De Novo Transcriptome Assembly, Gene Annotations, and Characterization of Functional Profiling Reveal Key Genes for Lead Alleviation in the Pb Hyperaccumulator Greek Mustard (Hirschfeldia incana L.)
Abstract
1. Introduction
2. Materials and Methods
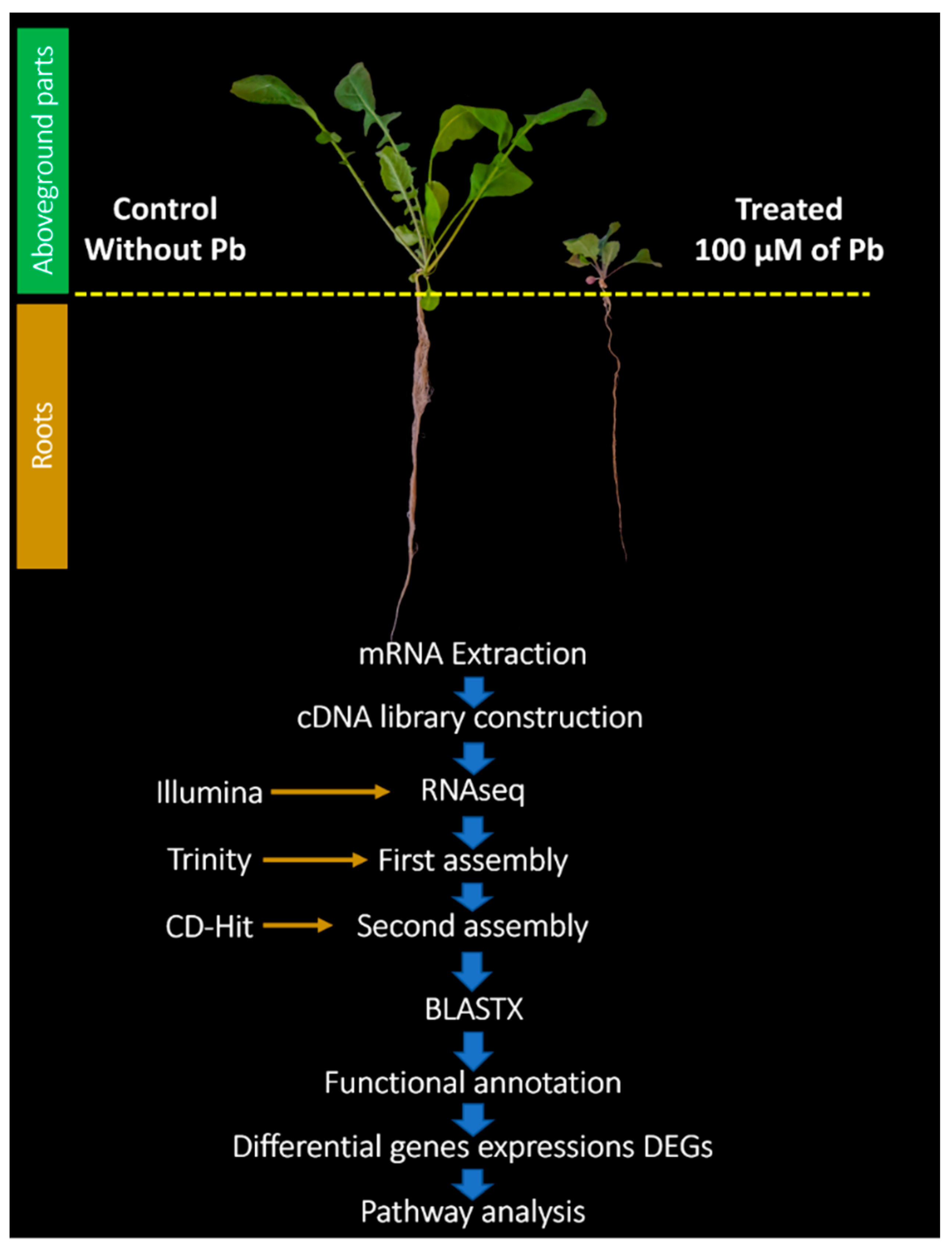
2.1. Plant Material
2.2. mRNA Extraction
2.3. cDNA Construction and Sequencing
2.4. De Novo Assembly of Transcriptome
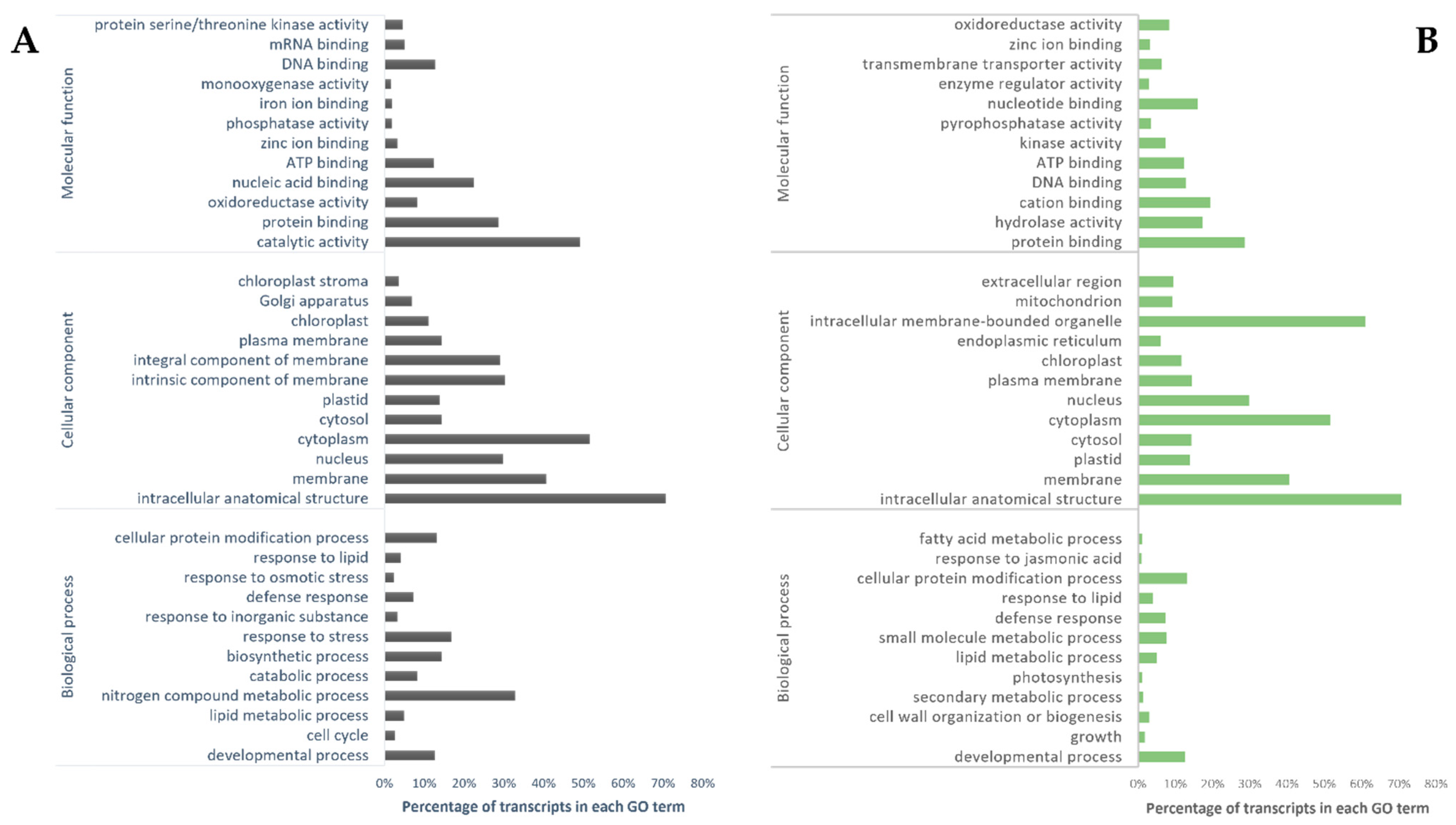
2.5. Annotation and Function Profiling
2.6. Transcript Estimation and DEG Analysis
3. Results and Discussion
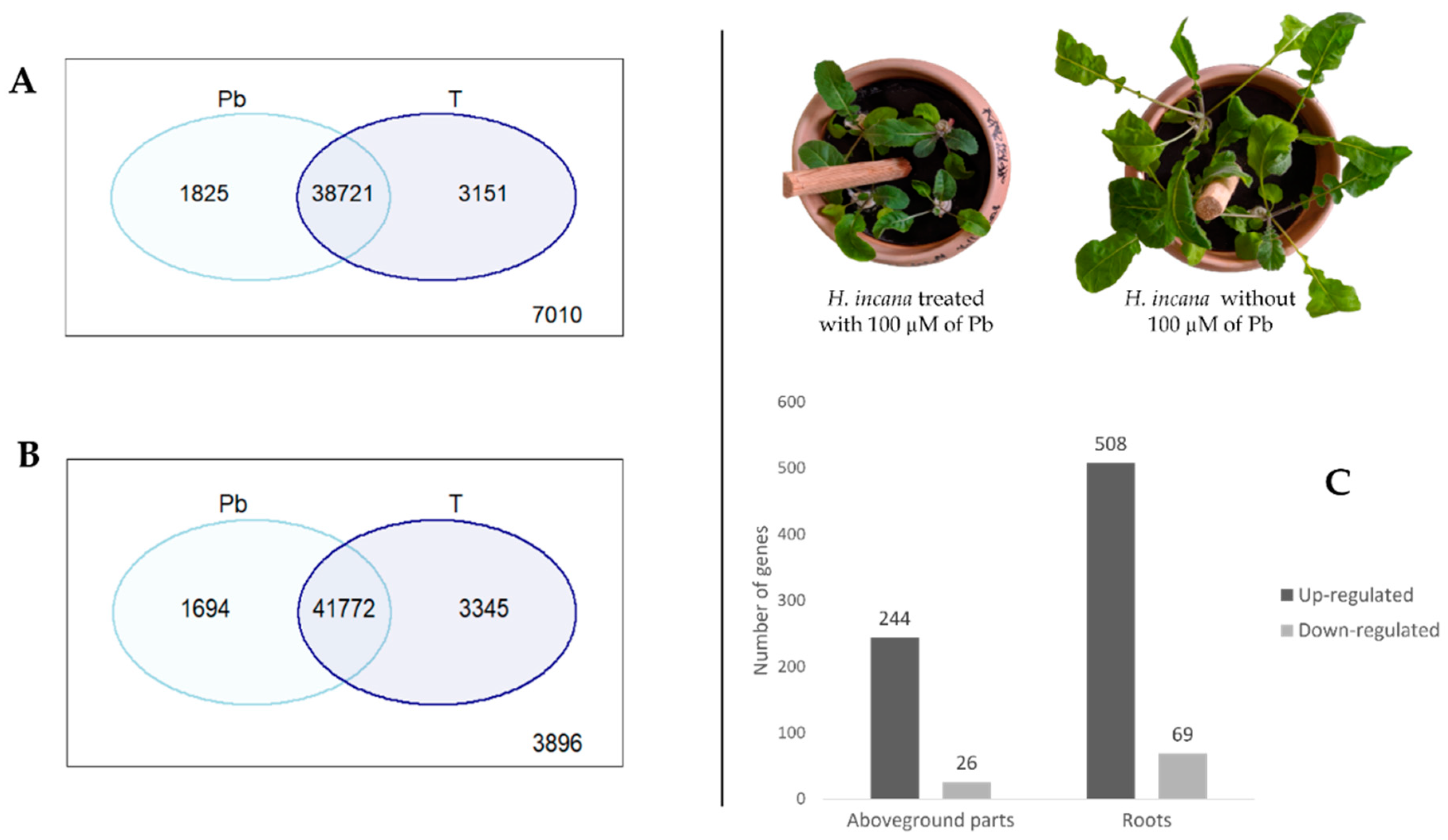
3.1. mRNA-seq and De Novo Transcriptome Assembly
3.2. Differential Expressed Genes (DEGs) under Pb Exposure
3.3. Jasmonates (JAs) and Plant Response to Pb—A Case Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACX1 | Acyl-CoA oxidase 1; |
| AOC4 | allene oxide cyclase 4; |
| AOS | allene oxide synthase; |
| BUSCO | Benchmarking Universal Single-Copy Orthologs; |
| CD-HIT | Cluster Database at High Identity with Tolerance; |
| DEGs | differentially gene expressions; |
| DE | differential expressed; |
| GO | Gene Ontology; |
| GTH | glutathione; |
| JAZ10 | JASMONATE ZIM-domain protein 10; |
| JAs | jasmonates; |
| KAT5 | 3-keto-acyl-CoA thiolase 2; |
| KEGG | Kyoto Encyclopedia of Genes and Genomes; |
| LOX2 | lipoxygenase; |
| HMs | heavy metals; |
| MTs | metallothioneins; |
| PCs | Phytochelatin; |
| REVIGO | reduce and visualize gene ontology; |
| RIN | RNA Integrity Number; |
| ROS | reactive oxygen species; |
| RSEM | RNA-Seq by Expectation-Maximization; |
| TFs | transcription factors; |
| 13(S)-HPOT | (9Z,11E,15Z) -(13S)-13-Hydroperoxyoctadeca-9,11,15-trienoic acid; |
| 12,13(S)-EOT | (9Z,15Z)-(13S)-12,13-Epoxyoctadeca-9,11,15-trienoic acid; |
| 12-OPDA | (15Z)-12-Oxophyto-10,15-dienoic acid; |
| OPC8 | 8-[(1R,2R)-3-Oxo-2-(Z)-pent-2-enylcyclopentyl] octanoate |
References
- Tangahu, B.V.; Abdullah, S.R.S.; Basri, H.; Idris, M.; Anuar, N.; Mukhlisin, M. A review on heavy metals (As, Pb, and Hg) uptake by plants through phytoremediation. Int. J. Chem. Eng. 2011, 2011, 939161. [Google Scholar] [CrossRef]
- Fahr, M.; Laplaze, L.; Bendaou, N.; Hocher, V.; El Mzibri, M.; Bogusz, D.; Smouni, A. Effect of lead on root growth. Front. Plant Sci. 2013, 4, 175. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zheng, B.; Yuan, Y.; Xu, Q.; Chen, P. Transcriptome profiling of Fagopyrum tataricum leaves in response to lead stress. BMC Plant Biol. 2020, 20, 54. [Google Scholar] [CrossRef] [PubMed]
- Mourato, M.; Reis, R.; Martins, L.L. Characterization of plant antioxidative system in response to abiotic stresses: A focus on heavy metal toxicity. Adv. Sel. Plant Physiol. Asp. 2012, 12, 23–44. [Google Scholar]
- John, R.; Ahmad, P.; Gadgil, K.; Sharma, S. Cadmium and lead-induced changes in lipid peroxidation, antioxidative enzymes and metal accumulation in Brassica juncea L. at three different growth stages. Arch. Agron. Soil Sci. 2009, 55, 395–405. [Google Scholar] [CrossRef]
- Mahdavian, K.; Ghaderian, S.M.; Schat, H. Pb accumulation, Pb tolerance, antioxidants, thiols, and organic acids in metallicolous and non-metallicolous Peganum harmala L. under Pb exposure. Environ. Exp. Bot. 2016, 126, 21–31. [Google Scholar] [CrossRef]
- Mohtadi, A.; Ghaderian, S.M.; Schat, H. A comparison of lead accumulation and tolerance among heavy metal hyperaccumulating and non-hyperaccumulating metallophytes. Plant Soil 2012, 352, 267–276. [Google Scholar] [CrossRef]
- Auguy, F.; Fahr, M.; Moulin, P.; Brugel, A.; Laplaze, L.; Mzibri, M.E.; Filali-Maltouf, A.; Doumas, P.; Smouni, A. Lead tolerance and accumulation in Hirschfeldia incana, a Mediterranean Brassicaceae from metalliferous mine spoils. PLoS ONE 2013, 8, e61932. [Google Scholar] [CrossRef]
- Siemens, J.; González, M.C.; Wolf, S.; Hofmann, C.; Greiner, S.; Du, Y.; Rausch, T.; Roitsch, T.; Ludwig-Müller, J. Extracellular invertase is involved in the regulation of clubroot disease in Arabidopsis thaliana. Mol. Plant Pathol. 2011, 12, 247–262. [Google Scholar] [CrossRef]
- Fahr, M.; Laplaze, L.; El Mzibri, M.; Doumas, P.; Bendaou, N.; Hocher, V.; Bogusz, D.; Smouni, A. Assessment of lead tolerance and accumulation in metallicolous and non-metallicolous populations of Hirschfeldia incana. Environ. Exp. Bot. 2015, 109, 186–192. [Google Scholar] [CrossRef]
- Pourrut, B.; Shahid, M.; Douay, F.; Dumat, C.; Pinelli, E. Molecular mechanisms involved in lead uptake, toxicity and detoxification in higher plants. In Heavy Metal Stress in Plants; Springer: Berlin/Heidelberg, Germany, 2013; pp. 121–147. [Google Scholar]
- Arazi, T.; Sunkar, R.; Kaplan, B.; Fromm, H. A tobacco plasma membrane calmodulin-binding transporter confers Ni2+ tolerance and Pb2+ hypersensitivity in transgenic plants. Plant J. 1999, 20, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Gravot, A.; Lieutaud, A.; Verret, F.; Auroy, P.; Vavasseur, A.; Richaud, P. AtHMA3, a plant P1B-ATPase, functions as a Cd/Pb transporter in yeast. FEBS Lett. 2004, 561, 22–28. [Google Scholar] [CrossRef]
- Morel, M.; Crouzet, J.; Gravot, A.; Auroy, P.; Leonhardt, N.; Vavasseur, A.; Richaud, P. AtHMA3, a P1B-ATPase allowing Cd/Zn/co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009, 149, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Lee, K.; Lee, J.; Noh, E.W.; Lee, Y. AtPDR12 contributes to lead resistance in Arabidopsis. Plant Physiol. 2005, 138, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-Y.; Bovet, L.; Kushnir, S.; Noh, E.W.; Martinoia, E.; Lee, Y. AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 2006, 140, 922–932. [Google Scholar] [CrossRef]
- Kim, D.Y.; Bovet, L.; Maeshima, M.; Martinoia, E.; Lee, Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007, 50, 207–218. [Google Scholar] [CrossRef]
- Cobbett, C.; Goldsbrough, P. Phytochelatins and metallothioneins: Roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 2002, 53, 159–182. [Google Scholar] [CrossRef]
- Auguy, F.; Fahr, M.; Moulin, P.; El Mzibri, M.; Smouni, A.; Filali-Maltouf, A.; Béna, G.; Doumas, P. Transcriptome changes in Hirschfeldia incana in response to lead exposure. Front. Plant Sci. 2016, 6, 1231. [Google Scholar] [CrossRef]
- Evangelistella, C.; Valentini, A.; Ludovisi, R.; Firrincieli, A.; Fabbrini, F.; Scalabrin, S.; Cattonaro, F.; Morgante, M.; Mugnozza, G.S.; Keurentjes, J.J. De novo assembly, functional annotation, and analysis of the giant reed (Arundo donax L.) leaf transcriptome provide tools for the development of a biofuel feedstock. Biotechnol. Biofuels 2017, 10, 138. [Google Scholar] [CrossRef]
- Hasnaoui, S.E.; Fahr, M.; Keller, C.; Levard, C.; Angeletti, B.; Chaurand, P.; Triqui, Z.E.A.; Guedira, A.; Rhazi, L.; Colin, F. Screening of native plants growing on a Pb/Zn mining area in eastern Morocco: Perspectives for phytoremediation. Plants 2020, 9, 1458. [Google Scholar] [CrossRef]
- Broughton, W.; Dilworth, M. Control of leghaemoglobin synthesis in snake beans. Biochem. J. 1971, 125, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Sharma, R.; Mishra, M.; Gupta, B.; Parsania, C.; Singla-Pareek, S.L.; Pareek, A. De novo assembly and characterization of stress transcriptome in a salinity-tolerant variety CS52 of Brassica juncea. PLoS ONE 2015, 10, e0126783. [Google Scholar] [CrossRef]
- Yong, H.-Y.; Zou, Z.; Kok, E.-P.; Kwan, B.-H.; Chow, K.; Nasu, S.; Nanzyo, M.; Kitashiba, H.; Nishio, T. Comparative transcriptome analysis of leaves and roots in response to sudden increase in salinity in Brassica napus by RNA-seq. BioMed Res. Int. 2014, 2014, 395–467. [Google Scholar] [CrossRef]
- Seol, Y.-J.; Kim, K.; Kang, S.-H.; Perumal, S.; Lee, J.; Kim, C.-K. The complete chloroplast genome of two Brassica species, Brassica nigra and B. oleracea. Mitochondrial DNA Part A 2017, 28, 167–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, L.; Chen, Y.; Shen, H.; Gong, Y.; Limera, C.; Liu, L. Transcriptome profiling of radish (Raphanus sativus L.) root and identification of genes involved in response to lead (Pb) stress with next generation sequencing. PLoS ONE 2013, 8, e66539. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Choudhary, S.; Bhardwaj, P. Comparative transcriptome profiling under cadmium stress reveals the uptake and tolerance mechanism in Brassica juncea. J. Plant Growth Regul. 2019, 38, 1141–1152. [Google Scholar] [CrossRef]
- Di Toppi, L.S.; Gabbrielli, R. Response to cadmium in higher plants. Environ. Exp. Bot. 1999, 41, 105–130. [Google Scholar] [CrossRef]
- Allan, D.L.; Jarrell, W.M. Proton and copper adsorption to maize and soybean root cell walls. Plant Physiol. 1989, 89, 823–832. [Google Scholar] [CrossRef]
- Peng, H.-Y.; Yang, X.-E.; Tian, S.-K. Accumulation and ultrastructural distribution of copper in Elsholtzia splendens. J. Zhejiang Univ. Sci. B 2005, 6, 311. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, S.; Wang, C.; Lu, J. Effects of Pb on the oxidative stress and antioxidant response in a Pb bioaccumulator plant Vallisneria natans. Ecotoxicol. Environ. Saf. 2012, 78, 28–34. [Google Scholar] [CrossRef]
- Satoh-Nagasawa, N.; Mori, M.; Nakazawa, N.; Kawamoto, T.; Nagato, Y.; Sakurai, K.; Takahashi, H.; Watanabe, A.; Akagi, H. Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 2012, 53, 213–224. [Google Scholar] [CrossRef]
- Hasan, M.; Alabdallah, N.M.; Alharbi, B.M.; Waseem, M.; Yao, G.; Liu, X.-D.; El-Gawad, A.; Hany, G.; El-Yazied, A.A.; Ibrahim, M.F. GABA: A Key Player in Drought Stress Resistance in Plants. Int. J. Mol. Sci. 2021, 22, 10136. [Google Scholar] [CrossRef]
- Daş, Z.A.; Dimlioğlu, G.; Bor, M.; Özdemir, F. Zinc induced activation of GABA-shunt in tobacco (Nicotiana tabaccum L.). Environ. Exp. Bot. 2016, 122, 78–84. [Google Scholar] [CrossRef]
- Mahmud, J.A.; Hasanuzzaman, M.; Nahar, K.; Rahman, A.; Hossain, M.; Fujita, M. γ-aminobutyric acid (GABA) confers chromium stress tolerance in Brassica juncea L. by modulating the antioxidant defense and glyoxalase systems. Ecotoxicology 2017, 26, 675–690. [Google Scholar] [CrossRef] [PubMed]
- Khoudi, H.; Maatar, Y.; Gouiaa, S.; Masmoudi, K. Transgenic tobacco plants expressing ectopically wheat H+-pyrophosphatase (H+-PPase) gene TaVP1 show enhanced accumulation and tolerance to cadmium. J. Plant Physiol. 2012, 169, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Monné, M.; Miniero, D.V.; Obata, T.; Daddabbo, L.; Palmieri, L.; Vozza, A.; Nicolardi, M.C.; Fernie, A.R.; Palmieri, F. Functional characterization and organ distribution of three mitochondrial ATP–Mg/Pi carriers in Arabidopsis thaliana. Biochim. Biophys. Acta BBA Bioenerg. 2015, 1847, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Block, M.D.; Verduyn, C.; Brouwer, D.D.; Cornelissen, M. Poly (ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J. 2005, 41, 95–106. [Google Scholar] [CrossRef]
- Singh, H.P.; Mahajan, P.; Kaur, S.; Batish, D.R.; Kohli, R.K. Chromium toxicity and tolerance in plants. Environ. Chem. Lett. 2013, 11, 229–254. [Google Scholar] [CrossRef]
- Sharma, A.; Kumar, V.; Shahzad, B.; Ramakrishnan, M.; Sidhu, G.P.S.; Bali, A.S.; Handa, N.; Kapoor, D.; Yadav, P.; Khanna, K. Photosynthetic response of plants under different abiotic stresses: A review. J. Plant Growth Regul. 2020, 39, 509–531. [Google Scholar] [CrossRef]
- Drummond, R.; Tutone, A.; Li, Y.-C.; Gardner, R. A putative magnesium transporter AtMRS2-11 is localized to the plant chloroplast envelope membrane system. Plant Sci. 2006, 170, 78–89. [Google Scholar] [CrossRef]
- Jha, S.K.; Sharma, M.; Pandey, G.K. Role of cyclic nucleotide gated channels in stress management in plants. Curr. Genom. 2016, 17, 315–329. [Google Scholar] [CrossRef]
- Moon, J.Y.; Belloeil, C.; Ianna, M.L.; Shin, R. Arabidopsis CNGC family members contribute to heavy metal ion uptake in plants. Int. J. Mol. Sci. 2019, 20, 413. [Google Scholar] [CrossRef]
- Nieves-Cordones, M.; Alemán, F.; Martínez, V.; Rubio, F. K+ uptake in plant roots. The systems involved, their regulation and parallels in other organisms. J. Plant Physiol. 2014, 171, 688–695. [Google Scholar] [CrossRef]
- Kohli, S.K.; Handa, N.; Gautam, V.; Bali, S.; Sharma, A.; Khanna, K.; Arora, S.; Thukral, A.K.; Ohri, P.; Karpets, Y.V. ROS Signaling in Plants Under Heavy Metal Stress. In Reactive Oxygen Species and Antioxidant Systems in Plants: Role and Regulation under Abiotic Stress; Khan, M., Khan, N., Eds.; Springer: Singapore, 2017. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.; Nahar, K.; Hossain, M.; Mahmud, J.A.; Hossen, M.; Masud, A.A.C.; Fujita, M. Potassium: A vital regulator of plant responses and tolerance to abiotic stresses. Agronomy 2018, 8, 31. [Google Scholar] [CrossRef]
- Akram, W.; Khan, W.U.; Shah, A.A.; Yasin, N.A.; Li, G. Liquiritoside Alleviated Pb Induced Stress in Brassica rapa subsp. Parachinensis: Modulations in Glucosinolate Content and Some Physiochemical Attributes. Front. Plant Sci. 2021, 12, 1799. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, P. Glucosinolates and its role in mitigating abiotic and biotic stress in Brassicaceae. In Plant Stress Physiology—Perspectives in Agriculture; IntechOpen: London, UK, 2022. [Google Scholar]
- Wu, H.; Chen, C.; Du, J.; Liu, H.; Cui, Y.; Zhang, Y.; He, Y.; Wang, Y.; Chu, C.; Feng, Z. Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 2012, 158, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, Z.; Hassan, Z.; Ul-Allah, S.; Khan, A.A.; Sattar, A.; Shahzad, U.; Amin, H.; Hussain, M. Transcription factors involved in plant responses to heavy metal stress adaptation. In Plant Perspectives to Global Climate Changes; Elsevier: Amsterdam, The Netherlands, 2022; pp. 221–231. [Google Scholar]
- Dutta, S.; Mitra, M.; Agarwal, P.; Mahapatra, K.; De, S.; Sett, U.; Roy, S. Oxidative and genotoxic damages in plants in response to heavy metal stress and maintenance of genome stability. Plant Signal. Behav. 2018, 13, e1460048. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [PubMed]
- Van de Mortel, J.E.; Schat, H.; Moerland, P.D.; Van Themaat, E.V.L.; Van Der Ent, S.; Blankestijn, H.; Ghandilyan, A.; Tsiatsiani, S.; Aarts, M.G. Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 2008, 31, 301–324. [Google Scholar] [CrossRef]
- Skibbe, M.; Qu, N.; Galis, I.; Baldwin, I.T. Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 2008, 20, 1984–2000. [Google Scholar] [CrossRef]
- Raza, A.; Charagh, S.; Zahid, Z.; Mubarik, M.S.; Javed, R.; Siddiqui, M.H.; Hasanuzzaman, M. Jasmonic acid: A key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 2021, 40, 1513–1541. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, W.; Tong, T.; Chen, G.; Zeng, F.; Jang, S.; Gao, W.; Li, Z.; Mak, M.; Deng, F. Molecular interaction and evolution of jasmonate signaling with transport and detoxification of heavy metals and metalloids in plants. Front. Plant Sci. 2021, 12, 625. [Google Scholar] [CrossRef]
- Lei, G.J.; Sun, L.; Sun, Y.; Zhu, X.F.; Li, G.X.; Zheng, S.J. Jasmonic acid alleviates cadmium toxicity in Arabidopsis via suppression of cadmium uptake and translocation. J. Integr. Plant Biol. 2020, 62, 218–227. [Google Scholar] [CrossRef]
- Liu, T.; Liu, S.; Guan, H.; Ma, L.; Chen, Z.; Gu, H.; Qu, L.-J. Transcriptional profiling of Arabidopsis seedlings in response to heavy metal lead (Pb). Environ. Exp. Bot. 2009, 67, 377–386. [Google Scholar] [CrossRef]
- Ahmad, P.; Raja, V.; Ashraf, M.; Wijaya, L.; Bajguz, A.; Alyemeni, M.N. Jasmonic acid (JA) and gibberellic acid (GA3) mitigated Cd-toxicity in chickpea plants through restricted cd uptake and oxidative stress management. Sci. Rep. 2021, 11, 19768. [Google Scholar] [CrossRef] [PubMed]
- Acosta, I.F.; Farmer, E.E. Jasmonates. In The Arabidopsis Book; The American Society of Plant Biologists: Rockville, MD, USA, 2010; pp. 8–129. [Google Scholar]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, G.; Xu, C.; Lee, G.I.; Bauer, P.; Ling, H.-Q.; Ganal, M.W.; Howe, G.A. The tomato suppressor of prosystemin-mediated responses 2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 2003, 15, 1646–1661. [Google Scholar] [CrossRef]
- Li, Q.; Lei, S.; Du, K.; Li, L.; Pang, X.; Wang, Z.; Wei, M.; Fu, S.; Hu, L.; Xu, L. RNA-seq based transcriptomic analysis uncovers α-linolenic acid and jasmonic acid biosynthesis pathways respond to cold acclimation in Camellia japonica. Sci. Rep. 2016, 6, 36463. [Google Scholar] [CrossRef]
- Upchurch, R.G. Fatty acid unsaturation, mobilization, and regulation in the response of plants to stress. Biotechnol. Lett. 2008, 30, 967–977. [Google Scholar] [CrossRef]
- Beisson, F.; Li, Y.; Bonaventure, G.; Pollard, M.; Ohlrogge, J.B. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 2007, 19, 351–368. [Google Scholar] [CrossRef]
- Kachroo, P.; Kachroo, A.; Lapchyk, L.; Hildebrand, D.; Klessig, D.F. Restoration of defective cross talk in ssi2 mutants: Role of salicylic acid, jasmonic acid, and fatty acids in SSI2-mediated signaling. Mol. Plant Microbe Interact. 2003, 16, 1022–1029. [Google Scholar] [CrossRef][Green Version]



| Initial Transcripts (TRINITY) | H. incana | B. napus | B. nigra | B. juncea |
|---|---|---|---|---|
| Total transcripts | 216,315 | - | - | - |
| Total assembled bases | 211,894,927 | - | - | - |
| Average length (pb) | 979.57 | - | - | - |
| N50 | 1304 | - | - | - |
| Percent GC | 44.47 | - | - | - |
| Secondary transcripts (CD-HIT) | ||||
| Total transcripts | 77,491 | 161,537 | NA | 53,669 |
| Total assembled bases | 74,378,239 | 111,953,629 | NA | 51,151,545 |
| Average length (pb) | 959.83 | 693 | 1173 | 953 |
| N50 | 1330 | 1093 | 1482 | 1282 |
| Percent GC | 43.83 | NA | 37 | 44.38 |
| BUSCO % | 94.2% | NA | 97% | NA |
| Number of genes | 50,707 | NA | 56,331 | NA |
| (A) | ||||
| Transcript ID | Transcript Name | Log2FC | p-Value | N° of GO Terms |
| Up-Regulated Transcripts in aboveground Parts of H. incana | ||||
| HiInc_DN6940_c0_g2 | Noc2p family | 10.56 1.94 | 5.43 × 10−8 | 8 |
| HiInc_DN64666_c0_g1 | RNA-directed DNA polymerase (Reverse transcriptase)-related family protein | 8.55 1.47 | 6.97 × 10−9 | 3 |
| HiInc_DN54630_c0_g1 | Translation initiation factor IF3-4 | 7.31 1.40 | 2.02 × 10−7 | 8 |
| HiInc_DN38797_c0_g1 | Peroxidase | 6.07 1.84 | 1.57 × 10−11 | 4 |
| HiInc_DN8233_c0_g1 | Inorganic pyrophosphatase 2 | 5.65 0.86 | 6.74 × 10−11 | 6 |
| HiInc_DN250_c0_g1 | Syringolide-induced protein | 5.28 0.88 | 2.89 × 10−9 | 7 |
| HiInc_DN35271_c0_g1 | Beta-glucosidase 32 | 5.13 1.64 | 3.11 × 10−12 | 6 |
| HiInc_DN5802_c0_g2 | Monogalactosyldiacylglycerol synthase 2 | 5.10 0.57 | 4.03 × 10−19 | 9 |
| HiInc_DN12789_c0_g1 | REF/SRPP-like protein | 5.02 0.69 | 3.25 × 10−13 | 7 |
| HiInc_DN1096_c0_g4 | Salicylate/benzoate carboxyl methyltransferase | 4.89 0.92 | 1.36 × 10−7 | 5 |
| Up-regulated transcripts in roots of H. incana | ||||
| HiInc_DN6451_c0_g2 | Endochitinase | 11.68 1.75 | 2.60 × 10−11 | 7 |
| HiInc_DN64488_c0_g1 | ATPase 9 | 10.29 1.96 | 1.52 × 10−7 | 4 |
| HiInc_DN5848_c0_g1 | Accelerated cell death 11 | 9.89 1.96 | 5.01 × 10−7 | 7 |
| HiInc_DN27683_c0_g1 | Putative endonuclease or glycosyl hydrolase | 9.74 1.32 | 2.38 × 10−13 | 2 |
| HiInc_DN6940_c0_g1 | Noc2p family | 9.47 1.49 | 2.56 × 10−10 | 4 |
| HiInc_DN10792_c0_g1 | Nucleic acid-binding | 8.82 1.48 | 3.06 × 10−9 | 3 |
| HiInc_DN62699_c2_g1 | BED zinc finger and hAT dimerization domain-containing protein DAYSLEEPER | 8.70 1.63 | 1.03 × 10−7 | 1 |
| HiInc_DN63847_c0_g1 | Alpha carbonic anhydrase 2 | 8.36 1.29 | 1.20 × 10−10 | 4 |
| HiInc_DN13103_c0_g1 | 60S ribosomal protein L10a-2 | 7.81 1.33 | 5.24 × 10−9 | 2 |
| HiInc_DN8575_c1_g1 | Disease resistance protein (TIR-NBS class) | 7.77 1.39 | 2.71 × 10−8 | 13 |
| (B) | ||||
| Transcript ID | Transcript name | Log2FC | p-value | N° of GO terms |
| Down-regulated transcripts in aboveground parts of H. incana | ||||
| HiInc_DN38456_c0_g1 | transmembrane protein, putative (DGR2) | −2.10 0.25 | 1.20 × 10−16 | 1 |
| HiInc_DN19265_c0_g1 | Chaperone DnaJ-domain superfamily protein | −2.17 0.43 | 7.06 × 10−7 | 5 |
| HiInc_DN12488_c0_g1 | Thioredoxin-like protein CXXS1 | −2.19 0.34 | 3.79 × 10−6 | 2 |
| HiInc_DN54188_c0_g1 | Tetratricopeptide repeat (TPR)-like | −2.24 0.45 | 1.02 × 10−6 | 2 |
| HiInc_DN9080_c0_g1 | Tubulin alpha chain | −2.29 0.39 | 4.30 × 10−9 | 15 |
| HiInc_DN33913_c0_g1 | Probable aquaporin PIP2-8 | −2.37 0.42 | 2.20 × 10−8 | 3 |
| HiInc_DN7675_c0_g1 | Gamma vacuolar processing enzyme | −2.51 0.35 | 2.21 × 10−12 | 12 |
| HiInc_DN10024_c0_g1 | RNA-binding (RRM/RBD/RNP motifs) | −2.75 0.55 | 8.13 × 10−7 | 6 |
| HiInc_DN18907_c0_g1 | RmlC-like cupins superfamily protein | −2.79 0.52 | 7.56 × 10−8 | 2 |
| HiInc_DN20249_c0_g1 | Ribosomal protein S3Ae | −3.27 0.53 | 8.09 × 10−10 | 8 |
| Down-regulated transcripts in roots of H. incana | ||||
| HiInc_DN1121_c0_g2 | RmlC-like cupins superfamily protein | −2.02 0.22 | 8.16 × 10−19 | 3 |
| HiInc_DN2301_c0_g1 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | −2.06 0.33 | 5.82 × 10−10 | 4 |
| HiInc_DN315_c5_g1 | Receptor-like protein 30 | −2.07 0.39 | 1.50 × 10−7 | 6 |
| HiInc_DN6047_c0_g1 | Protein kinase superfamily protein | −2.10 0.39 | 1.22 × 10−7 | 2 |
| HiInc_DN10269_c1_g2 | UDP-glycosyltransferase 76B1 | −2.42 0.49 | 1.07 × 10−6 | 8 |
| HiInc_DN38838_c0_g1 | 60S acidic ribosomal protein P2-3 | −2.45 0.33 | 2.33 × 10−13 | 2 |
| HiInc_DN16964_c0_g1 | Probable LRR receptor-like serine/threonine-protein kinase | −2.48 0.34 | 8.71 × 10−10 | 2 |
| HiInc_DN22651_c0_g3 | Glutathione S-transferase F3 | −2.51 0.26 | 7.18 × 10−7 | 5 |
| HiInc_DN24519_c1_g1 | Probable LRR receptor-like serine/threonine-protein kinase | −2.51 0.50 | 5.27 × 10−7 | 7 |
| HiInc_DN13802_c0_g1 | Tricyclene synthase | −2.64 0.26 | 5.76 × 10−24 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasnaoui, S.E.; Fahr, M.; Zouine, M.; Smouni, A. De Novo Transcriptome Assembly, Gene Annotations, and Characterization of Functional Profiling Reveal Key Genes for Lead Alleviation in the Pb Hyperaccumulator Greek Mustard (Hirschfeldia incana L.). Curr. Issues Mol. Biol. 2022, 44, 4658-4675. https://doi.org/10.3390/cimb44100318
Hasnaoui SE, Fahr M, Zouine M, Smouni A. De Novo Transcriptome Assembly, Gene Annotations, and Characterization of Functional Profiling Reveal Key Genes for Lead Alleviation in the Pb Hyperaccumulator Greek Mustard (Hirschfeldia incana L.). Current Issues in Molecular Biology. 2022; 44(10):4658-4675. https://doi.org/10.3390/cimb44100318
Chicago/Turabian StyleHasnaoui, Said El, Mouna Fahr, Mohamed Zouine, and Abdelaziz Smouni. 2022. "De Novo Transcriptome Assembly, Gene Annotations, and Characterization of Functional Profiling Reveal Key Genes for Lead Alleviation in the Pb Hyperaccumulator Greek Mustard (Hirschfeldia incana L.)" Current Issues in Molecular Biology 44, no. 10: 4658-4675. https://doi.org/10.3390/cimb44100318
APA StyleHasnaoui, S. E., Fahr, M., Zouine, M., & Smouni, A. (2022). De Novo Transcriptome Assembly, Gene Annotations, and Characterization of Functional Profiling Reveal Key Genes for Lead Alleviation in the Pb Hyperaccumulator Greek Mustard (Hirschfeldia incana L.). Current Issues in Molecular Biology, 44(10), 4658-4675. https://doi.org/10.3390/cimb44100318







