Losartan Attenuates Insulin Resistance and Regulates Browning Phenomenon of White Adipose Tissue in ob/ob Mice
Abstract
1. Introduction
2. Results
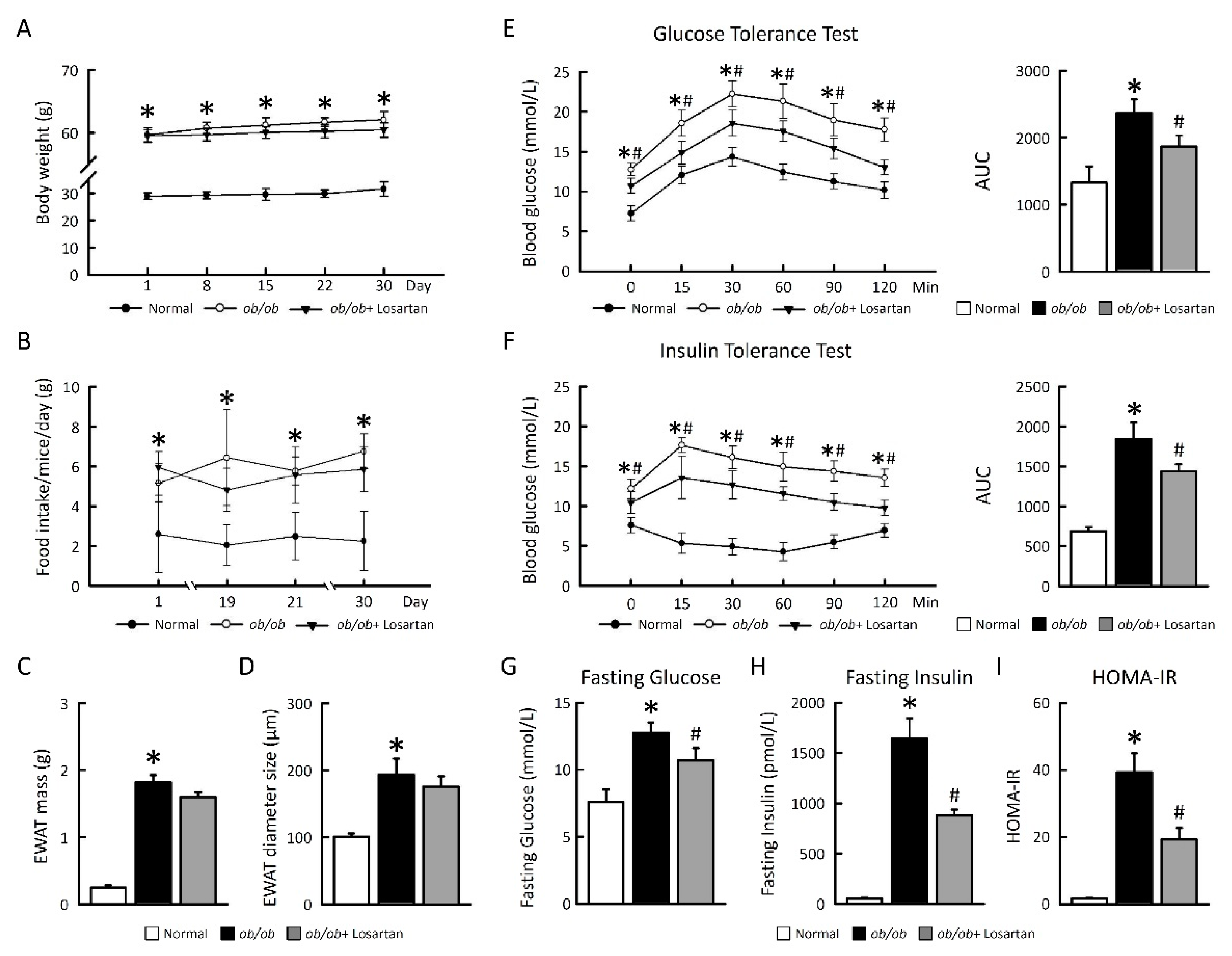
2.1. Losartan Improves Glucose Tolerance and Insulin Sensitivity in ob/ob Mice
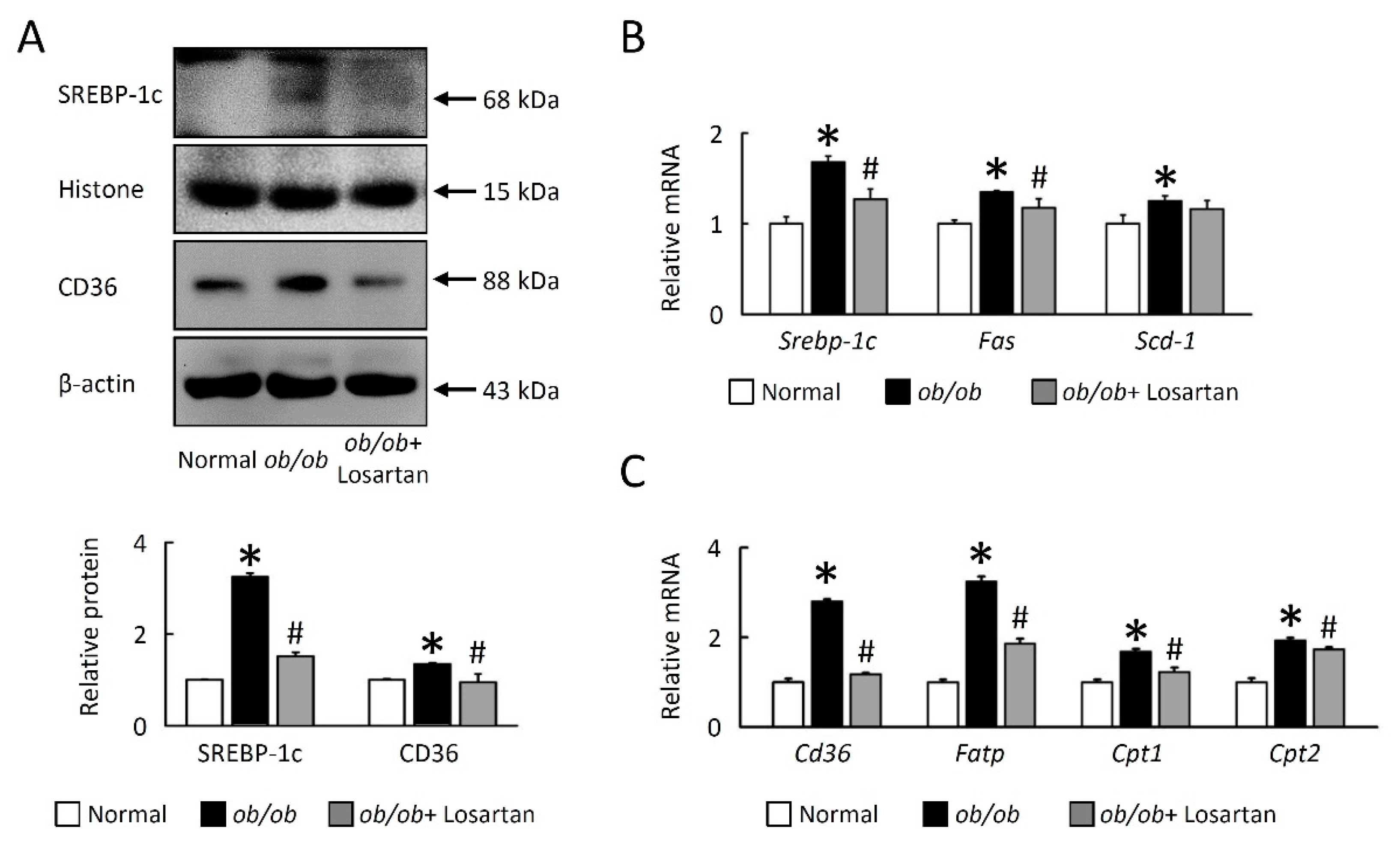
2.2. Losartan Reduces Lipogenesis in EWAT of ob/ob Mice
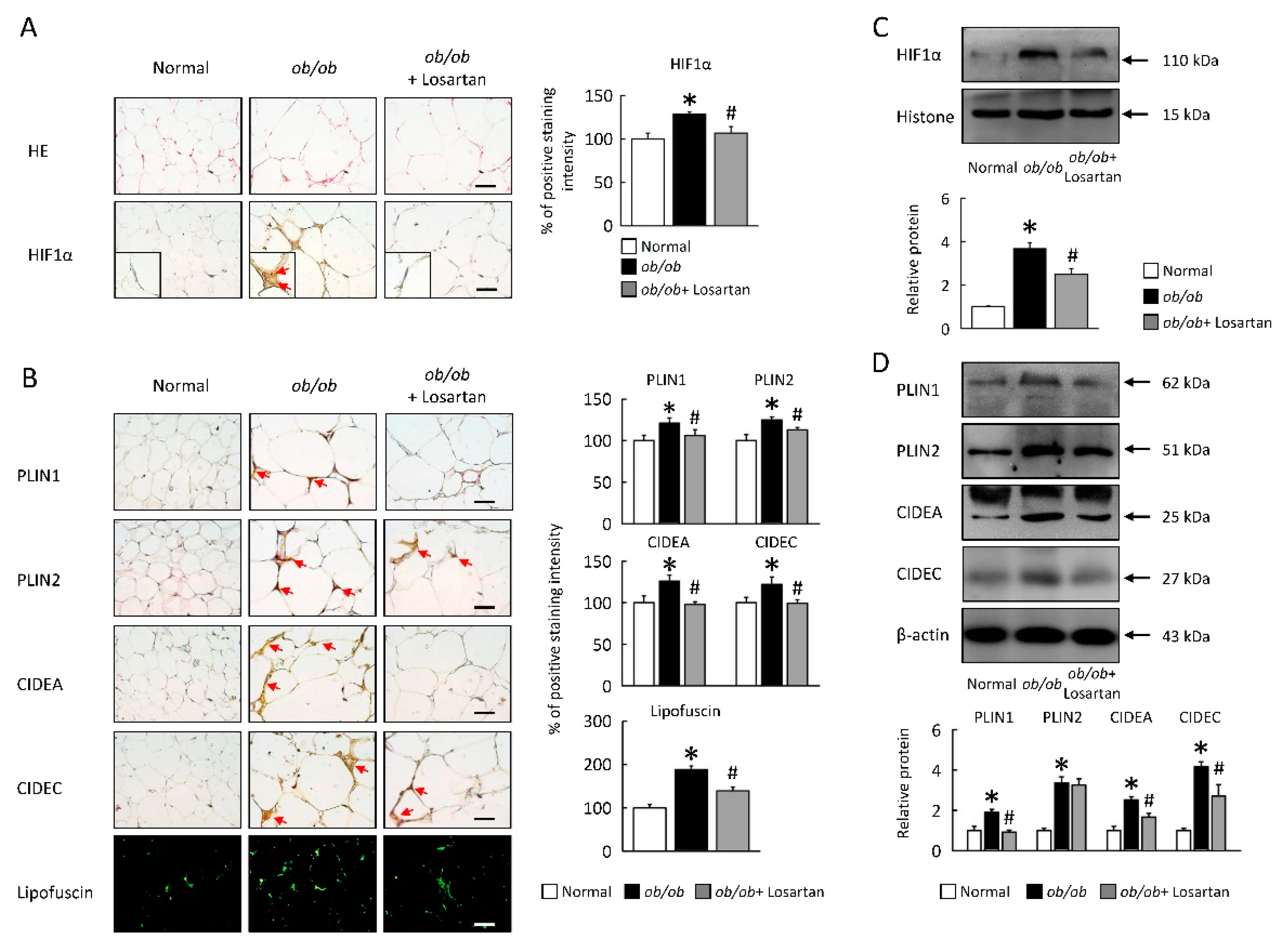
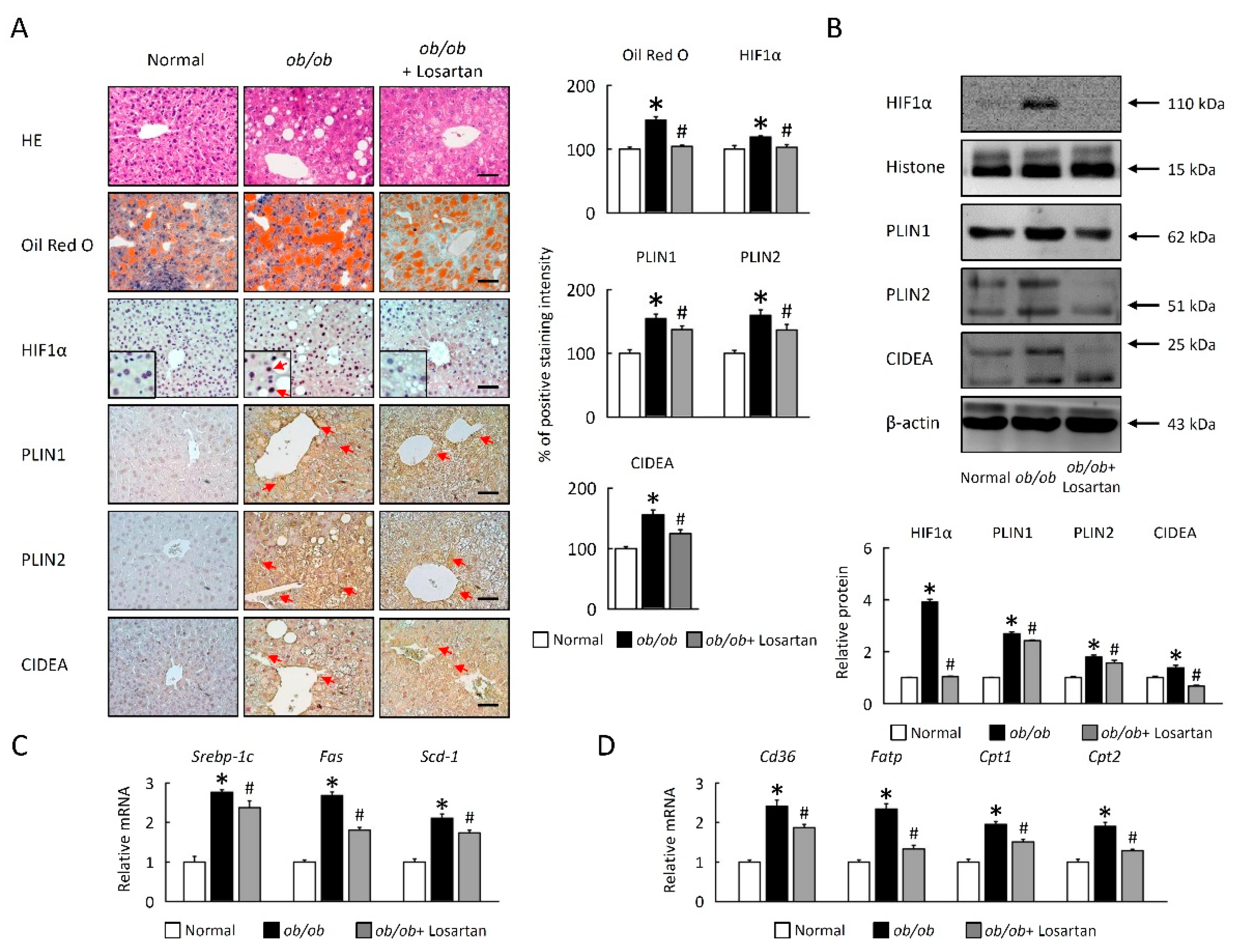
2.3. Losartan Attenuates HIF1α Expression and LDs Formation in EWAT
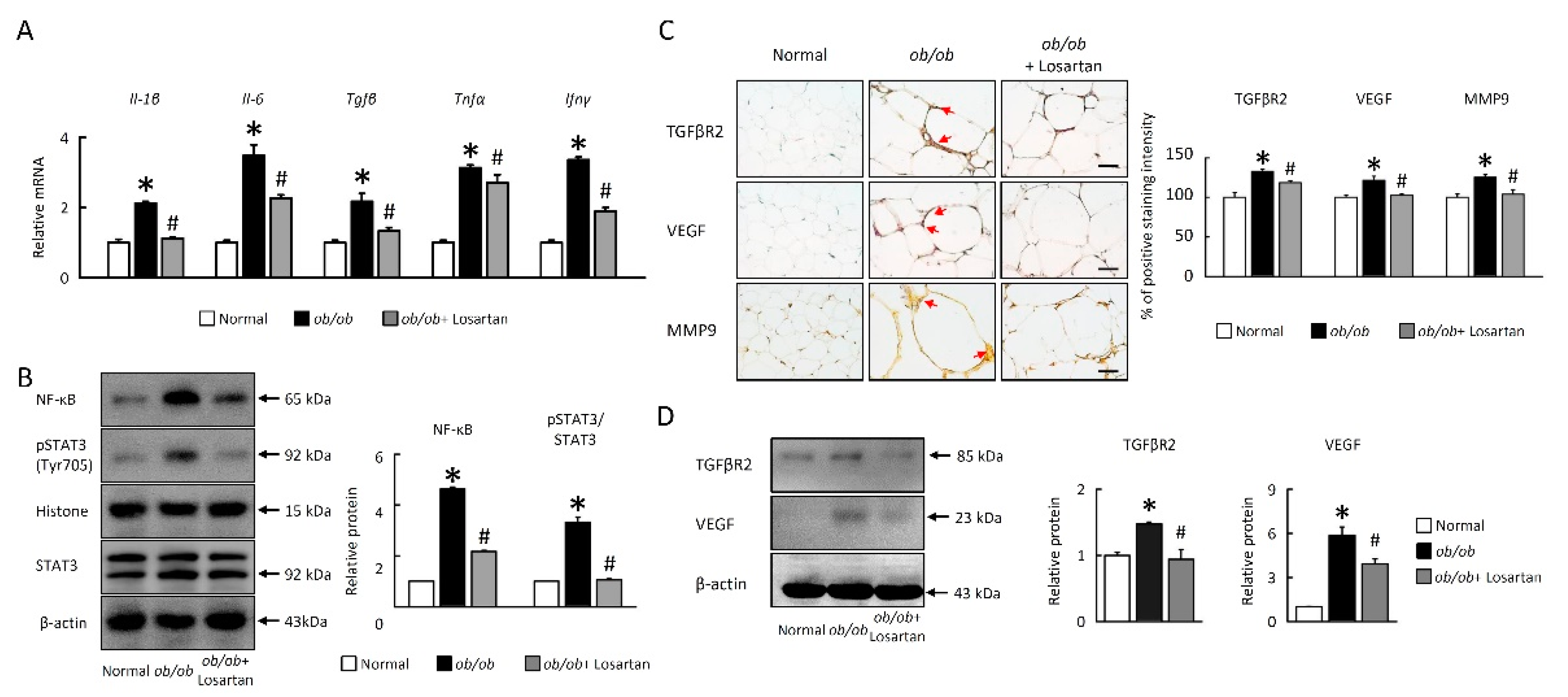
2.4. Losartan Attenuates NF-κB/HIF1α/STAT3 and Angiogenesis Pathways in EWAT
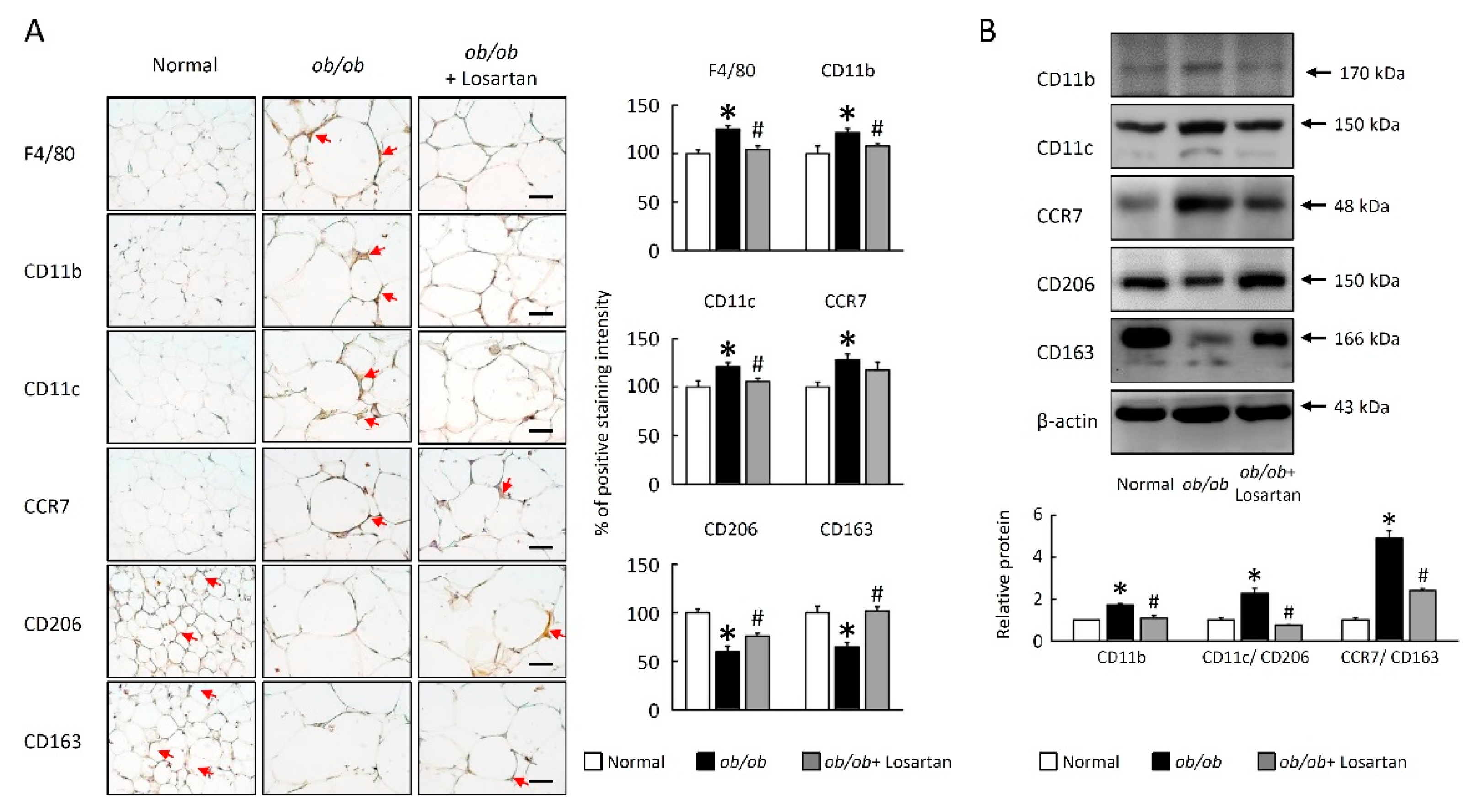
2.5. Losartan Decreased M1 Macrophage Polarization in EWAT
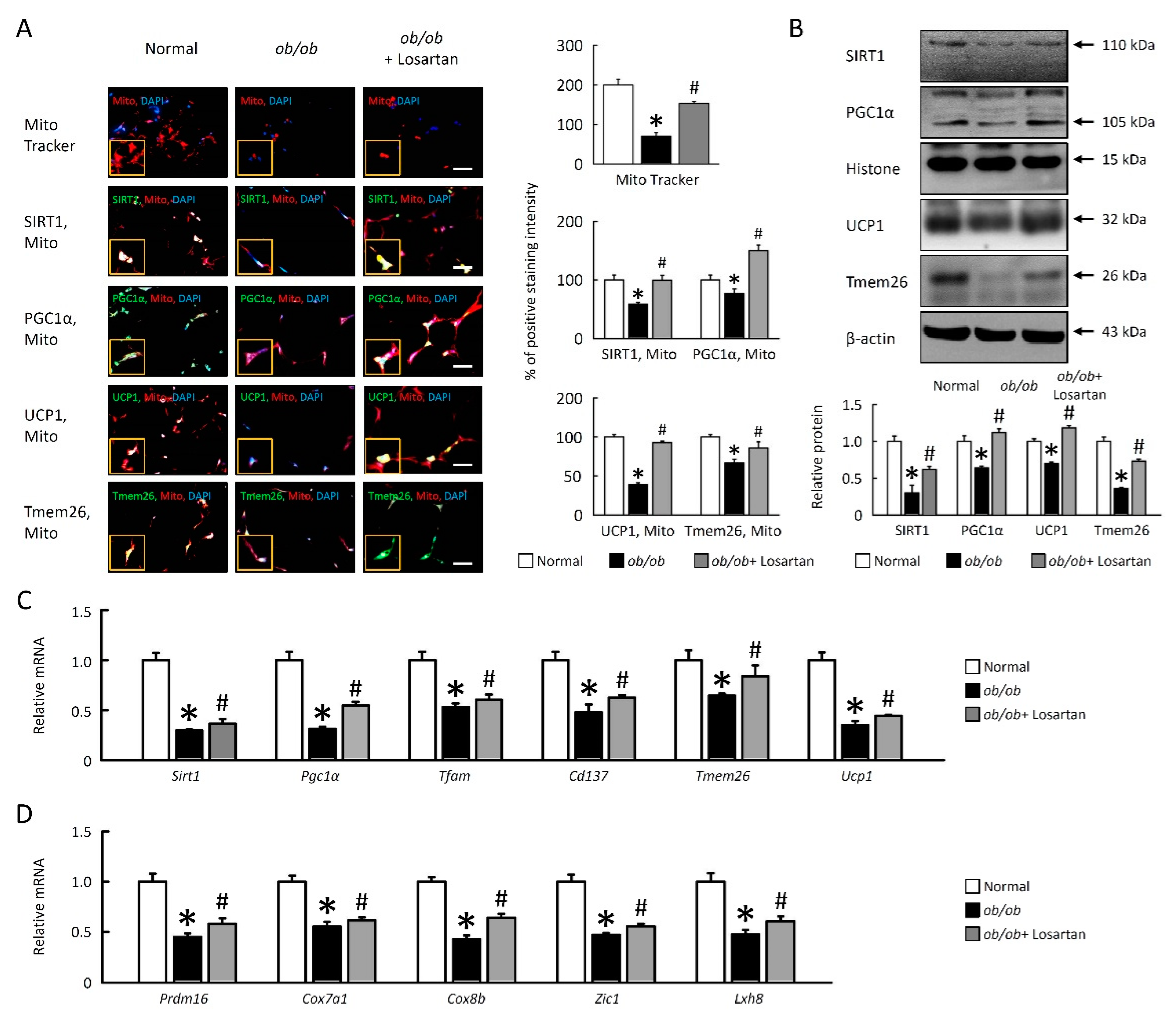
2.6. Losartan Improves Mitochondrial Dysfunction and Enhances Browning in EWAT
2.7. Losartan Suppresses LDs Accumulation and Lipogenesis in Liver
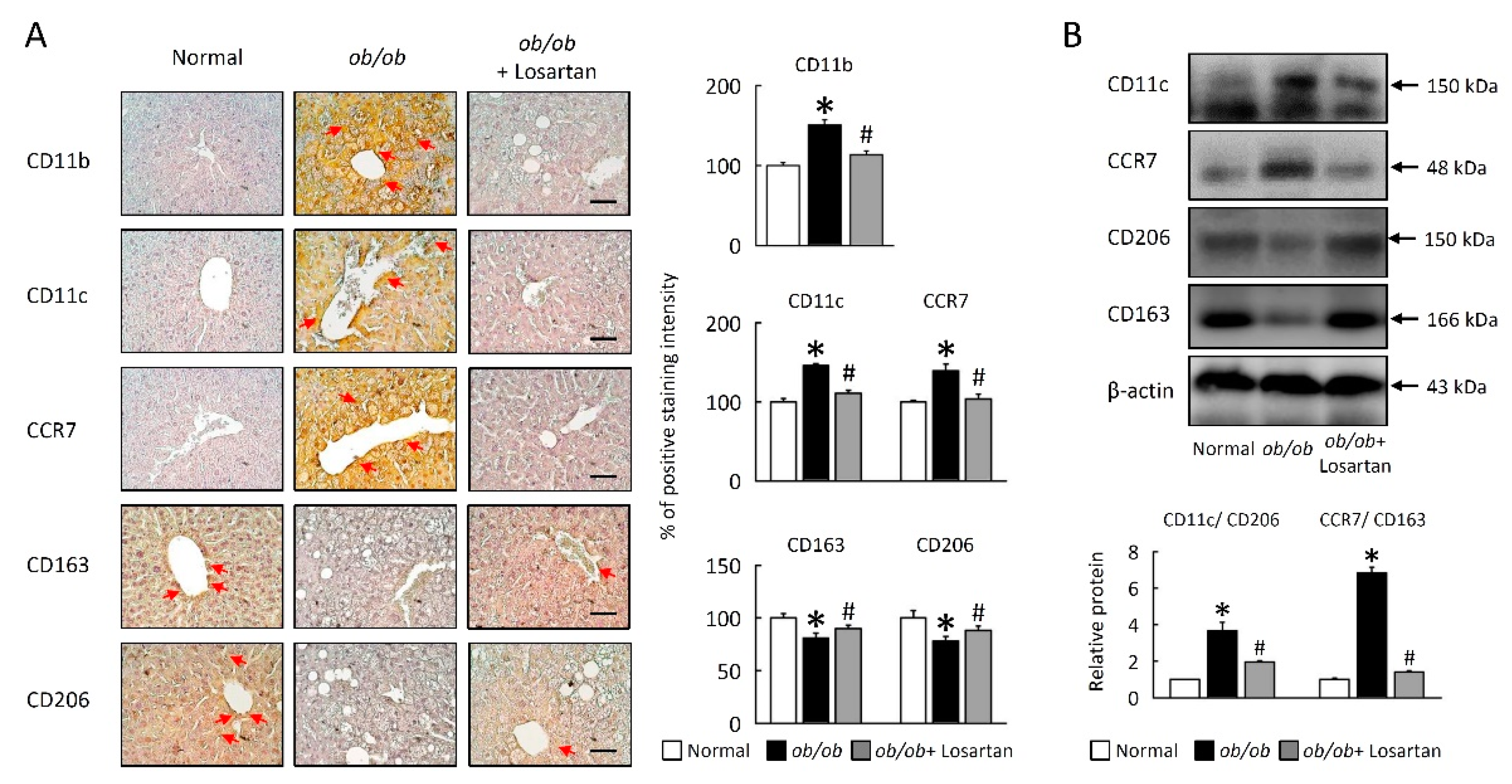
2.8. Losartan Attenuates M1/M2 Macrophage Ratio in Liver
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Glucose Tolerance Test (GTT)
4.3. Insulin Tolerance Test (ITT)
4.4. HOMA-IR
4.5. Histology, Immunohistochemistry, and Immunofluorescence
4.6. Oil Red O (ORO) Staining
4.7. Auto-Fluorescence Detection of Lipofuscin
4.8. RNA Isolation and Quantitative Real-Time PCR
4.9. Protein Isolation and Western Blot Analysis
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kahn, B.B.; Flier, J.S. Obesity and insulin resistance. J. Clin. Investig. 2000, 106, 473–481. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, S.; O’Driscoll, L. Metabolic syndrome: A closer look at the growing epidemic and its associated pathologies. Obes. Rev. 2015, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Black, P.H. The inflammatory response is an integral part of the stress response: Implications for atherosclerosis, insulin resistance, type II diabetes and metabolic syndrome X. Brain Behav. Immun. 2003, 17, 350–364. [Google Scholar] [CrossRef]
- Samuel, V.T.; Shulman, G.I. Mechanisms for insulin resistance: Common threads and missing links. Cell 2012, 148, 852–871. [Google Scholar] [CrossRef] [PubMed]
- Rohm, M.; Sommerfeld, A.; Strzoda, D.; Jones, A.; Sijmonsma, T.P.; Rudofsky, G.; Wolfrum, C.; Sticht, C.; Gretz, N.; Zeyda, M.; et al. Transcriptional cofactor TBLR1 controls lipid mobilization in white adipose tissue. Cell Metab. 2013, 17, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.S.; Coleman, R.A.; Kraemer, F.B.; McManaman, J.L.; Obin, M.S.; Puri, V.; Yan, Q.W.; Miyoshi, H.; Mashek, D.G. The role of lipid droplets in metabolic disease in rodents and humans. J. Clin. Investig. 2011, 121, 2102–2110. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, S.; Buzkova, J.; Muniandy, M.; Kaksonen, R.; Ollikainen, M.; Ismail, K.; Hakkarainen, A.; Lundbom, J.; Lundbom, N.; Vuolteenaho, K.; et al. Impaired Mitochondrial Biogenesis in Adipose Tissue in Acquired Obesity. Diabetes 2015, 64, 3135–3145. [Google Scholar] [CrossRef] [PubMed]
- Itabe, H.; Yamaguchi, T.; Nimura, S.; Sasabe, N. Perilipins: A diversity of intracellular lipid droplet proteins. Lipids Health Dis. 2017, 16, 83. [Google Scholar] [CrossRef] [PubMed]
- Carr, R.M.; Ahima, R.S. Pathophysiology of lipid droplet proteins in liver diseases. Exp. Cell Res. 2016, 340, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S.; Shargill, N.S.; Spiegelman, B.M. Adipose expression of tumor necrosis factor-alpha: Direct role in obesity-linked insulin resistance. Science 1993, 259, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Poulain, L.; Mathieu, H.; Thomas, A.; Borel, A.L.; Remy, C.; Levy, P.; Arnaud, C.; Dematteis, M. Intermittent hypoxia-induced insulin resistance is associated with alterations in white fat distribution. Sci. Rep. 2017, 7, 11180. [Google Scholar] [CrossRef] [PubMed]
- Engin, A. Adipose Tissue Hypoxia in Obesity and Its Impact on Preadipocytes and Macrophages: Hypoxia Hypothesis. Adv. Exp. Med. Biol. 2017, 960, 305–326. [Google Scholar] [PubMed]
- Ye, J.; Gao, Z.; Yin, J.; He, Q. Hypoxia is a potential risk factor for chronic inflammation and adiponectin reduction in adipose tissue of ob/ob and dietary obese mice. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E1118–E1128. [Google Scholar] [CrossRef]
- He, Q.; Gao, Z.; Yin, J.; Zhang, J.; Yun, Z.; Ye, J. Regulation of HIF-1α activity in adipose tissue by obesity-associated factors: Adipogenesis, insulin, and hypoxia. Am. J. Physiol. Endocrinol. Metab. 2011, 300, E877–E885. [Google Scholar] [CrossRef]
- Wentworth, J.M.; Naselli, G.; Brown, W.A.; Doyle, L.; Phipson, B.; Smyth, G.K.; Wabitsch, M.; O’Brien, P.E.; Harrison, L.C. Pro-inflammatory CD11c+CD206+ adipose tissue macrophages are associated with insulin resistance in human obesity. Diabetes 2010, 59, 1648–1656. [Google Scholar] [CrossRef] [PubMed]
- Heilbronn, L.K.; Campbell, L.V. Adipose tissue macrophages, low grade inflammation and insulin resistance in human obesity. Curr. Pharm. Des. 2008, 14, 1225–1230. [Google Scholar] [CrossRef]
- Warfel, J.D.; Bermudez, E.M.; Mendoza, T.M.; Ghosh, S.; Zhang, J.; Elks, C.M.; Mynatt, R.; Vandanmagsar, B. Mitochondrial fat oxidation is essential for lipid-induced inflammation in skeletal muscle in mice. Sci. Rep. 2016, 6, 37941. [Google Scholar] [CrossRef]
- Aon, M.A.; Bhatt, N.; Cortassa, S.C. Mitochondrial and cellular mechanisms for managing lipid excess. Front. Physiol. 2014, 5, 282. [Google Scholar] [CrossRef] [PubMed]
- Puthanveetil, P. FoxO1-miRNA interacting networks as potential targets for mitochondrial diseases. Drug Discov. Today 2019, 24, 342–349. [Google Scholar] [CrossRef]
- Barbagallo, I.; Vanella, L.; Cambria, M.T.; Tibullo, D.; Godos, J.; Guarnaccia, L.; Zappalà, A.; Galvano, F.; Li Volti, G. Silibinin regulates lipid metabolism and differentiation in functional human adipocytes. Front. Pharmacol. 2015, 6, 309. [Google Scholar] [CrossRef]
- Jun, J.C.; Devera, R.; Unnikrishnan, D.; Shin, M.K.; Bevans-Fonti, S.; Yao, Q.; Rathore, A.; Younas, H.; Halberg, N.; Scherer, P.E.; et al. Adipose HIF-1α causes obesity by suppressing brown adipose tissue thermogenesis. J. Mol. Med. 2017, 95, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Bharati, S.M.; Singh, N. Effect of losartan and atenolol on insulin sensitivity in nondiabetic hypertensive patients. J. Pharmacol. Pharmacother. 2016, 7, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.H.; Liu, H.M.; Chang, Z.Y.; Huang, T.H.; Lee, T.Y. Losartan Prevents Hepatic Steatosis and Macrophage Polarization by Inhibiting HIF-1α in a Murine Model of NAFLD. Int. J. Mol. Sci. 2021, 22, 7841. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, Q.; Yang, J.; Ma, Y.; Ding, G. Angiotensin II induces cholesterol accumulation and injury in podocytes. Sci. Rep. 2017, 7, 10672. [Google Scholar] [CrossRef]
- Sagae, S.C.; Lubaczeuski, C.; Zacharias, P.; Bonfleur, M.L.; Franci, C.R.; Sanvitto, G.L. Prevention of metabolic disorders and reproductive performance deficits by the blockade of Angiotensin II AT1 receptor in female rats fed with cafeteria diet. Physiol. Behav. 2013, 119, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shum, M.; Pinard, S.; Guimond, M.O.; Labbé, S.M.; Roberge, C.; Baillargeon, J.P.; Langlois, M.F.; Alterman, M.; Wallinder, C.; Hallberg, A.; et al. Angiotensin II type 2 receptor promotes adipocyte differentiation and restores adipocyte size in high-fat/high-fructose diet-induced insulin resistance in rats. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E197–E210. [Google Scholar] [CrossRef] [PubMed]
- Graus-Nunes, F.; Rachid, T.L.; de Oliveira Santos, F.; Barbosa-da-Silva, S.; Souza-Mello, V. AT1 receptor antagonist induces thermogenic beige adipocytes in the inguinal white adipose tissue of obese mice. Endocrine 2017, 55, 786–798. [Google Scholar] [CrossRef] [PubMed]
- Aksnes, T.A.; Seljeflot, I.; Torjesen, P.A.; Höieggen, A.; Moan, A.; Kjeldsen, S.E. Improved insulin sensitivity by the angiotensin II-receptor blocker losartan is not explained by adipokines, inflammatory markers, or whole blood viscosity. Metabolism 2007, 56, 1470–1477. [Google Scholar] [CrossRef]
- Nishimura, H.; Sanaka, T.; Tanihata, Y.; Naito, T.; Higuchi, C.; Otsuka, K. Losartan elevates the serum high-molecular weight-adiponectin isoform and concurrently improves insulin sensitivity in patients with impaired glucose metabolism. Hypertens Res. 2008, 31, 1611–1618. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ruan, H.; Lodish, H.F. Insulin resistance in adipose tissue: Direct and indirect effects of tumor necrosis factor-alpha. Cytokine Growth Factor Rev. 2003, 14, 447–455. [Google Scholar] [CrossRef]
- Miyoshi, H.; Souza, S.C.; Endo, M.; Sawada, T.; Perfield, J.W., 2nd; Shimizu, C.; Stancheva, Z.; Nagai, S.; Strissel, K.J.; Yoshioka, N.; et al. Perilipin overexpression in mice protects against diet-induced obesity. J. Lipid Res. 2010, 51, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Kern, P.A.; Di Gregorio, G.; Lu, T.; Rassouli, N.; Ranganathan, G. Perilipin expression in human adipose tissue is elevated with obesity. J. Clin. Endocrinol. Metab. 2004, 89, 1352–1358. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sullivan, S.; Trujillo, M.; Lee, M.J.; Schneider, S.H.; Brolin, R.E.; Kang, Y.H.; Werber, Y.; Greenberg, A.S.; Fried, S.K. Perilipin expression in human adipose tissues: Effects of severe obesity, gender, and depot. Obes. Res. 2003, 11, 930–936. [Google Scholar] [CrossRef]
- Tateya, S.; Kim, F.; Tamori, Y. Recent advances in obesity-induced inflammation and insulin resistance. Front. Endocrinol. (Lausanne) 2013, 4, 93. [Google Scholar] [CrossRef] [PubMed]
- Suganami, T.; Tanaka, M.; Ogawa, Y. Adipose tissue inflammation and ectopic lipid accumulation. Endocr. J. 2012, 59, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Liu, J.; Shi, L.; Tang, Y.; Gao, D.; Long, J.; Liu, J. Mitochondrial dysfunction precedes depression of AMPK/AKT signaling in insulin resistance induced by high glucose in primary cortical neurons. J. Neurochem. 2016, 137, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Gao, H.; Hasegawa, Y.; Lu, X. Fight against fibrosis in adipose tissue remodeling. Am. J. Physiol. Endocrinol. Metab. 2021, 321, E169–E175. [Google Scholar] [CrossRef] [PubMed]
- Acín-Pérez, R.; Iborra, S.; Martí-Mateos, Y.; Cook, E.C.L.; Conde-Garrosa, R.; Petcherski, A.; Muñoz, M.D.M.; Martínez de Mena, R.; Krishnan, K.C.; Jiménez, C.; et al. Fgr kinase is required for proinflammatory macrophage activation during diet-induced obesity. Nat. Metab. 2020, 2, 974–988. [Google Scholar] [CrossRef]
- Jornayvaz, F.R.; Shulman, G.I. Regulation of mitochondrial biogenesis. Essays Biochem. 2010, 47, 69–84. [Google Scholar]
- Garcia, D.; Shaw, R.J. AMPK: Mechanisms of Cellular Energy Sensing and Restoration of Metabolic Balance. Mol. Cell 2017, 66, 789–800. [Google Scholar] [CrossRef]
- Qiang, L.; Wang, L.; Kon, N.; Zhao, W.; Lee, S.; Zhang, Y.; Rosenbaum, M.; Zhao, Y.; Gu, W.; Farmer, S.R.; et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell 2012, 150, 620–632. [Google Scholar] [CrossRef]
- Tang, B.L. Sirt1 and the Mitochondria. Mol. Cells. 2016, 39, 87–95. [Google Scholar] [PubMed]
- Roza, N.A.; Possignolo, L.F.; Palanch, A.C.; Gontijo, J.A. Effect of long-term high-fat diet intake on peripheral insulin sensibility, blood pressure, and renal function in female rats. Food Nutr. Res. 2016, 60, 28536. [Google Scholar] [CrossRef] [PubMed]
- Romijn, H.J.; van Uum, J.F.; Breedijk, I.; Emmering, J.; Radu, I.; Pool, C.W. Double immunolabeling of neuropeptides in the human hypothalamus as analyzed by confocal laser scanning fluorescence microscopy. J. Histochem. Cytochem. 1999, 47, 229–236. [Google Scholar] [CrossRef] [PubMed]
| Gene | Forward | Reverse |
|---|---|---|
| Srebp-1c | 5′ actgtcttggttgttgatgagctggagcat 3′ | 5′ atcggcgcggaagctgtcggggtagcgtc 3′ |
| Fas | 5′ tgtcattggcctcctcaaaaagggcgtcca 3′ | 5′ tcaccactgtgggctctgcagagaagcgag 3′ |
| Scd-1 | 5′ ccggagaccccttagatcga 3′ | 5′ tagcctgtaaaagatttctgcaaacc 3′ |
| Fatp | 5′ gcttcaacagccgtatcctc 3’ | 5′ tcttcttgttggtggcactg 3’ |
| Cd36 | 5′ gcaaaacgactgcaggtcaac 3′ | 5′ tggtcccagtctcatttagcca 3′ |
| Cpt1 | 5′ ggacagagactgtgcgttcct 3′ | 5′ gcgatatccaacagtgcttga 3′ |
| Cpt2 | 5′ caaggccctggctgatgatgtg 3′ | 5′ agtctctgtccgcccctctcg 3′ |
| Il-1β | 5′ aacctgctggtgtgtgacgttc 3′ | 5′ cagcacgaggcttttttgttgt 3′ |
| Il-6 | 5′-gtactccagaagaccagagg-3′ | 5′-tgctggtgacaaccacggcc-3′ |
| Tgfβ | 5′ tatagcaacaattcctggcg 3′ | 5′ tgctgtcacaggagcagtg 3′ |
| Tnfα | 5′ ttgacctcagcgctgagttg 3′ | 5′ cctgtagcccacgtcgtagc 3′ |
| Ifnγ | 5′ cctcaaacttggcaatactc 3′ | 5′ agcaacaacataagcgtcat 3′ |
| Sirt1 | 5’ gcaacagcatcttgcctgat 3’ | 5′ gtgctactggtctcactt 3′ |
| Pgc1α | 5′ gactcagtgtcaccaccgaaa-3′ | 5′ tgaacgagagcgcatcctt 3′ |
| Tfam | 5′ ggaatgtggagcgtgctaaaa 3′ | 5′-tgctggaaaaacacttcggaata 3′ |
| Ucp1 | 5′ cctgcctctctcggaaacaa 3′ | 5′-tgtaggctgcccaatgaaca 3′ |
| Tmem26 | 5′ accctgtcatcccacagag 3′ | 5′ tgtttggtggagtcctaaggtc 3′ |
| Cd137 | 5′ cgtgcagaactcctgtgataac 3′ | 5′ gtccacctatgctggagaagg 3′ |
| Prdm16 | 5′ cagcacggtgaagccattc 3′ | 5′ gcgtgcatccgcttgtg 3′ |
| Cox7a1 | 5′ cagcgtcatggtcagtctgt 3′ | 5′ agaaaaccgtgtggcagaga 3′ |
| Cox8b | 5′ gaaccatgaagccaacgact 3′ | 5′ gcgaagttcacagtggttcc 3′ |
| Zic1 | 5′ ctgttgtgggagacacgatg 3′ | 5′ cctcttctcagggctcacag 3′ |
| Lxh8 | 5′ acacgagctgctacattaagga 3′ | 5′ cccagtcagtcgagtggatg 3′ |
| Gadph | 5′ tcaccaccatggagaaggc 3′ | 5′ gctaagcagttggtggtgca 3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.-M.; Wang, C.-H.; Chang, Z.-Y.; Huang, T.-H.; Lee, T.-Y. Losartan Attenuates Insulin Resistance and Regulates Browning Phenomenon of White Adipose Tissue in ob/ob Mice. Curr. Issues Mol. Biol. 2021, 43, 1828-1843. https://doi.org/10.3390/cimb43030128
Liu H-M, Wang C-H, Chang Z-Y, Huang T-H, Lee T-Y. Losartan Attenuates Insulin Resistance and Regulates Browning Phenomenon of White Adipose Tissue in ob/ob Mice. Current Issues in Molecular Biology. 2021; 43(3):1828-1843. https://doi.org/10.3390/cimb43030128
Chicago/Turabian StyleLiu, Hsuan-Miao, Cheng-Hui Wang, Zi-Yu Chang, Tse-Hung Huang, and Tzung-Yan Lee. 2021. "Losartan Attenuates Insulin Resistance and Regulates Browning Phenomenon of White Adipose Tissue in ob/ob Mice" Current Issues in Molecular Biology 43, no. 3: 1828-1843. https://doi.org/10.3390/cimb43030128
APA StyleLiu, H.-M., Wang, C.-H., Chang, Z.-Y., Huang, T.-H., & Lee, T.-Y. (2021). Losartan Attenuates Insulin Resistance and Regulates Browning Phenomenon of White Adipose Tissue in ob/ob Mice. Current Issues in Molecular Biology, 43(3), 1828-1843. https://doi.org/10.3390/cimb43030128







