1. Introduction
Cryo-electron microscopy (cryo-EM) has become a recognized powerful technique in structural biology for three-dimensional (3D) structure determination of biological macromolecules, supramolecular complexes, and subcellular structures [
1,
2,
3]. It does not need crystallization and has been widely used to study large macromolecular complexes that are difficult to be crystallized. The goal of cryo-EM 3D reconstruction is to reconstruct a high-resolution estimation of the 3D structure of the molecule from a set of micrographs [
4,
5,
6]. Cryo-EM can be used to investigate complete and fully functional macromolecular complexes in different functional states, providing a richness of biological insight [
7,
8]. Cryo-EM has made tremendous progress in the past few years [
9,
10]. Owing to these exciting new developments, cryo-EM was selected by Nature Methods as the “Method of the Year 2015”, and the Nobel Prize in Chemistry 2017 was awarded to Jacques Dubochet, Joachim Frank, and Richard Henderson “for developing cryo-electron microscopy for the high-resolution structure determination of biomolecules in solution” [
5].
As one of the major cryo-EM techniques, single-particle reconstruction has become one of the most successful techniques for structural biology [
11,
12,
13]. Single-particle reconstruction using cryo-EM has been undergoing fast transformations, leading to an abundance of new high-resolution structures and reaching close to atomic resolution [
14,
15]. In the single-particle reconstruction, the same macromolecule is projected from various unknown directions, and the final 3D structure of the macromolecule can be reconstructed from the two-dimensional (2D) projection images using the estimated projection directions in 3D space [
16,
17]. One of the major challenges to be overcome in the single-particle reconstruction of biological samples is to estimate the projection directions of the projection images [
18,
19]. However, due to the very low signal-to-noise ratio (SNR) of the projection images caused by low-dose electron radiation, it is usually difficult to obtain the correct estimation of the projection directions. Consequently, the single-particle 3D reconstruction of cryo-EM is a very challenging task [
20,
21].
Class averaging in single-particle cryo-EM is an important procedure for producing high-quality initial 3D structures and discarding invalid particles or contaminants [
22]. It organizes a dataset by grouping together the particles corresponding to the same (or quite similar) projection directions. Each group of cryo-EM projection images is regarded as a class and is averaged to produce an averaged image called a class average. By averaging, the random noise in the background tends to be canceled, and the features of interest in the projection images are reinforced by each other as the number of superimposed projection images becomes large [
23,
24]. Class averages can be used to improve ab initio modeling in cryo-EM. They can also be applied for discovering heterogeneity or symmetricity as well as for separating particles into subgroups for additional analysis [
25].
Different solutions have been proposed for solving the 2D class averaging problem in cryo-EM [
26,
27,
28,
29,
30,
31]. Some popular cryo-EM software packages, such as cryoSPARC [
32] and RELION [
33,
34,
35] have implemented 2D class averaging. RELION uses a maximum likelihood expectation maximization (ML-EM) 2D classification procedure to infer parameters for a statistical model from the data. The ML-EM scheme has suffered less from initial reference bias, but it is computationally expensive. The iterative stable alignment and clustering (ISAC) algorithm [
36] is another famous 2D class averaging method. ISAC relies on a modified k-means clustering method and the concepts of stability and reproducibility, which can extract validated, homogeneous subsets of projection images. ISAC is also time consuming.
Image alignment is a fundamental step in the class averaging procedure [
37,
38]. The cryo-EM projection images are required to be identified and rotationally and translationally aligned to distinguish among different classes. After alignment, the cryo-EM projection images with nearly the same projection directions are grouped in the 2D classification step. Well-aligned cryo-EM projection images with correct in-plane rotations and translational shifts in the x-axis and y-axis directions can improve the accuracy of the 2D classification [
39]. Correctly classifying the cryo-EM projection images into homogeneous groups renders the satisfactory determination of the preliminary 3D structures [
40]. Although translational invariant and rotational invariant image representation methods have been used in cryo-EM, they usually are not powerful enough to discover subtle differences between projection images [
41]. It is necessary to design efficient image alignment algorithms to find the best alignment parameters and generate high-quality class averages.
Image alignment is aimed at estimating three alignment parameters: a rotation angle and two translational shifts in the x-axis and y-axis directions. Image rotational alignment and translational alignment in real space need too many iterations to compute the alignment parameters, and the calculated alignment parameters are integers. In Fourier space, alignment parameters can be computed directly without enumeration. In this paper, an efficient image alignment algorithm using the 2D interpolation in the frequency domain of images is proposed to improve the estimation accuracy of alignment parameters, which can obtain subpixel and subangle accuracy. Specifically: (1) for image rotational alignment, two images are transformed by polar fast Fourier transform (PFFT) to calculate a discrete cross-correlation matrix, and then the 2D interpolation is performed around the maximum value in the cross-correlation matrix. The rotation angle between the two images is directly determined according to the position of the maximum value in the cross-correlation matrix after interpolation. (2) For image translational alignment, all operation steps are consistent with image rotational alignment, where fast Fourier transform (FFT) is used instead of PFFT. (3) For image alignment with rotation and translation, only a few iterations of combined rotational and translational alignment are needed to align images. Furthermore, the proposed algorithm and a spectral clustering algorithm [
42] are used to compute class averages for single-particle 3D reconstruction. The main contributions of this paper are summarized as follows:
2D interpolation in the frequency domain is used to improve the estimation accuracy of the alignment parameters, which can obtain subpixel and subangle accuracy.
The alignment parameters of rotation angles and translational shifts in the x-axis and y-axis directions can be computed directly in Fourier space without enumeration, which is very fast.
A spectral clustering algorithm is used for the unsupervised 2D classification of single-particle cryo-EM projection images.
The rest of this paper is organized as follows: In
Section 2, the proposed image alignment algorithm is described in detail, including the image rotational alignment, the image translational alignment, and image alignment with rotation and translation. The unsupervised 2D classification of cryo-EM projection images performed by using a spectral clustering algorithm is also introduced. In
Section 3, the flexibility and performance of the proposed image alignment algorithm are demonstrated through three datasets, including a Lena image, a simulated dataset of cryo-EM projection images, and a real dataset of cryo-EM projection images. The single-particle 3D reconstruction using produced class averages is also performed and compared with RELION. Finally, this paper is concluded in
Section 4.
2. Materials and Methods
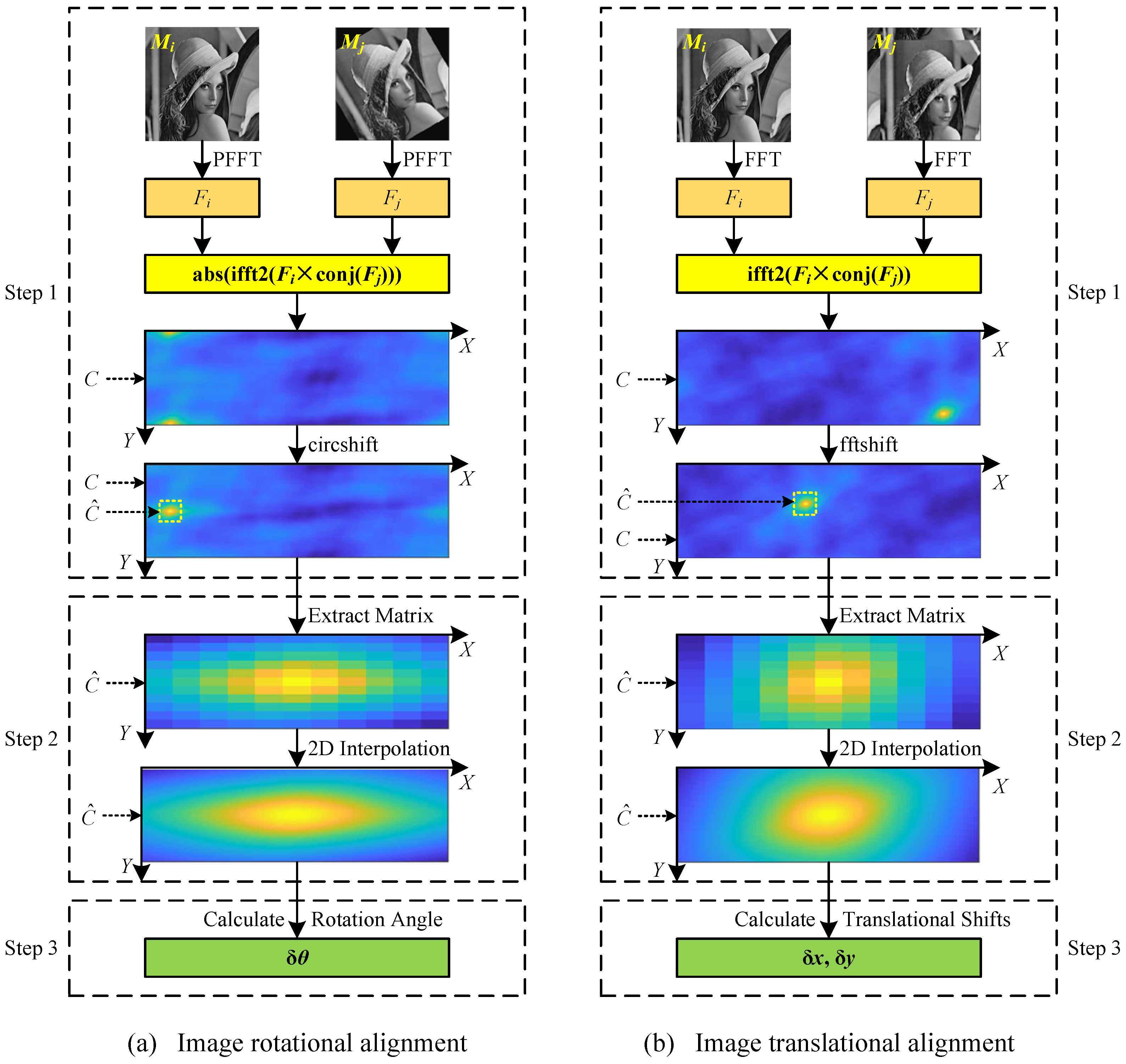
In this section, the proposed image alignment algorithm is demonstrated in detail, including (1) image rotational alignment; (2) image translational alignment; and (3) image alignment with rotation and translation. The diagrams of the proposed image rotational and translational alignment algorithms using 2D interpolation in the frequency domain of images are shown in
Figure 1. Then the proposed algorithm and a spectral clustering algorithm are used to compute class averages.
2.1. Image Rotational Alignment
Image rotational alignment is one of the basic operations in image processing. The rotation angle between two images can be estimated either in real space or in Fourier space. In real space, image rotational alignment is a rotation-matching process, that is, an exhaustive search. An image is rotated in a certain step size, and the similarity between the rotated image and the reference image is calculated. When the image is rotated for one circle, the index corresponding to the maximum similarity is the final estimated rotation angle between the two images. This method is simple, but it is time consuming and inaccurate. Assuming the search step size is p, image rotational alignment in real space requires rotation-matching calculations. Although the coarse-to-fine search method can be used, it still needs to be calculated many times.
In this paper, the image rotational alignment is implemented in Fourier space without rotation-matching iteration, which is a direct calculation method. In general, the cryo-EM projection images are square; therefore, only the rotational alignment of the square image is considered. For two images
and
of size
, the proposed image rotational alignment method is illustrated in
Figure 1a. In the rest of this paper, the proposed image rotational alignment algorithm is represented as function
. There are three key steps in the image rotational alignment algorithm:
Step 1: Calculate a cross-correlation matrix using PFFT. Firstly, images
and
are transformed by PFFT to obtain two corresponding spectrum maps
and
with the size of
. Then, the cross-correlation matrix
C is calculated according to:
where
is an absolute value function,
is a 2D inverse fast Fourier transform function, and
is a complex conjugate function. These functions have been implemented in MATLAB. The values in matrix
C need to be circularly shifted by
positions to exchange rows to horizontally center the large values in matrix
C, where the function
implemented in MATLAB can be used. The size of the cross-correlation matrix
C is
.
Step 2: 2D interpolation around the maximum value in the cross-correlation matrix
C. The rotation angle
of the image
relative to the image
can be roughly determined according to the position of the maximum value in the cross-correlation matrix
C on the x-axis. The rotation angle calculated by this method is an integer. In order to calculate the rotation angle more accurately, the 2D interpolation is performed around the maximum value in the cross-correlation matrix
C. Specifically, an 11 * 11 matrix
centered on the maximum value in the matrix
C is extracted from the matrix
C (see the dotted box in
Figure 1a), and then the 2D interpolation is performed in the matrix
. Theoretically, any interpolation method can be used in the proposed algorithm. In this paper, the spline interpolation is used to perform the 2D interpolation, which has been implemented in MATLAB as function
with parameter ‘spline’. After 2D interpolation, the size of the matrix
becomes
.
Step 3: Calculate the rotation angle. The rotation angle
can be directly calculated according to the position of the maximum value in the matrix
after interpolation on the x-axis. Generally, the rotation angle
of an image is in the range of
, so
needs to be corrected according to:
2.2. Image Translational Alignment
Image translational alignment can also be realized in real space or Fourier space. In real space, image translational alignment is also an exhaustive search, and it is more complex than image rotational alignment. For two images and of size , it needs to compute the similarity between each row (column) of and each row (column) of and then determines the translational shift in the x-axis direction and the translational shift in the y-axis direction according to the maximum similarity. Therefore, the image translational alignment in real space requires similarity calculations. In addition, the translational shifts estimated in real space are integers, which are not accurate enough.
Similar to image rotational alignment, in this paper, the image translational alignment is implemented in Fourier space. It is a direct calculation method without enumeration. For two images
and
of size
, the proposed image translational alignment method is illustrated in
Figure 1b. In the rest of this paper, the proposed image translational alignment algorithm is represented as function
. There are three key steps in the image translational alignment algorithm:
2.3. Image Alignment with Rotation and Translation
Image alignment with rotation and translation is a fundamental but challenging step in class averaging. It is the coupling of image rotational alignment and image translational alignment and generally requires iterations. In this paper, an efficient image alignment algorithm using the 2D interpolation in the frequency domain of images is proposed, which is listed in Algorithm 1. The functions
and
represent the image rotational alignment algorithm described in
Section 2.1 and the image translational alignment algorithm described in
Section 2.2, respectively. The functions
and
represent the image rotation operation and image translation operation, respectively. For each iteration, the test image
M is first rotationally aligned and then translationally aligned to calculate the three alignment parameters. The final alignment parameters
,
,
, and the original test image
M are used to calculate the final aligned image
, reducing the error accumulation caused by interpolation calculation in image rotation and translation. When the alignment parameters
,
, and
during the iteration remain unchanged, the iteration can be terminated ahead of time. According to our experience, the program converges within ten iterations in the majority of cases. In addition, there is no complicated operation in the algorithm. It can efficiently and effectively align two images. Therefore, the proposed algorithm can be used to align a large number of cryo-EM projection images.
| Algorithm 1: Image alignment algorithm using 2D interpolation in the frequency domain. |
![Cimb 43 00117 i001 Cimb 43 00117 i001]() |
2.4. Class Averaging
In this paper, the proposed image alignment algorithm and a spectral clustering algorithm [
42] are used to implement class averaging. The calculation process of the class averaging is shown in
Figure 2. First of all, all the cryo-EM projection images are aligned by the proposed image alignment algorithm to calculate the similarity matrix
S between them. As in many studies [
5,
25], the 2D correlation coefficient is used to compute the similarity:
where
and
are the pixel values of images
and
, respectively.
and
are the mean values of the pixel values of images
and
, respectively.
m is the size of the projection image in one dimension.
Afterward, the similarity matrix
S is converted into an adjacency matrix
using a k-nearest neighbor (kNN) algorithm [
43] and a shared nearest neighbor (SNN) algorithm [
44]. Specifically, the matrix
, which is used to represent the number of shared near neighbors between projection images
and
, is calculated as follows:
where
and
are the sets of
k nearest neighbors of projection images
and
, respectively, which can be found according to the similarity matrix
S. The matrix
is converted into an adjacency matrix
by binarization:
where
is the threshold parameter used to represent at least
shared nearest neighbors between projection images
and
. The empirical value of parameter
k in the kNN algorithm can be calculated adaptively according to the total number of projection images
N:
Finally, the adjacency matrix
is used as the input of the normalized spectral clustering algorithm [
45] to perform unsupervised classification. Projection images grouped in a class are aligned and weighted averaged to produce a class average. Assuming that the
jth class contains
projection images, the class average
can be calculated as:
where
is the similarity between the projection image
that is closest to the cluster center of the
jth class and the projection image
that is aligned with
in the
jth class.