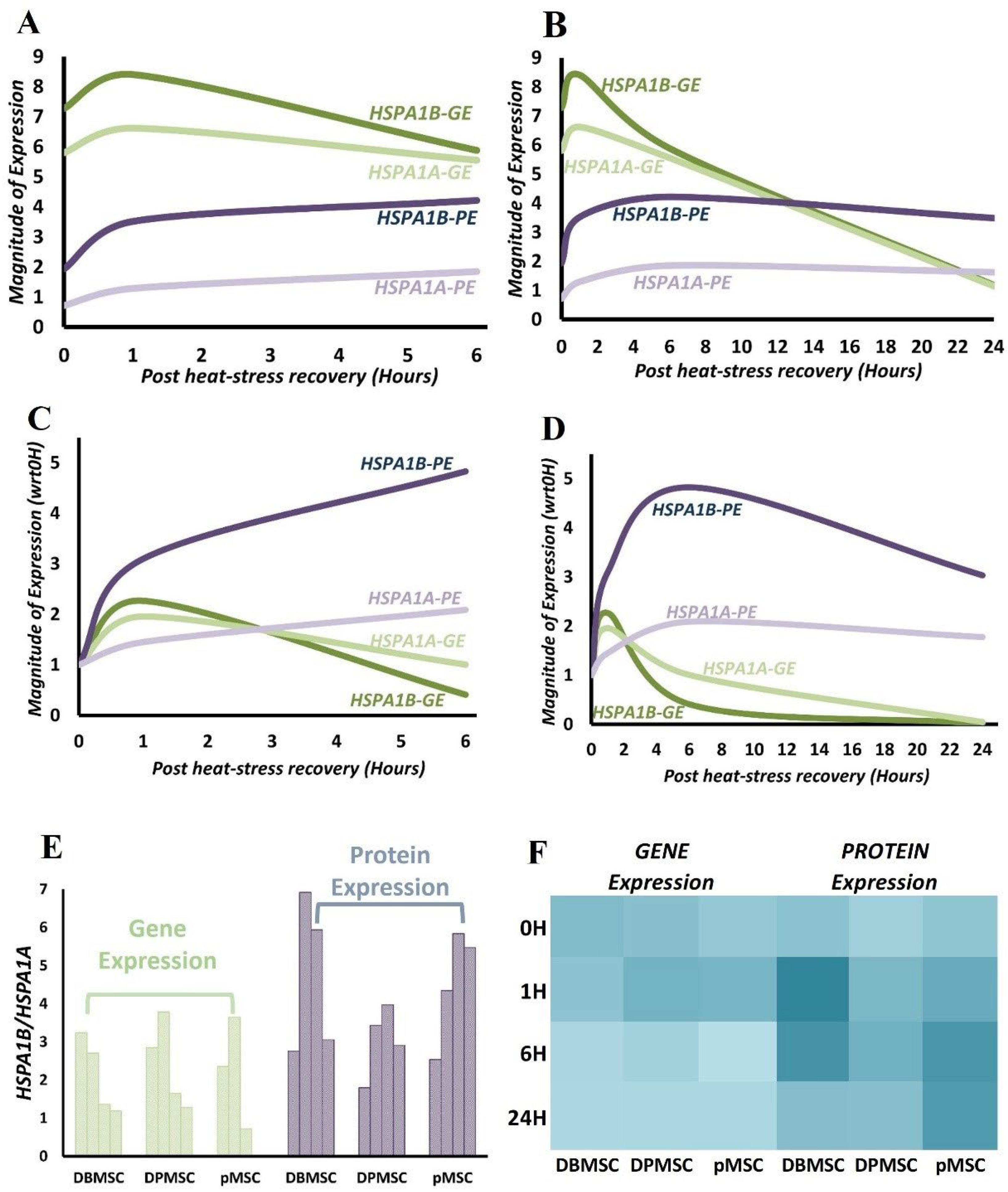
3.6. HSPA1B Expression Dominates HSPA1A Expression: A Unified Model
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Chia, W.K.; Cheah, F.C.; Abdul Aziz, N.H.; Kampan, N.C.; Shuib, S.; Khong, T.Y.; Tan, G.C.; Wong, Y.P. A review of placenta and umbilical cord-derived stem cells and the immunomodulatory basis of their therapeutic potential in bronchopulmonary dysplasia. Front. Pediatr. 2021, 9, 615508. [Google Scholar] [CrossRef] [PubMed]
- Pipino, C.; Shangaris, P.; Resca, E.; Zia, S.; Deprest, J.; Sebire, N.J.; David, A.L.; Guillot, P.V.; De Coppi, P. Placenta as a reservoir of stem cells: An underutilized resource? Br. Med. Bull. 2013, 105, 43–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, O.J. Ethical issues in stem cell therapy. Endocr. Rev. 2009, 30, 204–213. [Google Scholar] [CrossRef] [Green Version]
- Pogozhykh, O.; Prokopyuk, V.; Figueiredo, C.; Pogozhykh, D. Placenta and placental derivatives in regenerative therapies: Experimental studies, history, and prospects. Stem Cells Int. 2018, 2018, 4837930. [Google Scholar] [CrossRef]
- Oliveira, M.S. Placental-derived stem cells: Culture, differentiation and challenges. World J. Stem Cells 2015, 7, 769. [Google Scholar] [CrossRef]
- Hipp, M.S.; Kasturi, P.; Hartl, F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019, 20, 421–435. [Google Scholar] [CrossRef]
- Vilchez, D.; Simic, M.S.; Dillin, A. Proteostasis and aging of stem cells. Trends Cell Biol. 2014, 24, 161–170. [Google Scholar] [CrossRef]
- Yan, P.; Ren, J.; Zhang, W.; Qu, J.; Liu, G.H. Protein quality control of cell stemness. Cell Regen. 2020, 9, 22. [Google Scholar] [CrossRef]
- De Lima Fernandes, C.F.; Iglesia, R.P.; Melo-Escobar, M.I.; Prado, M.B.; Lopes, M.H. Chaperones and beyond as key players in pluripotency maintenance. Front. Cell Dev. Biol. 2019, 7, 150. [Google Scholar] [CrossRef]
- Shende, P.; Bhandarkar, S.; Prabhakar, B. Heat shock proteins and their protective roles in stem cell biology. Stem Cell Rev. Rep. 2019, 15, 637–651. [Google Scholar] [CrossRef]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Ulrich Hartl, F. Molecular Chaperone Functions in Protein Folding and Proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The HSP70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- Llamas, E.; Alirzayeva, H.; Loureiro, R.; Vilchez, D. The intrinsic proteostasis network of stem cells. Curr. Opin. Cell Biol. 2020, 67, 46–55. [Google Scholar] [CrossRef]
- Saibil, H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef] [Green Version]
- Waudby, C.A.; Dobson, C.M.; Christodoulou, J. Nature and regulation of protein folding on the ribosome. Trends Biochem. Sci. 2019, 44, 914–926. [Google Scholar] [CrossRef] [Green Version]
- Radons, J. The human HSP70 family of chaperones: Where do we stand ? Cell Stress Chaperones 2016, 21, 379–404. [Google Scholar] [CrossRef] [Green Version]
- Qi, W.; White, M.C.; Choi, W.; Guo, C.; Dinney, C.; McConkey, D.J.; Siefker-Radtke, A. Inhibition of inducible heat shock protein-70 (Hsp72) enhances bortezomib-induced cell death in human bladder cancer cells. PLoS ONE 2013, 8, e69509. [Google Scholar] [CrossRef] [Green Version]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [Green Version]
- Seo, J.H.; Park, J.H.; Lee, E.J.; Vo, T.T.L.; Choi, H.; Kim, J.Y.; Jang, J.K.; Wee, H.J.; Lee, H.S.; Jang, S.H.; et al. ARD1-mediated HSP70 acetylation balances stress-induced protein refolding and degradation. Nat. Commun. 2016, 7, 12882. [Google Scholar] [CrossRef] [Green Version]
- Richter, K.; Haslbeck, M.; Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef]
- Guan, Y.; Zhu, X.; Liang, J.; Wei, M.; Huang, S.; Pan, X. Upregulation of HSPA1A/HSPA1B/HSPA7 and downregulation of HSPA9 were related to poor survival in colon cancer. Front. Oncol. 2021, 11, 4395. [Google Scholar] [CrossRef]
- Abumaree, M.H.; Al Jumah, M.A.; Kalionis, B.; Jawdat, D.; Al Khaldi, A.; AlTalabani, A.A.; Knawy, B.A. Phenotypic and functional characterization of mesenchymal stem cells from chorionic villi of human term placenta. Stem Cell Rev. Rep. 2013, 9, 16–31. [Google Scholar] [CrossRef]
- Abomaray, F.M.; Al Jumah, M.A.; Alsaad, K.O.; Jawdat, D.; Al Khaldi, A.; Alaskar, A.S.; Al Harthy, S.; Al Subayyil, A.M.; Khatlani, T.; Alawad, A.O.; et al. Phenotypic and functional characterization of mesenchymal stem/multipotent stromal cells from decidua basalis of human term placenta. Stem Cells Int. 2016, 2016, 5184601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abumaree, M.H.; Abomaray, F.M.; Alshehri, N.A.; Almutairi, A.; Alaskar, A.S.; Kalionis, B.; Al Jumah, M.A. Phenotypic and functional characterization of mesenchymal stem/multipotent stromal cells from decidua parietalis of human term placenta. Reprod. Sci. 2016, 23, 1193–1207. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Yang, Z.M.; Chen, X.H.; Tan, M.Y.; Wang, J.; Li, X.Q.; Xie, H.Q.; Deng, L. Isolation of mesenchymal stem cells from human placental decidua basalis and resistance to hypoxia and serum deprivation. Stem Cell Rev. Rep. 2009, 5, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Jinwal, U.K.; Miyata, Y.; Iii, J.K.; Jones, J.R.; Trotter, J.H.; Chang, L.; Leary, J.O.; Morgan, D.; Lee, D.C.; Shults, C.L.; et al. Chemical manipulation of HSP70 ATPase activity regulates tau stability. Neurobiol. Dis. 2009, 29, 12079–12088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, A.K.; Pratt, W.B.; Lieberman, A.P.; Osawa, Y.; Arbor, A.; Arbor, A. Targeting HSP70 facilitated protein quality control for treatment of polyglutamine diseases Amanda. Cell Mol. Life Sci. 2021, 77, 977–996. [Google Scholar] [CrossRef]
- Wang, A.M.; Miyata, Y.; Klinedinst, S.; Peng, H.; Jason, P.; Komiyama, T.; Li, X.; Morishima, Y.; Merry, D.E.; Pratt, W.B.; et al. Activation of HSP70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat. Chem. Biol. 2013, 9, 112–118. [Google Scholar] [CrossRef] [Green Version]
- Hamidi, H.; Lilja, J.; Ivaska, J. Using xCELLigence RTCA Instrument to Measure Cell Adhesion. Bio-Protocol 2017, 7, e2646. [Google Scholar] [CrossRef] [Green Version]
- Spandidos, A.; Wang, X.; Wang, H.; Seed, B. PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2009, 38, 792–799. [Google Scholar] [CrossRef]
- Daugaard, M.; Rohde, M.; Jäättelä, M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007, 581, 3702–3710. [Google Scholar] [CrossRef] [Green Version]
- Su, W.; Zhou, Q.; Wang, Y.; Chishti, A.; Li, Q.Q.; Dayal, S.; Shiehzadegan, S.; Cheng, A.; Moore, C.; Bi, X.; et al. Deletion of the Capn1 gene results in alterations in signaling pathways related to Alzheimer’s disease, protein quality control and synaptic plasticity in mouse brain. Front. Genet. 2020, 11, 334. [Google Scholar] [CrossRef]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with imageJ. Biophotonics Int. 2004, 11, 36–41. [Google Scholar] [CrossRef]
- Mayer, M.P. Gymnastics of molecular chaperones. Mol. Cell 2010, 39, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, A.; Bhattacharya, A.; McDonald, R.A.; Cordes, M.; Ellington, B.; Bertelsen, E.B.; Zuiderweg, E.R.P. Heat shock protein 70 kDa chaperone/DnaJ cochaperone complex employs an unusual dynamic interface. Proc. Natl. Acad. Sci. USA 2011, 108, 18966–18971. [Google Scholar] [CrossRef] [Green Version]
- Schuermann, J.P.; Jiang, J.; Cuellar, J.; Llorca, O.; Wang, L.; Gimenez, L.E.; Jin, S.; Taylor, A.B.; Demeler, B.; Morano, K.A.; et al. Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol. Cell 2008, 23, 232–243. [Google Scholar] [CrossRef] [Green Version]
- Stürner, E.; Behl, C. The role of the multifunctional bag3 protein in cellular protein quality control and in disease. Front. Mol. Neurosci. 2017, 10, 177. [Google Scholar] [CrossRef] [Green Version]
- Behl, C. Breaking BAG: The co-chaperone BAG3 in health and disease. Trends Pharmacol. Sci. 2016, 37, 672–688. [Google Scholar] [CrossRef]
- Lackner, D.H.; Schmidt, M.W.; Wu, S.; Wolf, D.A.; Bähler, J. Regulation of transcriptome, translation, and proteome in response to environmental stress in fission yeast. Genome Biol. 2012, 13, R25. [Google Scholar] [CrossRef] [Green Version]
- Wallace, E.W.J.; Kear-Scott, J.L.; Pilipenko, E.V.; Schwartz, M.H.; Laskowski, P.R.; Rojek, A.E.; Katanski, C.D.; Riback, J.A.; Dion, M.F.; Franks, A.M.; et al. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 2015, 162, 1286–1298. [Google Scholar] [CrossRef]
- Niforou, K.; Cheimonidou, C.; Trougakos, I.P. Molecular chaperones and proteostasis regulation during redox imbalance. Redox Biol. 2014, 2, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Gong, W.; Wu, S.; Perrett, S. HSP70 in redox homeostasis. Cells 2022, 11, 829. [Google Scholar] [CrossRef]
- Lamech, L.T.; Haynes, C.M. The unpredictability of prolonged activation of stress response pathways. J. Cell Biol. 2015, 209, 781–787. [Google Scholar] [CrossRef] [Green Version]
- Fan, G.C. Role of heat shock proteins in stem cell behavior. Prog. Mol. Biol. Transl. Sci. 2012, 111, 305–322. [Google Scholar] [CrossRef]
- Bradley, E.; Bieberich, E.; Mivechi, N.F.; Tangpisuthipongsa, D.; Wang, G. Regulation of embryonic stem cell pluripotency by heat shock protein 90. Stem Cells 2012, 30, 1624–1633. [Google Scholar] [CrossRef] [Green Version]
- Setati, M.M.; Prinsloo, E.; Longshaw, V.M.; Murray, P.A.; Edgar, D.H.; Blatch, G.L. Leukemia inhibitory factor promotes Hsp90 association with STAT3 in mouse embryonic stem cells. IUBMB Life 2010, 62, 61–66. [Google Scholar] [CrossRef]
- Setroikromo, R.; Wierenga, P.K.; Van Waarde, M.A.W.H.; Brunsting, J.F.; Vellenga, E.; Kampinga, H.H. Heat shock proteins and Bcl-2 expression and function in relation to the differential hyperthermic sensitivity between leukemic and normal hematopoietic cells. Cell Stress Chaperones 2007, 12, 320–330. [Google Scholar] [CrossRef]
- Luo, S.; Mao, C.; Lee, B.; Lee, A.S. GRP78/BiP is required for cell proliferation and protecting the inner cell mass from apoptosis during early mouse embryonic development. Mol. Cell. Biol. 2006, 26, 5688–5697. [Google Scholar] [CrossRef] [Green Version]
- Baharvand, H.; Fathi, A.; van Hoof, D.; Salekdeh, G.H. Concise review: Trends in stem cell proteomics. Stem Cells 2007, 25, 1888–1903. [Google Scholar] [CrossRef]
- Sørensen, J.G.; Nielsen, M.M.; Kruhøffer, M.; Justesen, J.; Loeschcke, V. Full genome gene expression analysis of the heat stress response in Drosophila melanogaster. Cell Stress Chaperones 2005, 10, 312–328. [Google Scholar] [CrossRef]
- Vihervaara, A.; Duarte, F.M.; Lis, J.T. Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet. 2018, 19, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Mahat, D.B.; Salamanca, H.H.; Duarte, F.M.; Danko, C.G.; Lis, J.T. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol. Cell 2016, 62, 63–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jovic, K.; Sterken, M.G.; Grilli, J.; Bevers, R.P.J.; Rodriguez, M.; Riksen, J.A.G.; Allesina, S.; Kammenga, J.E.; Snoek, L.B. Temporal dynamics of gene expression in heat-stressed Caenorhabditis elegans. PLoS ONE 2017, 12, e0189445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Z.; Teo, G.; Krueger, S.; Rock, T.M.; Koh, H.W.; Choi, H.; Vogel, C. Differential dynamics of the mammalian mRNA and protein expression response to misfolding stress. Mol. Syst. Biol. 2016, 12, 855. [Google Scholar] [CrossRef]
- Bouchama, A.; Aziz, M.A.; Mahri, S.A.; Gabere, M.N.; Al Dlamy, M.; Mohammad, S.; Al Abbad, M.; Hussein, M. A Model of exposure to extreme environmental heat uncovers the human transcriptome to heat stress. Sci. Rep. 2017, 7, 9429. [Google Scholar] [CrossRef] [Green Version]
- Jovanovic, M.; Rooney, M.S.; Mertins, P.; Przybylski, D.; Chevrier, N.; Satija, R.; Rodriguez, E.H.; Fields, A.P.; Schwartz, S.; Raychowdhury, R.; et al. Dynamic profiling of the protein life cycle in response to pathogens. Science 2015, 347, 664–667. [Google Scholar] [CrossRef] [Green Version]
- Fournier, M.L.; Paulson, A.; Pavelka, N.; Mosley, A.L.; Gaudenz, K.; Bradford, W.D.; Glynn, E.; Li, H.; Sardiu, M.E.; Fleharty, B.; et al. Delayed correlation of mRNA and protein expression in rapamycin-treated cells and a role for Ggc1 in cellular sensitivity to rapamycin. Mol. Cell. Proteom. 2010, 9, 271–284. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, J.; Larson, D.R. Transcription in living cells: Molecular mechanisms of bursting. Annu. Rev. Biochem. 2020, 89, 189–212. [Google Scholar] [CrossRef] [Green Version]
- Tunnacliffe, E.; Chubb, J.R. What is a transcriptional burst? Trends Genet. 2020, 36, 288–297. [Google Scholar] [CrossRef]
- Alagar Boopathy, L.R.; Jacob-Tomas, S.; Alecki, C.; Vera, M. Mechanisms tailoring the expression of heat shock proteins to proteostasis challenges. J. Biol. Chem. 2022, 298, 101796. [Google Scholar] [CrossRef]
- Anckar, J.; Sistonen, L. Regulation of HSF1 function in the heat stress response: Implications in aging and disease. Annu. Rev. Biochem. 2011, 80, 1089–1115. [Google Scholar] [CrossRef]
- Åkerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2011, 11, 545–555. [Google Scholar] [CrossRef]
- Zheng, X.; Krakowiak, J.; Patel, N.; Beyzavi, A.; Ezike, J.; Khalil, A.S.; Pincus, D. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. eLife 2016, 90, e18638. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Guo, Y.; Guettouche, T.; Smith, D.F.; Voellmy, R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 Complex) that forms a stress-sensitive complex with HSF1. Cell 1998, 94, 471–480. [Google Scholar] [CrossRef] [Green Version]
- Gomez-pastor, R.; Burchfiel, E.T.; Thiele, D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 4–19. [Google Scholar] [CrossRef]
- Liu, Y.; Beyer, A.; Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Aebersold, R. The interdependence of transcript and protein abundance: New data—New complexities. Mol. Syst. Biol. 2016, 12, 856. [Google Scholar] [CrossRef]
- Zander, G.; Hackmann, A.; Bender, L.; Becker, D.; Lingner, T.; Salinas, G.; Krebber, H. MRNA quality control is bypassed for immediate export of stress-responsive transcripts. Nature 2016, 540, 593–596. [Google Scholar] [CrossRef]
- Silver, J.T.; Noble, E.G. Regulation of survival gene HSP70. Cell Stress Chaperones 2012, 17, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, D.C.; Silva, R.; Rondinelli, E.; Ürményi, T.P. Trypanosoma cruzi: Modulation of HSP70 mRNA stability by untranslated regions during heat shock. Exp. Parasitol. 2010, 126, 245–253. [Google Scholar] [CrossRef]
- Lv, H.; Yuan, X.; Zhang, J.; Lu, T.; Yao, J.; Zheng, J.; Cai, J.; Xiao, J.; Chen, H.; Xie, S.; et al. Heat shock preconditioning mesenchymal stem cells attenuate acute lung injury via reducing NLRP3 inflammasome activation in macrophages. Stem Cell Res. Ther. 2021, 12, 290. [Google Scholar] [CrossRef]
- Shimoni, C.; Goldstein, M.; Ribarski-Chorev, I.; Schauten, I.; Nir, D.; Strauss, C.; Schlesinger, S. Heat shock alters mesenchymal stem cell identity and induces premature senescence. Front. Cell Dev. Biol. 2020, 8, 956. [Google Scholar] [CrossRef]
- Alekseenko, L.L.; Zemelko, V.I.; Domnina, A.P.; Lyublinskaya, O.G.; Zenin, V.V.; Pugovkina, N.A.; Kozhukharova, I.V.; Borodkina, A.V.; Grinchuk, T.M.; Fridlyanskaya, I.I.; et al. Sublethal heat shock induces premature senescence rather than apoptosis in human mesenchymal stem cells. Cell Stress Chaperones 2013, 19, 355–366. [Google Scholar] [CrossRef]
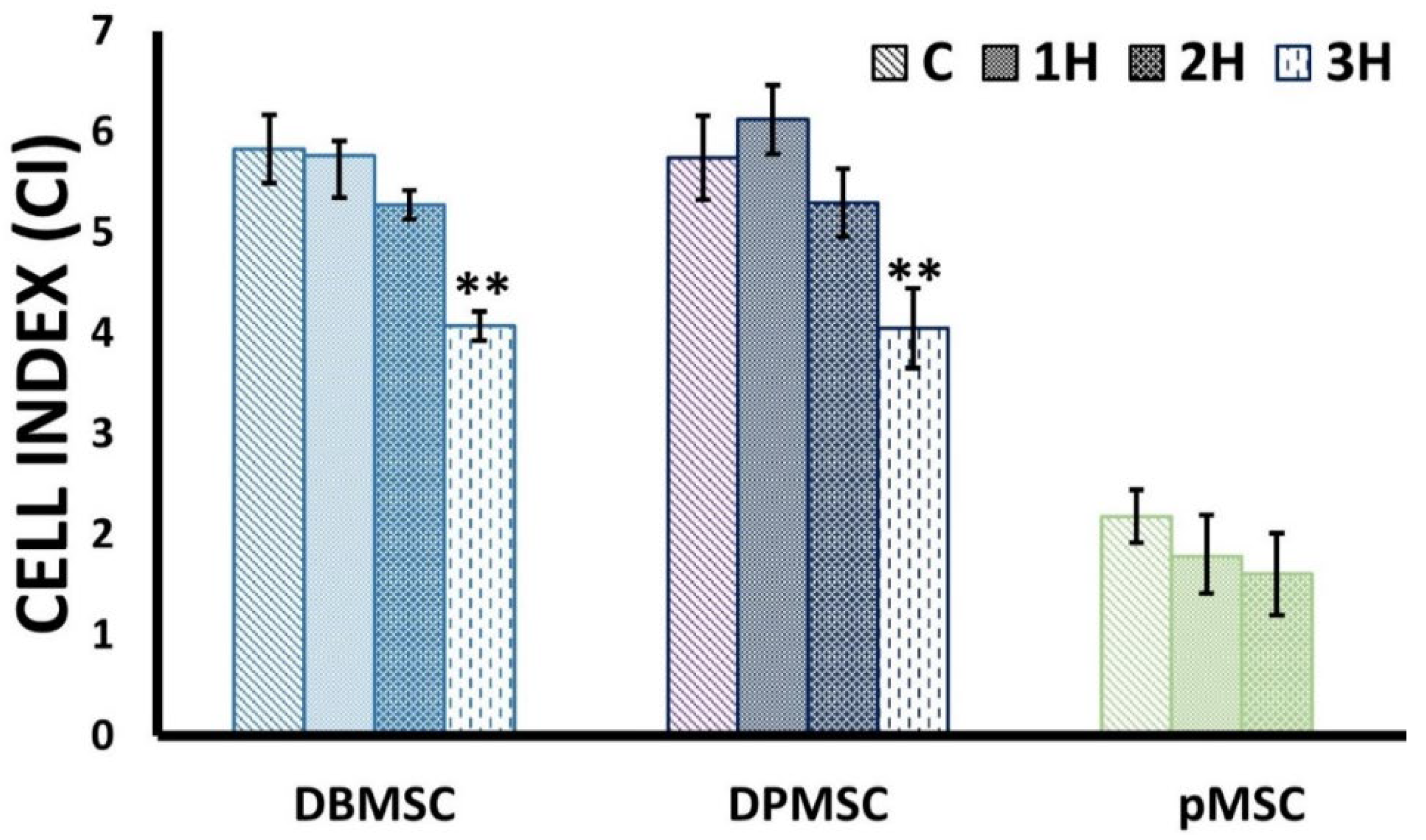
| Cell Type | Exposure Duration [ED] | % Difference Cell Index (CI) |
|---|---|---|
| DBMSCs | 1 h | 1.13 |
| 2 h | 9.45 | |
| 3 h | 30.15 | |
| DPMSCs | 1 h | −6.61 |
| 2 h | 7.74 | |
| 3 h | 29.48 | |
| pMSCs | 1 h | 18.28 |
| 2 h | 26.40 |
| Exposure Temperature [ET] | Exposure Duration [ED] | Hsp70 Induction |
|---|---|---|
| 42 °C | 1 h | 40.67 ± 7.7 |
| 2 h | 41.04 ± 6.3 | |
| 44 °C | 1 h | 56.07 ± 8.4 |
| 2 h | 68.77 ± 2.38 | |
| 3 h | 108.13 ± 1.54 |
| Chaperone Family | Overexpressed Genes | Proportion |
|---|---|---|
| Small Heat Shock Protein | HSPB1, HSPB8, CRYAB | 3/8 |
| 60 kDa heat shock protein, mitochondrial | HSPD1 | 1/1 |
| Heat shock 70 kDa protein | HSPA1A, HSPA1B, HSPA1L, HSPA4L, HSPA5, HSPA6 | 6/11 |
| Heat shock protein 90 kDa alpha (cytosolic), class A | HSP90AA1 | 1/1 |
| Heat shock protein 105 kDa | HSPH1 | 1/1 |
| DnaJ homolog subfamilyA | DNAJA1, DNAJA4 | 2/4 |
| DnaJ homolog subfamily B | DNAJB1, DNAJB9 | 2/11 |
| BAG family molecular chaperone regulator 3 | BAG3 | 1/1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alharbi, B.M.; Bugshan, A.; Almozel, A.; Alenzi, R.; Bouchama, A.; Khatlani, T.; Mohammad, S.; Malik, S.S. Heat-Induced Proteotoxic Stress Response in Placenta-Derived Stem Cells (PDSCs) Is Mediated through HSPA1A and HSPA1B with a Potential Higher Role for HSPA1B. Curr. Issues Mol. Biol. 2022, 44, 4748-4768. https://doi.org/10.3390/cimb44100324
Alharbi BM, Bugshan A, Almozel A, Alenzi R, Bouchama A, Khatlani T, Mohammad S, Malik SS. Heat-Induced Proteotoxic Stress Response in Placenta-Derived Stem Cells (PDSCs) Is Mediated through HSPA1A and HSPA1B with a Potential Higher Role for HSPA1B. Current Issues in Molecular Biology. 2022; 44(10):4748-4768. https://doi.org/10.3390/cimb44100324
Chicago/Turabian StyleAlharbi, Bothina Mohammed, Aisha Bugshan, Azhaar Almozel, Reem Alenzi, Abderrezak Bouchama, Tanvir Khatlani, Sameer Mohammad, and Shuja Shafi Malik. 2022. "Heat-Induced Proteotoxic Stress Response in Placenta-Derived Stem Cells (PDSCs) Is Mediated through HSPA1A and HSPA1B with a Potential Higher Role for HSPA1B" Current Issues in Molecular Biology 44, no. 10: 4748-4768. https://doi.org/10.3390/cimb44100324
APA StyleAlharbi, B. M., Bugshan, A., Almozel, A., Alenzi, R., Bouchama, A., Khatlani, T., Mohammad, S., & Malik, S. S. (2022). Heat-Induced Proteotoxic Stress Response in Placenta-Derived Stem Cells (PDSCs) Is Mediated through HSPA1A and HSPA1B with a Potential Higher Role for HSPA1B. Current Issues in Molecular Biology, 44(10), 4748-4768. https://doi.org/10.3390/cimb44100324





