Abstract
The leucine-rich repeat receptor-like protein kinase (LRR-RLK) gene family is the largest family of the receptor-like protein kinases (RLKs) superfamily in higher plants, which is involved in regulating the plant growth and development, stress responses, signal transduction and so on. However, no comprehensive analyses of LRR-RLKs have been reported in sugarcane. Here, we performed a comprehensive analysis of the LRR-RLK gene family in sugarcane ancestor species Saccharum spontaneum. A total of 437 LRR-RLK genes were identified and categorized into 14 groups based on a maximum likelihood phylogenetic tree. The chromosome location showed an uneven distribution on all 32 chromosomes in sugarcane. Subsequently, the exon–intron organization structure and conserved motif arrangement were relatively conserved among the same groups or subgroups and between Arabidopsis and S. spontaneum genomes. Furthermore, the promoter sequences analyses showed that sugarcane LRR-RLK genes (SsLRR-RLKs) were strongly regulated by various environmental stimuli, phytohormonal factors and transcription factors (TFs). Eventually, the expression profiles of SsLRR-RLK genes at different stresses were analyzed based on RNA-seq data, suggesting their potential roles in the regulation of sugarcane responses to diverse abiotic and biotic stress. Overall, the findings provide insight into the potential functional roles and lay the foundation for further functional study.
1. Introduction
During plant growth and development, plant cells are capable of sensing the stimulation of biotic and abiotic factors through extracellular cell surface receptors. These receptors mediate cell-to-cell and cell-to-environment interactions by interacting with polysaccharides, proteins, lipids and other ligands, which trigger a series of signal transductions [1,2,3]. Receptor-like protein kinases (RLKs) are a class of protein kinases that are widespread in plants, which are the key components in a variety of signal recognition pathways [1]. In 1990, the first RLK gene of higher plant species was identified in maize (Zea mays), and, since then, more RLK genes have been isolated from other plant species [4,5]. According to the classification based on the N-terminal extracellular domains (ECD) amino acid sequences, the RLKs superfamily can be further classified into 17 subfamilies, including leucine-rich repeat receptor-like protein kinases (LRR-RLKs), pathogenesis-related protein 5-like receptor kinases (PR5K-RLKs), S-domain RLKs (S-RLKs) and epidermal growth factor-like RLKs (EGF-RLKs) [4,5,6]. Among all of these subfamilies, the major and largest group of plant RLKs is the LRR-RLKs subfamily. The structural features of the LRR-RLK gene family include an intracellular protein kinase catalytic domain (KD), a single-pass transmembrane domain (TM) and a typical leucine-rich repeat (LRR) domain [6,7,8]. In terms of their structural features, previous evidence shows that plant LRR-RLKs play an essential role in various important processes [9,10,11,12,13,14].
In plants, previous studies have testified that LRR-RLKs genes play an essential role in diverse processes during both the plant growth cycle and responses to multiple stresses [10,11,12,13]. In the previous work, the best characterized LRR-RLKs are CLAVATA1 (CLV1) and BRASSINOSTEROID INSENSITIVE1 (BRI1) in Arabidopsis thaliana [15,16,17]. BRs are a class of plant growth regulator substances in plants that promote plant growth, plant meristems, flowering time and vascular differentiation [18,19]. In addition, many other aspects are regulated by LRR-RLKs, such as extra sporogenous cells (EXS) promoting the germination of plant seeds [20], HAESA (HAE) promoting plant floral organ abscission [21], CRINKLY4 (cr4) participating in epidermal cell differentiation [22], PXY (phloem intercalated with xylem) regulating the activity of procambium in plant vascular tissues [23], Xa21 being an effective bacterial blight resistance gene [24] and receptor-like kinase1 (RPK1) regulating ABA-induced sensitivity but being unchanged in jasmonic acid- and ethylene-induced sensitivity [25,26]. In addition, ERECTA has been confirmed not only to regulate the Arabidopsis ovule development process but also to be involved in resistance to bacterial wilt disease in Arabidopsis [27,28,29].
With the rapid development of genome sequencing technology, it has become possible to identify a whole gene family using bioinformatics tools at the plant genome level. Previous reports suggested that the members, structural features and expression profiles at the transcriptional level of the LRR-RLK gene family have been isolated in plants, including A. thaliana, Solanum lycopersicum, Glycine max and other species [9,11,13,14], but no information is available in sugarcane species. In most of these species, it is well known that LRR-RLKs are the largest group of the transmembrane receptor-like kinase subfamily and perform a variety of biochemical functions. For LRR-RLKs, according to the results of the phylogenic tree and gene structure, approximately 225 identified LRR-RLK genes were grouped into 15 groups in Arabidopsis [14], 234 identified were classified into 10 groups in S. lycopersicum [13], 309 identified were grouped into five groups in Oryza sativa [30], 467 identified were grouped into 14 groups in G. max [11], 303 identified were categorized into 15 groups in Brassica rapa [31], 379 identified in Populus trichocarpa were categorized into 14 groups [10] and 298, 511, 515 and 317 identified in Gossypium arboreum, G. barbadense, G. hirsutum and G. raimondii were classified into 15 groups [12], respectively. Apparently, there are a large number of LRR-RLKs in plant species, but detailed studies on their biochemical functions have only been performed for a few genes. Therefore, comprehensive analyses of the evolution, gene structure and expression patterns of the LRR-RLK gene family are necessary.
Sugarcane (Saccharum spp. hybrids) is now an important sugar and biofuel crop in the world, particularly in tropical and subtropical countries, producing 80% of the world’s sugar and 40% of the world’s biofuel. Considering the important resource value of sugarcane Saccharum genus, genome sequencing has been widely concerned. However, the complexity of the genetic background in sugarcane blocks the decoding of the genome sequence of modern sugarcane hybrids. Fortunately, Ray Ming’s research team, for the first time, deciphered the genome of one accession of the sugarcane ancestor species (S. spontaneum L.) [32]. Genomic deciphering is a milestone in the basic biological research of sugarcane, which can also provide the genome data for identification of the interesting family genes. In the present study, we first performed genome-wide identification, characterization and a phylogenetic and gene structure analysis of LRR-RLK genes. Moreover, the expression pattern of sugarcane LRR-RLK genes in several stress response processes was carried out. Our findings provide a foundation for the further functional characterization of the SsLRR-RLK gene family in regulating networks during plant growth and development and stress defense.
2. Materials and Methods
2.1. Arabidopsis LRR-RLKs and Saccharum spontaneum Genome Resources
All Arabidopsis LRR-RLKs sequences were obtained from TAIR database v10.0 (http://www.arabidopsis.org/, accessed on 21 April 2021) [33]. The classification of different AtLRR-RLKs and nomenclature of groups were known from the PlantsP server v.2011 Arabidopsis 2010 project (http://plantsp.genomics.purdue.edu/projects/lrr/Clouse2010.htm, accessed on 21 April 2021) [14]. Meanwhile, the detailed genome information of S. spontaneum was based on public database (http://www.life.illinois.edu/ming/downloads/Spontaneum_genome/, accessed on 21 April 2021) [32].
2.2. Identification of LRR-RLKs in Saccharum spontaneum Genome
Plant LRR-RLK gene family is featured with LRR domain, KD domain and TM domain, and the corresponding hidden Markov models (HMM) of LRR and HMM models of KD were downloaded from Pfam database version 32.0 (http://pfam.xfam.org/, accessed on 23 April 2021) [34] and used to run a blast search with an E-value less than 10−10 against the proteome in S. spontaneum by using HMMER v3.1 software [35]. After initial screening, the resulting hits were collected for further filtering. In order to ensure we got as close as possible to the overall LRR-RLKs, all Arabidopsis LRR-RLKs members (AtLRR-RLKs) reported by Shiu et al. [6] were obtained from TAIR database v10.0 and served as the query to run a similarity search (E-value < 1 × 10−5, identity > 50%) against the proteome in S. spontaneum by using BLAST+ v.2.6.0 [36]. These resulting hit sequences were then analyzed by Pfam database [34] and SMART (http://smart.embl-heidelberg.de, accessed on 23 April 2021) [37] to confirm the presence of LRR domain and KD domain and other characteristic domains. Furthermore, TMHMM server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/, accessed on 24 April 2021) [38] was devoted for TM domain prediction. Protein sequences that contained both LRR domain, KD domain and TM domain were considered as LRR-RLK genes.
LRR-RLK proteins in S. spontaneum and their key features, such as length of amino acids, isoelectric point (pI) and molecular weights, were analyzed using ProtParam online tool (http://au.expasy.org/tools/protparam.html, accessed on 24 April 2021) [39], the subcellular localization of SsLRR-RLKs was carried out by ProtComp version 9.0 software on SoftBerry online database (http://linux1.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc, accessed on 24 April 2021) [40] and the result of signal peptides was calculated using Signalp v5.0 (http://www.cbs.dtu.dk/services/SignalP/, accessed on 24 April 2021) [41], respectively.
2.3. Multiple Sequence Alignments and Phylogenetic Tree Construction
The sequence of each SsLRR-RLK and previously reported AtLRR-RLK protein was selected for phylogenetic analysis. Multiple sequence comparison by log-expectation (MUSCLE) [42] was carried out a multiple sequence alignment of the complete amino acid sequences of identified SsLRR-RLKs and AtLRR-RLKs. Finally, an unrooted phylogenetic tree was generated for sugarcane/Arabidopsis together by FastTree 2 tool with default arguments using the maximum likelihood (ML) method [43].
2.4. The Analysis of the Chromosome Distribution, Gene Structure, and Conserved Motif of the Sugarcane LRR-RLK Genes
Chromosome locations of all SsLRR-RLK genes were mapped based on the physical position of sugarcane chromosomes. The figure of chromosomal distribution of the SsLRR-RLK genes was drawn at MG2C v2.0 (http://mg2c.iask.in/mg2c_v2.0/, accessed on 26 April 2021) [44]. The exon–intron structure distribution patterns of each SsLRR-RLK were determined using GSDS 2.0 (http://gsds.cbi.pku.edu.cn/, accessed on 26 April 2021) [45]. To reveal the structural divergence of individual SsLRR-RLKs, the conserved motifs in their encoded proteins were predicted with Multiple Em for Motif Elicitation (MEME) online program version 5.0.5 (http://meme-suite.org/tools/meme, accessed on 27 April 2021) [46]. The maximum number of motifs was set as 20.
2.5. Promoter and Regulatory Analysis of the Sugarcane LRR-RLK Genes
The upstream 2000 bp sequence of each SsLRR-RLK gene translation start codon (ATG) was extracted from their genome sequence as a promoter region (Supplementary Material S3). In order to predict potential cis-acting regulatory elements, all promoter sequences of each SsLRR-RLK genes were submitted to PlantCARE online tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 28 April 2021) [47]. In addition, transcription factor (TF) binding sites were predicted using the PlantTFDB v4.0 online tool (http://plantregmap.cbi.pku.edu.cn/binding_site_prediction.php, accessed on 28 April 2021) [48].
2.6. Expression Pattern Analysis of the Sugarcane LRR-RLK Genes
For a better understanding of the potential roles of sugarcane LRR-RLK expression pattern during different stress responses, the expression profiles of all LRR-RLK genes were carried out based on the RNA-seq data from five different datasets of sugarcane transcriptomes in response to different stress treatments, except for low nitrogen stress, which contained two different tissues i.e. leaves and roots, and all the other RNA-seq raw data were obtained from sugarcane leaves. The above five sets of transcriptomes were provided by our laboratory research team. Detailed information of the transcriptomes prepared are as follows: (1) leaf and root transcriptome in two sugarcane varieties, ROC22 (resistant) and Badila (susceptible), under low nitrogen condition [49], respectively; (2) leaf transcriptome in two sugarcane varieties, ROC22 (susceptible) and FN39 (resistant), under low temperature treatment (unpublished); (3) leaf transcriptome of two sugarcane varieties, FN12-047 (susceptible) and ROC22 (resistant), subjected to infection by sugarcane leaf blight disease (Leptosphaeria taiwanensis) at different stages (unpublished); (4) transcriptome data of two sugarcane varieties, YC05-179 (resistant) and ROC22 (susceptible), inoculated with sugarcane smut pathogen (Sporisorium scitaminea Syd.) at different time points (24, 48 and 120 h post-inoculation (hpi)) [50]. These transcriptome datasets were selected for further analysis. Quality assessment of raw RNA-Seq reads were processed using FastQC, and then the high-quality cleaned reads were aligned to the S. spontaneum genome using HISAT2. Following alignments, raw counts for each gene were normalized and derived into FPKM (fragments per kilobase of transcript per million fragments mapped) value [51]. The transcript abundance of each sugarcane LRR-RLK genes were calculated as FPKM. Finally, we used 1.0 FPKM threshold, which created a full list of SsLRR-RLK genes with significant expression level. In addition, the resulting expression data based on the log-transformed data using log2 (FPKM+offset) with an offset=1.0 were then used for generation of heatmap [51,52] by pheatmap one package of R [53]. Here, in order to better and more easily understand the gene family analysis process, a diagram (Supplementary Material S4) showing all of the steps developed (from the use of Arabidopsis and S. spontaneum genome resources to the qRT-PCR), and summarizing all of the tools used in the manuscript, was made
2.7. Plant Materials, RNA Extraction and qRT-PCR Analysis
The sugarcane variety FN12-047 (Saccharum spp. hybrids) was provided by the Key Laboratory of Sugarcane Biology and Genetic Breeding, Fujian Agriculture and Forestry University (Fuzhou, China). In this study, we selected the sugarcane maturity stage for sampling. The field was watched throughout the plant life cycle, especially after appearance of sugarcane leaf blight disease (SLB) symptoms, and five different disease development stages were defined according to the previous report [54]. The leaf samples that had no visible sign of the SLB symptom and without pathogenic spores or hyphae observed under the microscope were represented as control [54]; Supplementary Material S5 shows the details. Three biological replicates were performed for each infection stage and for control (healthy), and a total of 18 samples were obtained. For each sugarcane accession, three plants with similar growth vigor and disease severity were selected for sampling, and the leaf located at the same leaf position was collected [54]. The collected samples were immediately put into liquid nitrogen and stored at −80 °C for later use.
Total RNA of each sample was extracted using TRIzol Reagent (Invitrogen, Waltham, Massachusetts, USA) and RNase-free DNase (TaKaRa, Shimogyo-ku, Kyoto, Japan) was treated according to the manufacturer’s protocol. The quality of the RNA was tested by using NanoDrop-1000 and RNA integrity was checked by electrophoresis. First strand cDNA synthesis was performed using the Prime-ScriptTM RT Reagent Kit (TaKaRa, Shimogyo-ku, Kyoto, Japan). Plant LRR-RLK gene family is the largest family in the plant RLK superfamily. In this study, a total of 437 LRR-RLK genes were identified in sugarcane. According to the results of the expression pattern analysis of the SsLRR-RLK genes in the transcriptome dataset (leaf transcriptome of two sugarcane varieties, FN12-047 (susceptible) and ROC22 (resistant), subjected to infection by sugarcane leaf blight disease), on the one hand, based on the transcriptome data (leaf transcriptome of sugarcane varieties FN12-047 (susceptible) subjected to infection by sugarcane leaf blight disease) FPKM values of 437 SsLRR-RLK genes, we found that approximately 24% of SsLRR-RLK genes (106 out of 437) were not expressed, approximately 49% of SsLRR-RLK genes (213 out of 437) were down-regulated and the remaining 27% of SsLRR-RLK genes (118 out of 437) were up-regulated. On the other hand, based on the FPKM value of 437 SsLRR-RLK genes depending on transcriptome data (leaf transcriptome of ROC22 (resistant) subjected to infection by sugarcane leaf blight disease), we found that approximately 22% of SsLRR-RLK genes (95 out of 437) were not expressed, approximately 29% of SsLRR-RLK genes (126 out of 437) were down-regulated and the remaining 41% of SsLRR-RLK genes (216 out of 437) were up-regulated. The detailed data are shown in Supplementary Material Table S8. Among the up-regulated SsLRR-RLK genes, we selected nine SsLRR-RLK genes with significant expression levels (FPKM values ranging from 5.4 to 36.9) to both verify the accuracy of the transcription dataset through RT-qPCR experimental results and show the transcriptional response of SsLRR-RLK gene in different periods. These qRT-PCR primers designed by Primer Premier 6 are listed in Supplementary Material Table S1. Furthermore, as an internal control primer, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) is unanimously recognized for its stability in academic research, such as in different sugarcane tissues and various adversity stress, and the primer sequence of GAPDH is universal [54]. Therefore, this housekeeping gene was used as the only internal control gene. The qRT-PCR amplification was performed using SYBR Green Master Mix Reagent (TaKaRa, Shimogyo-ku, Kyoto, Japan), according to the manufacturer’s instructions on an Applied Biosystems 7500 Real-Time PCR system. The standard qRT-PCR program refers to reports by Wang et al. [54]. Three technical replicates for each sample were performed and the relative expression level of each gene was calculated from the 2−ΔΔCt value [55].
3. Results
3.1. Identification and Distribution of LRR-RLK Genes in Saccharum spontaneum
The LRR-RLK amino acid sequences are composed of 437 members, which were identified from the whole genome of one accession of sugarcane ancestor species S. spontaneum, and all contain LRR-, TM- and KD-domains simultaneously. They accounted for 1.23% of the whole coding genes in S. spontaneum. Detailed information of the sugarcane LRR-RLK family is listed in Supplementary Material Table S2 and the summarized information of each group or subgroup is shown in Table 1. The results showed that the encoded protein molecular weight (MW) of all predicted LRR-RLKs ranged from 39.00 to 349.15 kDa and that their pIs varied from 5.06 to 10.63. In addition, the number of amino acids (AA) in encoded proteins varies from 450 to 1750 AA for approximately 97.7% of the aforementioned proteins, except for ten with different lengths, including the SsLRR-RLK305 encoding protein with 436 AA, SsLRR-RLK353 encoding protein with 358 AA and SsLRR-RLK72/92/25/244/348/355/407/427 encoding proteins with 1775, 2069, 2678, 3171, 1829, 1930, 1830 and 2005 AA, respectively. Supplementary Material Table S2 provides the results of the number of TM by TMHMM server v.2.0. A total of 422 SsLRR-RLKs had at least one TM domain, while 15 proteins had more than three TM domains. Meanwhile, physical positions of SsLRR-RLKs were used to map them onto corresponding chromosomes of this species. Results indicated that 437 LRR-RLK genes could be mapped onto all 32 chromosomes, from chromosome 1A (Chr1A) to Chr8D (Figure 1). In spite of every chromosome containing a certain number of SsLRR-RLK genes, their distribution appeared to be uneven across different chromosomes. The distribution ratio for each chromosome ranged from 1.37% to 5.72%. The largest number of SsLRR-RLK genes was found on Chr2C (25 genes); on the contrary, just six members were found on Chr6C. In addition, 22 SsLRR-RLK genes were located on Chr4D, followed by 19 on Chr1A, 2B, 2D and 4A, and the least (seven) on Chr5D and Chr7C. The results indicate that the distribution pattern of LRR-RLK in S. spontaneum is unlike the other gene family, such as the potato PLD gene family [56] and the maize JHDM gene family [34], while it is similar with LRR-RLK gene families in other plant species, such as soybeans [11].

Table 1.
LRR-RLK proteins in Saccharum spontaneum and their key features.
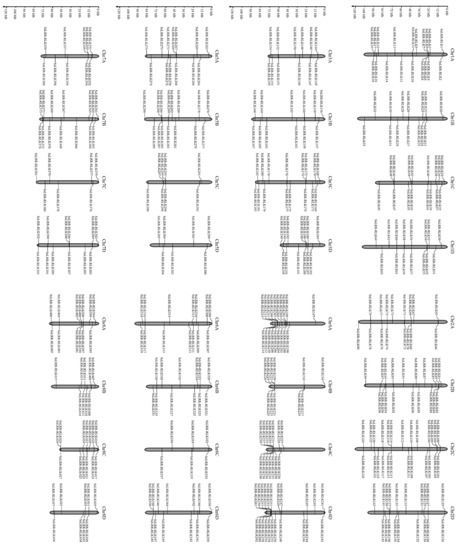
Figure 1.
Chromosome distribution of LRR-RLK family in S. spontaneum. 437 SsLRR-RLK alleles were located in all 32 chromosomes. The figure was drawn using MG2C v2.0.
3.2. Signal Peptide and Subcellular Localization Analysis of the LRR-RLK Family Proteins
The signal peptide analysis of the 437 LRR-RLK proteins was carried out using SignalP 5.0 Server. A total of 117 (26.8%) of LRR-RLK proteins do not contain a signal peptide sequence. Subsequently, in order to predict the location of the above proteins, we performed a subcellular localization prediction. The results show that 89.5% (391/437) of LRR-RLK proteins are likely to be located at the plasma membrane, whereas the other 10.5% (46/437) are more likely to be located in the extracellular domain (shown in Supplementary Material Tables S3 and S4).
3.3. Phylogenetic Analysis of Sugarcane LRR-RLKs
The 437 LRR-RLKs identified in this work and the previously reported 225 A. thaliana [57] LRR-RLKs were aligned (Additional data S1) for the maximum likelihood (ML) phylogenetic tree construction. The results revealed that sugarcane LRR-RLKs were classified into 14 groups and 21 distinct clades (Figure 2 and Additional data S2). It is noteworthy that not a single SsLRR-RLK protein is placed into the I-b and IX subfamily, which might be the result of gene loss during the evolutionary process of LRR-RLK family genes in S. spontaneum. In summary, group III, X-b, XI-a and XII show the largest number (75, 59, 124 and 51, respectively) of SsLRR-RLKs, whereas group X-c, XI-b, XIII-a and XIII-b contain only three, one, one and two in the phylogeny, respectively. Meanwhile, the majority of LRR-RLKs in A. thaliana is distributed in group I, III and XI-a, which contain 48, 46 and 33 LRR-RLKs, respectively. The phylogenetic analysis suggested that the 437 SsLRR-RLK proteins may have similar functions to their Arabidopsis orthologs, and provided insight into the evolution of gene family members and gene family diversity in sugarcane. The detailed classification of Arabidopsis and sugarcane LRR-RLKs is described in Supplementary Material Table S5.
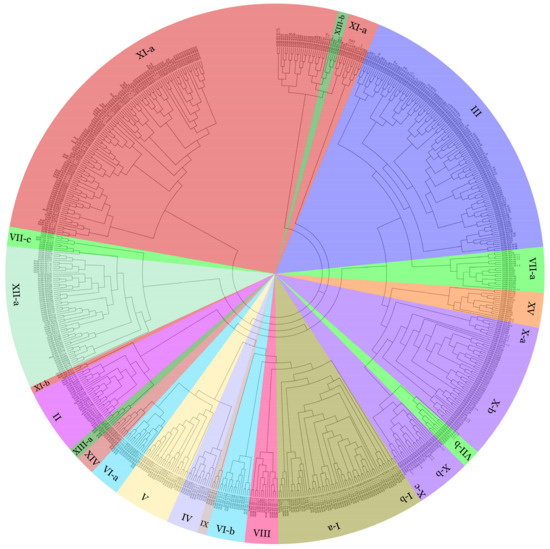
Figure 2.
Phylogenetic tree of the LRR-RLK genes between Arabidopsis and S. spontaneum. The phylogenetic tree was generated using FastTree 2.0 using the maximum likelihood method. All sugarcane LRR-RLKs were classified into 14 distinct groups and different groups were shown by different color.
3.4. Exon–Intron Organization and Conserved Motifs of SsLRR-RLKs
To identify the diversification of the SsLRR-RLK genes, the exon–intron organizations and conserved motifs were analyzed. Supplementary Material Figure S1 reveals the detailed information of each SsLRR-RLK gene and Figure 3 describes the representative exon–intron distributions in sugarcane. Among the LRR-RLKs, nearly one third (157 out of 437) contained only one intron, while 53 genes were without any introns. Two, three, four, five and six introns were found in 64, 48, 18, 11 and 9 genes, respectively. Furthermore, a total of 77 SsLRR-RLKs contained more than six introns, and, out of them, 19 had more than 15 introns. In terms of the results of exon–intron number and distribution, the same groups or subgroups have a very close organization, which strongly supports their evolutionary relationship. For instance, a large number of SsLRR-RLKs in groups VII-b, VII-c, X-c and XV contain zero, one, two and three introns, except for the gene SsLRR-RLK82, which contains six introns. Nevertheless, the members of groups I, III and XI exhibited a high changeability in the number of introns. Interestingly, the members of group XIII-b contained 24 introns, which was approximately twice as much as group XIII-a.
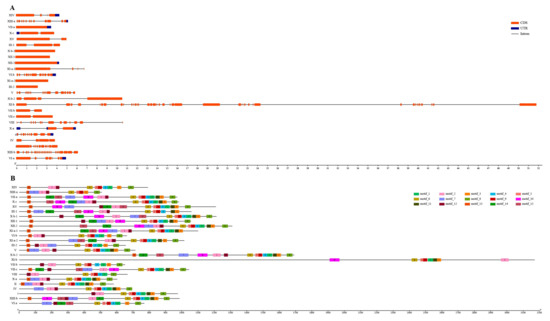
Figure 3.
Representative exon–intron (A) and conserved motifs structure (B) of each S. spontaneum LRR-RLK subfamily. (A) Exon–intron structures of SsLRR-RLKs. Untranslated regions (UTRs), exons and introns are represented by blue boxes, red boxes and black lines, respectively. (B) The motif distribution of SsLRR-RLK proteins. The 15 predicted motifs are represented by distinct color boxes using the MEME program.
Meanwhile, the conserved motifs and diversification of all SsLRR-RLK proteins were generated from the results of the MEME motif analysis (Supplementary Material Figure S2). As exhibited in Figure 3, a total of 15 distinct conserved motifs were found: motifs 1 to 15, which are widely distributed across all subfamilies, except for SsLRR-RLK386/265/338. This specific motifs arrangement might contribute to the functional divergence of LRR-RLK genes in sugarcane. SsLRR-RLKs members within the same groups or subgroups were usually found to have shared a composition. For instance, the clustered SsLRR-RLK pairs, i.e. SsLRR-RLK81/101, SsLRR-RLK398/433 and SsLRR-RLK179/314, were demonstrated to have a highly similar motif distribution. It is noteworthy that differences within the group and subgroups were found for not only the number of motifs but also the types of motifs in one SsLRR-RLK protein (Supplementary Material Table S6).
3.5. A cis-Acting Regulatory Elements and TF Binding Sites Analysis of SsLRR-RLKs
The cis-acting regulatory elements of the promoter region (2000 bp sequence upstream of translation start site) among 437 LRR-RLK members were detected using PlantCARE, and the results indicated that a total of 128 putative cis-acting elements were widespread in SsLRR-RLKs. Detailed information of all genes is shown in Supplementary Material Table S7 and summarized in Table 2. Briefly, cis-acting regulatory elements can be mainly classified into seven major categories. By contrast, four types of cis-acting regulatory elements related to phytohormones, environmental stress, photoreactions and development were particularly abundant. The first type of photoreaction-related elements included G-Box, TCCC-motif, ACA-motif, Box 4, GT1-motif, LS7 and Sp1 etc., of which, SP1 and G-Box appeared to be the most widespread, with more than five copies. The phytohormone regulation-related cis-acting elements of the second type responds to auxin, abscisic acid (ABA), jasmonic acid (JA), gibberellins (GA), salicylic acid (SA) and ethylene (ETH). AuXRR-core and AuxRE are involved in auxin responsiveness, GARE-motif and P-box are considered to be gibberellin responsive elements involved in the regulation of gibberellin and other cis-elements participated in a specific stress response. The third type of widely distributed regulatory elements is associated with environmental stimuli, including abiotic and biotic stresses; of which, ARE, MYB, MYC, GC-motif, STRE, LTR and MBS were the most abundant. In particular, MYB and MYC were the most abundant, both having more than 15 copies in all subfamilies, except for X-c and VII-b. Finally, cis-acting elements of the fourth type, including CAT-box, O2-site, ACII, Myb-binding site, CCGTCC-box and CCGTCC-motif etc., are generally related to plant growth and development, which is either related to the cell cycle and cell proliferation response or required for tissue-specific expression. Overall, the cis-acting regulatory elements SP1, G-BOX, CCGTCC-box/motif, ABRE, CGTCA-motif, MYB, MYC and STRE are prominently distributed in these four categories. Captivatingly, two very specific cis-elements, OCT and JERE, could be prominently found in the X-c subfamily (containing SsLRR-RLK66/84/104 genes), with 8.0 and 10.7 copies, respectively. Thus, considering the distribution of cis-acting regulatory elements in the promoter regions, we speculate that the expression of SsLRR-RLKs was extensively regulated, implying the important roles in plant meristem maintenance and organogenesis, as well as in mediating defense responses to environmental stresses.

Table 2.
Statistics analysis of cis-acting elements detected in promoter of SsLRR-RLK genes.
In addition, transcription factors (TFs) play an essential role in many biological processes by regulating and mediating related target gene expression. To explore the potential regulation relationship among TFs and SsLRR-RLKs, TFs binding sites were predicted using PlantTFDB. As shown in Supplementary Material Figure S3, NAC, TALE, G2-like, WRKY, ERF and MYB were the most widely functioning TF family and could regulate the majority of SsLRR-RLKs. Meanwhile, the abundant TF families were concerned in various aspects of plant growth and development, stress defense and signal transduction, suggesting that SsLRR-RLKs with TF binding sites were considered to participate in these key processes by TF-mediated regulation.
3.6. Expression Profiling of SsLRR-RLK Genes Based on RNA-seq Datasets
For a better understanding of the SsLRR-RLKs functions, the expression patterns of all SsLRR-RLKs in different abiotic stresses (i.e., cold and low nitrogen treatments) and biotic stresses (i.e., challenged by sugarcane leaf blight disease and sugarcane smut) were investigated based on RNA-seq datasets. The expression profiles of SsLRR-RLK genes under a low temperature are presented in Table S8 (Supplementary Material Table S8) and Figure 4E,F. Our results showed that 27.0% (118 out of 437) of genes were expressed in the relatively tolerant variety FN39 under cold stress, and 28.0% (33/118) genes among them were induced and shown to be highly expressed, such as SsLRR-RLK88/131/263. However, 72.0% (85/118) of genes were suppressed after cold stress, and SsLRR-RLK169/197/246/266/420 among them was shown to be more strongly suppressed. By contrast, the expression level of 27.9% (122 out of 437) of LRR-RLK genes in the variety ROC22 was expressed, and 51.6% (63 out of 122) with a high expression, such as SsLRR-RLK88/131/263/409. In contrast, 48.4% of genes showed a low expression, such as SsLRR-RLK169/197/246/266/396/420.
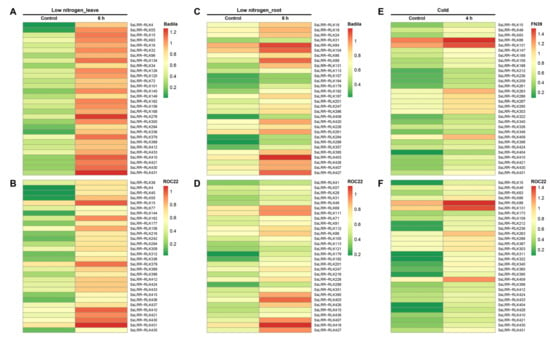
Figure 4.
Heatmaps of the expression analysis of SsLRR-RLKs in response to diverse abiotic stress. The differential expression profiles in two different sugarcane varieties is shown by heatmap under (A,B) low nitrogen (leaf) stress, (C,D) low nitrogen (root) stress and (E,F) cold stress. Heatmap was generated based on FPKM values. The color scale represents expression values of each sample, with red showing high levels and green showing low levels of transcript abundance.
Subsequently, we further investigated the expression profiles of SsLRR-RLKs in root and leaf transcriptome in two sugarcane varieties, ROC22 and Badila, under low nitrogen stress, respectively (Supplementary Material Table S8). The results indicated that 51.5% (225 out of 437) and 53.3% (233 out of 437) of genes were expressed, while 33.3% (75 out of 225) and 62.7% (146 out of 233) of genes were shown to be highly expressed in the roots in the varieties Badila and ROC22 under a low nitrogen stress. Besides, 66.7% (150 out of 225) and 37.3% (87 out of 233) of genes were shown to have a low expression in Badila and ROC22, respectively. Among these genes, SsLRR-RLK84/88/104/403 and SsLRR-RLK69/111/403/407/418/427 had the highest expression level, whereas SsLRR-RLK161/286/394/417 and SsLRR-RLK12/161/295 had the weakest expression in roots in Badila and ROC22, respectively (Figure 4CD). In addition, The RNA-seq datasets of the leaves transcriptome in ROC22 and Badila under low nitrogen stress were used to generate a heatmap (Supplementary Material Table S8 and Figure 4AB). It suggested that 52.9% (231 out of 437) and 46.9% (205 out of 437) of genes were expressed, and 77.9% (180 out of 231) and 73.7% (151 out of 205) of genes had a high expression in Badila and ROC22, respectively. However, 22.1% (51 out of 231) and 26.3% (54 out of 205) of genes were suppressed in Badila and ROC22, respectively. Among these genes, SsLRR-RLK15/276/379/410/431 and SsLRR-RLK15/379/410/431 were strongly expressed, whereas SsLRR-RLK141/169/396/420 and SsLRR-RLK169/420 showed the weakest expression in Badila and ROC22, respectively.
Additionally, the datasets obtained from leaves of FN12-047 and ROC22 subjected to pathogenic infection of sugarcane leaf blight disease at two disease-developed stages were used for analysis of the expression profiles. As shown in Figure 5AB, expression heatmaps of the SsLRR-RLK genes responsive to sugarcane leaf blight disease in susceptible variety FN12-047 and resistant variety ROC22 revealed that 34.1% (149 out of 437) and 34.6% (151 out of 437) of genes were expressed, and 64 out of 149 genes and 105 out of 151 genes were shown to be highly expressed at early stages of disease development in FN12-047 and ROC22, respectively. Among them, SsLRR-RLK64/359/391 and SsLRR-RLK64/146/173 showed significant expression, particularly in FN12-047 and ROC22, respectively. In contrast, 34.3% (150 out of 437) and 35.7% (156 out of 437) of genes showed the weakest expression, whereas 86 out of 150 and 114 out of 156 genes were highly expressed at medium to late stages of the disease development. Moreover, five (SsLRR-RLK18/64/352/359/391) and six LRR-RLKs (SsLRR-RLK18/64/88/131/146/173) were strongly expressed at medium to late stages in FN12-047 and ROC22, respectively. Furthermore, expression profiles of the SsLRR-RLKs response to the varieties YC05-179 (resistant) and ROC22 (susceptible) challenged by smut pathogen Sporisorium scitaminea in the samples collected at different time points (24, 48 and 120 h post-inoculation (hpi)) were investigated, and the heatmap was constructed (Figure 5CD). Generally, for both investigated varieties, approximately half of the LRR-RLK genes appeared to be expressed across all three time points, and those significantly induced in variety YC05-179 were SsLRR-RLK120/141 at 24 hpi, SsLRR-RLK64/120/141/ at 48 hpi and SsLRR-RLK64/120/141 at 120 hpi. By contrast, those strongly induced in variety ROC22 were SsLRR-RLK17 at 24 hpi, SsLRR-RLK17/228 at 48 hpi and SsLRR-RLK6/17/203/228/246/249/346 at 120 hpi.
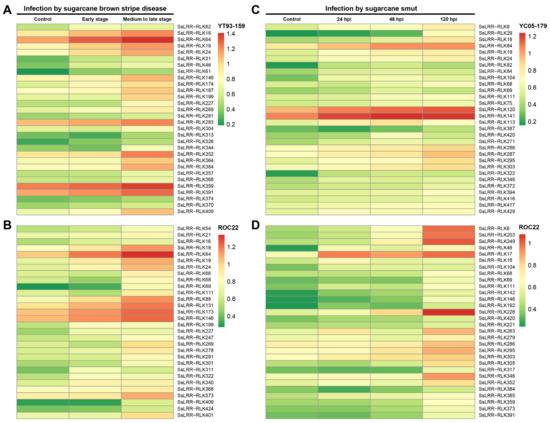
Figure 5.
Heatmaps of the expression analysis of SsLRR-RLKs under different biotic stresses. The differential expression patterns in two different sugarcane varieties were shown by heatmap under (A,B) infection by sugarcane leaf blight disease and (C,D) infection by sugarcane smut. Heatmap was generated based on FPKM values. The color scale represents expression values of each sample, with red showing high levels and green showing low levels of transcript abundance.
Overall, many SsLRR-RLK genes were strongly induced or suppressed by different stress treatments. For example, SsLRR-RLK88 significantly responded to the stresses of the cold, low nitrogen and sugarcane leaf blight disease infection. SsLRR-RLK141 was obviously induced by all of the investigated stress treatments. In contrast, some LRR-RLK genes were simultaneously induced in both varieties with a different tolerance/resistance by one treatment. For example, SsLRR-RLK88 and SsLRR-RLK131 were induced by the cold treatment and SsLRR-RLK18/64/146 was induced by the infection of sugarcane leaf blight disease. Most interestingly, the expression profiles for some SsLRR-RLK genes, such as SsLRR-RLK131, was significantly expressed in response to the infection of sugarcane leaf blight disease, whereas it was strongly repressed under cold stress. This indicates the potential roles in multiple biotic and abiotic stress responses of these differentially expressed SsLRR-RLK genes.
3.7. Expression Analysis of SsLRR-RLK Genes by Quantitative Real-Time PCR
According to the expression profiling of SsLRR-RLK genes, an overview of expressed SsLRR-RLKs under sugarcane leaf blight disease stress (SLB) is known. To further investigate their physiological functions, we first selected and analyzed nine SsLRR-RLK genes by qRT-PCR based on the expression analysis of SsLRR-RLK genes. As shown in Figure 6, the results showed that the expression pattern of each LRR-RLK gene is differentially expressed in the leaves. Meanwhile, on the basis of their expression levels, these nine SsLRR-RLKs were clearly divided into three groups (A–C). Among them, SsLRR-RLK88/146 and SsLRR-RLK346 genes of group A were significantly expressed in the more early and early stage. The genes of group B were specifically expressed in the medium stage, such as SsLRR-RLK19/24 and SsLRR-RLK391. Compared with the group A and B analyzed genes, the C group of genes (SsLRR-RLK203/249 and SsLRR-RLK359) was significantly expressed in the late and more latter stage. The qRT-PCR results were highly consistent with that of the RNA-seq results, and revealed that the identified LRR-RLK genes in sugarcane exhibited significantly up-regulated expression levels. In summary, the LRR-RLK family may be involved in the disease defense process of sugarcane.
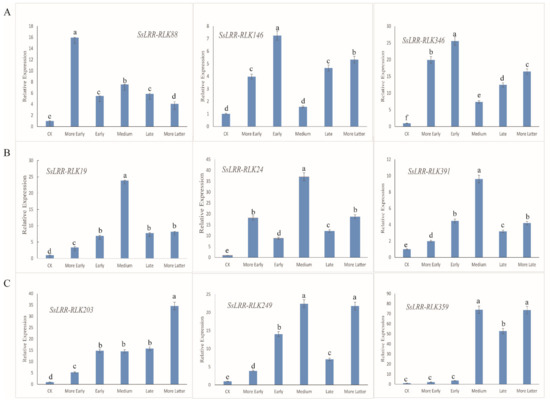
Figure 6.
Relative expression levels of nine selected LRR-RLK genes were analyzed using quantitative real-time PCR (qRT-PCR). Among them, three obvious periods were discovered, namely the early, medium and late periods. (A) contains three highly expressed genes that respond to early period (SsLRR-RLK88/146/346) that were identified. (B) contains three highly expressed genes that respond to medium period (SsLRR-RLK19/24/391) that were identified. (C) contains three highly expressed genes that respond to late period (SsLRR-RLK203/249/359) that were identified. X-axes show different stages and y-axes show relative expression level of each LRR-RLK gene. Bars represent the standard deviation (SD) (n = 3) of three technical replicates. Different lowercase letters above the bars in the figure indicate a significant difference (p < 0.05). Lowercase letters reflect the 5% significance level. As shown in the chart, a, b, c, d, e and f are different from each other, and they are all significantly different from each other. In addition, the same lowercase letter means no significantly different.
4. Discussion
Plant LRR-RLKs are the largest family of plant RLKs, which play various essential roles in plant life activities. The genome-wide identification and functional analysis of LRR-RLK gene families have been extensively carried out in many plant species whose genomes have been sequenced, such as Arabidopsis, P. trichocarpa, G. hirsutum, S. lycopersicum, G. max and other plants [9,10,11,12,13,14,30,31]. Previous studies identified 225, 234 and 467 LRR-RLK genes in soybean [11], tomato [13] and Arabidopsis [14], respectively. Sugarcane is subjected to various abiotic and biotic stresses during growth and development [54,58]. It is worth noting that the members of LRR-RLK genes in all of these species mentioned are larger than those in Arabidopsis, which might be due to their small genome. However, very little information is known about the LRR-RLK gene and its gene family in sugarcane because the modern sugarcane genome has been undeciphered until now. Fortunately, the genome-wide sequence of S. spontaneum AP85-441, one accession in the sugarcane ancestor wild species, has recently been published [32]. In order to better understand the characteristics and biological functions of SsLRR-RLK genes, we investigated the LRR-RLK gene family in sugarcane by bioinformatics means, together with a gene structure assay, and an expression pattern analysis was based on the several transcriptomic datasets. In the current study, a total of 437 SsLRR-RLKs members are successfully identified based on the genome of S. spontaneum. Compared with the LRR-RLK genes in other plants, the members in our study are smaller than those in soybean, but larger than those in tomato and Arabidopsis; this might be because the genome size of S. spontaneum AP85-441 is larger than that of tomato and Arabidopsis. When talking about the number of their categorization, 437 SsLRR-RLK genes are categorized into 14 groups (I, II, III, IV, V, VI, VII, VIII, X, XI, XII, XIII, XIV and XV) (Figure 2) and 10 subgroups (VI-a, VI-b, VII-a, VII-b, VII-c, X-a, X-b, X-c, XI-a, XI-b, XIII-a and XIII-b), of which, the group IX and the subgroup I-b are not found in sugarcane wild species accession AP85-441, indicating that the evolution of the LRR-RLKs in these two subfamilies may occur after the divergence of dicots and monocots or gene loss during evolution, according to the opinion of previous report [33], which also occurred in G. max [11] and G. hirsutum [12]. Previous reports showed that the LRR-RLKs in Arabidopsis, S. lycopersicum and P. trichocarpa were mainly categorized into 15, 10 and 14 groups, suggesting the same number of groups to that in P. trichocarpa [10], but less than that in Arabidopsis [14] and more than that in S. lycopersicum [13]. Furthermore, the LRR-RLK genes in G. max, P. trichocarpa and G. arboretum were mainly categorized into 26, 9 and 11 subgroups, suggesting a similar number of subgroups to that in P. trichocarpa and G. arboretum [12], but less than that in G. max. In contrast, the quantity of group II in Arabidopsis, G. max, P. trichocarpa, and strawberry is 14, 26, 29 and 8, respectively. Group V LRR-RLKs amount to 9, 18, 11 and 7, in the aforementioned four plants, respectively. Moreover, there are 33, 3, 28 and 56 members in group XI-a in the mentioned four species, respectively. By contrast, a total of 437 sugarcane LRR-RLK genes are identified in this study, and among them, 13 belonged to group II, 15 to group V and 124 to group XI-a, which is similar to G. max, but more than strawberry, and less than P. trichocarpa. Thus, group XI-a is relatively expanded in sugarcane in contrast to the other plant genomes mentioned. It was notable that LRR-RLKs were more closely related to the members of the same type than those of the different types in the same species, revealing a relatively high degree of homology similarity within the same type of SsLRR-RLKs. Furthermore, the diversification of exon–intron organizations and conserved motifs among the LRR-RLK gene members always plays an essential role in the evolution of this gene family. For instance, a previous study reported that the presence of multiple introns has been shown to be essential for ERECTA gene expression in Arabidopsis [59]. In addition, the findings further confirmed the classifications of the SsLRR-RLK genes, while the structure of SsLRR-RLKs in different groups or subgroups showed relatively lower identities [10,11]. Our research also identified the similar motif arrangements among the SsLRR-RLK proteins within groups or subgroups, which suggested that the protein structure is highly conserved within a specific subfamily. Indisputably, the roles of the vast majority of these conserved motifs remain to be clarified. Overall, the exon–intron structure and conserved motifs organization of the SsLRR-RLK members in the same group or subgroup, combined with the results of the phylogenetic analysis, could strongly support the consistency of the group or subgroup categories. As identified, cis-acting regulatory elements serve as an important molecular switch involved in the transcriptional regulation of the gene activities under various environmental factors, photoreactions and phytohormones, including the cold, drought, light, wounding, ethylene (ETH) and abscisic acid (ABA), etc. [10]. We found that the promoters of the SsLRR-RLK genes contain a large number of cis-acting regulatory elements related to environmental stress and hormonal signal responses, such as AAAC-motif, AACA-motif, ACA-motif, DRE, ABRE, LTR, MBS, I-box, WUN-motif, ERE, TGA-element and CGTCA-motif, etc. Two very interesting elements, OCT (participating in the cell proliferation response) and JERE (participating in the MeJA response), were significantly found in the X-c subfamily, with more than eight copies, implying that SsLRR-RLK genes within the X-c subfamily are mainly involved in the cell proliferation and response to MeJA (methyl jasmonate). In a previous study, the AACA-motif was shown to be an enhancer element that is required for the endosperm-specific expression of the glutelin gene from rice [60], and DRE and ABRE regulatory elements were shown to be interdependent in the ABA-responsive expression in response to ABA in Arabidopsis [61]. Hence, our results suggested that these cis-acting regulatory elements also play essential roles in the regulation of sugarcane LRR-RLKs expression in response to corresponding stress.
In recent years, although many LRR-RLK genes have been reported based on the whole-genome in the plant kingdom, their biological functions have been verified only in a small quantity of LRR-RLKs until now [14,16,19]. As the research progresses, increasing evidence suggests that LRR-RLKs are involved in the immune response, signal transduction and response to abiotic and biotic stresses [11,12,62]. In addition, many plant LRR-RLKs have been elucidated to play key roles in plant disease resistance. For example, FLAGELLIN-SENSING 2 (FLS2) and EF-Tu RECEPTOR (EFR) recognize the corresponding antigens and mediate the defense against pathogens in the processes of the plant immune response in Arabidopsis [63,64,65]. Besides, recent research indicates that somatic embryogenesis receptor-like kinases (SERKs) are essential in steroidal hormones BRs signaling, including the rice OsSERK1 against fungal pathogen infection by mediating defense signaling transduction [66,67,68].
Abiotic and biotic factors, such as the cold, low nitrogen and pathogens, pose a serious threat to the yield of sugarcane. LRR-RLKs are essential for plants to perceive and receive environmental signal factors as transmembrane receptor-like kinase proteins to perform a series of biological functions [1,3,6]. In Arabidopsis, LRR-RLK genes were confirmed to regulate plant growth and development, including to mediate the defense against pathogens [27], signal transduction [16,18], adjust the meristem size [20] and promote stem cell differentiation [17,21]. Recently, many plant LRR-RLKs have also been reported to respond to many environmental stresses, including dehydration [12], heat [12], pathogens [12,27], the cold [12], high salinity [12] and ABA treatments [26]. Additionally, the expression difference in SsLRR-RLK genes based on the genome-wide identification and expression analysis between one pair of varieties with a different tolerance or resistance may play an important role in stress response or disease resistance. In summary LRR-RLK genes in sugarcane are also likely to have diverse biological functions. However, there are no reports about LRR-RLK genes observed in sugarcane. Thus, to investigate the potential roles and expression levels of LRR-RLK genes in sugarcane, we comprehensively analyzed the expression patterns of SsLRR-RLKs in the sugarcane’s response to biotic and abiotic stress. Five different datasets of RNA-seq used in our study were carried out, and the value could be significantly changed due to highly expressed SsLRR-RLK genes. The results showed that the expression of the vast majority of SsLRR-RLK genes was distinctively regulated in response to a given stress, which forcefully reveals that they may be vital stress response genes. According to the heat map, we also found that the SsLRR-RLK genes in the same subfamily have no consistent expression patterns depending on the analyses of gene expression data. Interestingly, some SsLRR-RLK genes might be highly expressed in a subfamily, whereas other genes exhibit a low expression or no expression at all. In a previous study, Sun et al. [12] revealed that the expression pattern of Gossypium LRR-RLK genes was widely involved in stress defense, including dehydration, the cold and salt, which provides insight into potential functional diversity within every one subfamily. The expression profiling of the SsLRR-RLK genes indicated that the SsLRR-RLKs could be responsive not only to abiotic stresses but also to biotic stresses. Therefore, these SsLRR-RLK genes stand as strong functional candidates for follow-up research into environmental stress in sugarcane. Overall, these above findings first provide a part of extremely valuable biological information that will accelerate our understanding of the regulatory roles of LRR-RLK genes in sugarcane stress defense response processes. Meanwhile, our analyses were also helpful in selecting candidate LRR-RLK genes for the further functional verification and disease resistance breeding of sugarcane.
5. Conclusions
Here, we performed the first comprehensive analysis of the identification and characterization of the LRR-RLKs gene family in sugarcane. Using the latest available S. spontaneum genome sequence as a reference, a total of 437 SsLRR-RLK genes were identified and were classified into 14 groups based on a phylogenetic tree analysis. The distribution of SsLRR-RLKs was found to be a random but unbalanced distribution across all chromosomes. The result of the exon–intron structures and conserved motifs were considerably conserved among members within the same groups and the same subgroups. The cis-acting regulatory elements and TF binding sites analysis of SsLRR-RLKs also indicated that SsLRR-RLK genes were multi-functional. Subsequently, further expression profiles and a qRT-PCR expression analysis of these SsLRR-RLK genes that were responsive to stresses in sugarcane were carried out based on the transcriptome datasets. In summary, all of these findings provide more valuable tools and information for future research of the potential biological functions of the SsLRR-RLK genes in sugarcane.
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/article/10.3390/cimb43030116/s1. Additional data S1: Multiple sequences of LRR-RLK from A. thaliana and S. spontaneum were aligned by MUSCLE software with default parameters; Additional data S2: Phylogenetic tree constructed; Table S1: The primers used in the qRT-PCR analyses; Table S2: Detailed information of all identified SsLRR-RLK genes; Table S3: Signal peptide analysis; Table S4: Subcellular localization prediction of all LRR-RLKs proteins in sugarcane; Table S5: Statistics of A. thaliana and S. spontaneum LRR-RLKs distribution among different groups and subgroups; Table S6: Sequence logos of the conserved motifs of all SsLRR-RLKs among each group and subgroup predicted by MEME; Table S7: Cis-acting regulatory element analysis of the SsLRR-RLK genes; Table S8: FPKM values of 437 SsLRR-RLK genes based on five different stress RNA-seq data; Figure S1: Exon–intron structure of all identified LRR-RLK members in sugarcane; Figure S2: Detailed schematic illustrations of the conserved motifs identified in SsLRR-RLK proteins. Fifteen conserved motifs were distinguished by different colors; Figure S3: Detailed schematic illustrations of the TF binging sites predicted. Only shows the top 12 transcription factor families; Supplementary Material S3: The upstream 2000 bp sequence of 437 SsLRR-RLK genes; Supplementary Material S4: a diagram showing all of the steps developed; Supplementary Material S5: morphology of disease of sugarcane leaf blight (SLB).
Author Contributions
L.X. and W.C. conceived and designed the experiments. W.C., Z.W. and F.X. carried out the experiments and analyzed the data; Y.S., G.L. and W.A. contributed to data collection. W.C. wrote the draft manuscript. Y.S. and L.X. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (31571732) and the Sugar Crop Research System of China (CARS-17).
Acknowledgments
The authors are thankful to all team members in Xu’s lab, Key Laboratory of Sugarcane Biology and Genetic Breeding, Ministry of Agriculture, Fujian Agriculture and Forestry University, for helpful discussion and suggestions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Walker, J.C. Structure and function of the receptor-like protein kinases of higher plants. Plant Mol. Biol. 1994, 26, 1599–1609. [Google Scholar] [CrossRef]
- Geer, P.V.; Hunter, T.; Lindberg, R.A. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu. Rev. Cell Biol. 1994, 10, 251–337. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, J.C.; Zhang, R. Relationship of a putative receptor protein kinase from maize to the S-locus glycoproteins of Brassica. Nature 1990, 345, 743–746. [Google Scholar] [CrossRef]
- Xue, H.W. Global analysis of expression profiles of rice receptor-like kinase genes. Mol. Plant 2012, 5, 143–153. [Google Scholar]
- Shiu, S.H. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci Stke 2001, 113, re22. [Google Scholar] [CrossRef]
- Kobe, B.; Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef]
- Liu, P.L.; Du, L.; Huang, Y.; Gao, S.M.; Yu, M. Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 2017, 17, 47. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Li, L.; Wang, P.; Zhang, S.; Wu, J. Genome-wide characterization, evolution, and expression analysis of the leucine-rich repeat receptor-like protein kinase (LRR-RLK) gene family in Rosaceae genomes. BMC Genom. 2017, 18, 763. [Google Scholar] [CrossRef] [Green Version]
- Zan, Y.; Ji, Y.; Zhang, Y.; Yang, S.; Song, Y.; Wang, J. Genome-wide identification, characterization and expression analysis of populus leucine-rich repeat receptor-like protein kinase genes. BMC Genom. 2013, 14, 318. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Guo, Y.; Qiu, L.J. Genome-wide identification and evolutionary analysis of leucine-rich repeat receptor-like protein kinase genes in soybean. BMC Plant Biol. 2016, 16, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, R.; Wang, S.; Ma, D.; Liu, C. Genome-wide analysis of LRR-RLK gene family in four gossypium species and expression analysis during cotton development and stress responses. Genes 2018, 9, 592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Z.; Wang, J.; Yang, S.; Song, Y. Identification and expression analysis of the LRR-RLK gene family in tomato (Solanum lycopersicum) Heinz 1706. Genome 2015, 58, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Gou, X.; He, K.; Yang, H.; Yuan, T.; Lin, H.; Clouse, S.D.; Li, J. Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genom. 2010, 11, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, C.; Zhu, Y.F.; Cui, Y.W.; Cheng, K.L.; Liang, W.; Wei, Z.Y.; Zhu, M.S.; Yin, H.J.; Zeng, L.; Xiao, Y.; et al. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants 2018, 4, 205–211. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Seto, H.; Fujioka, S.; Yoshida, S.; Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 2001, 410, 380–383. [Google Scholar] [CrossRef]
- Somssich, M.; Je, B.I.; Simon, R.; Jackson, D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development 2016, 143, 3238–3248. [Google Scholar] [CrossRef] [Green Version]
- Nam, K.H.; Jianming, L. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Jiangqi, W.; Kevin, A.L.; Jason, T.D.; Frans, E.T.; John, C.W. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef] [Green Version]
- Canales, C.; Bhatt, A.M.; Scott, R.; Dickinson, H. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 2002, 12, 1718–1727. [Google Scholar] [CrossRef]
- Jinn, T.L.; Stone, J.M.; Walker, J.C. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 2000, 14, 108–117. [Google Scholar]
- Demko, V.; Ako, E.; Perroud, P.F.; Quatrano, R.; Olsen, O.A. The phenotype of the CRINKLY4 deletion mutant of Physcomitrella patens suggests a broad role in developmental regulation in early land plants. Planta 2016, 244, 275–284. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Kondo, Y.; Fukuda, H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in arabidopsis. Plant Cell 2010, 22, 2618–2629. [Google Scholar] [CrossRef] [Green Version]
- Song, W.Y.; Wang, G.L.; Chen, L.L.; Kim, H.S.; Pi, L.Y.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.X.; Zhu, L.H. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 1995, 270, 1804–1806. [Google Scholar] [CrossRef] [Green Version]
- Osakabe, Y.; Maruyama, K.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in arabidopsis. Plant Cell 2005, 17, 1105–1119. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.C.; Hong, S.W.; Whang, S.S.; Lim, P.O.; Nam, H.G.; Koo, J.C. Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 2011, 52, 651–662. [Google Scholar] [CrossRef] [Green Version]
- Godiard, L.; Sauviac, L.; Torii, K.U.; Grenon, O.; Marco, Y. ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 2003, 36, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Torii, K.U.; Mitsukawa, N.; Oosumi, T.; Matsuura, Y.; Yokoyama, R.; Whittier, R.F.; Komeda, Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 1996, 8, 735–746. [Google Scholar] [PubMed] [Green Version]
- Chinchilla, D.; Libo, S.; Ping, H.; Sacco de, V.; Birgit, K. One for all: The receptor-associated kinase BAK1. Trends Plant Sci. 2009, 14, 535–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Wang, G.L. Genome-wide identification, characterization and phylogenetic analysis of the rice LRR-kinases. PLoS ONE 2011, 6, e16079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rameneni, J.J.; Lee, Y.; Dhandapani, V.; Yu, X.; Su, R.C.; Oh, M.H.; Yong, P.L. Genomic and post-translational modification analysis of leucine-rich-repeat receptor-like kinases in brassica rapa. PLoS ONE 2015, 10, e0142255. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Tang, H.; Zhang, Q.; Hua, X.; Ma, X.; Zhu, F.; Jones, T.; Zhu, X.; Bowers, J.; et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 2018, 50, 1565–1573. [Google Scholar] [CrossRef] [Green Version]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Penelope, C.; Eberhardt, R.Y.; Eddy, S.R.; Jaina, M.; Mitchell, A.L.; Potter, S.C.; Marco, P.; Matloob, Q.; Amaia, S.V.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2014, 43, D257–D260. [Google Scholar] [CrossRef]
- Viklund, H.; Elofsson, A. Best alpha-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci. 2004, 13, 1908–1917. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy Server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Long, R.; Wang, H.; Shen, Y.; Kang, J.; Zhang, T.; Sun, Y.; Zhang, Y.; Li, M.; Yang, Q. Molecular cloning and functional analysis of a salt-induced gene encoding an RNA-binding protein in alfalfa. Mol. Breed. 2014, 34, 1465–1473. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; Heijne, G.V.; Nielsen, H.H. SIGNALP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Kong, Y.; Wang, Q.; Sun, Y.; Gong, D.; Lv, J.; Liu, G. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Hereditas 2015, 37, 91–97. [Google Scholar]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Nadya, W.; Chris, M.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Gao, S.; Shumway, S.Y.; Lin, Z.; Guo, J.; Li, M.; Wang, Z.; Que, Y.; Xu, L. Transcripts and low nitrogen tolerance: Regulatory and metabolic pathways in sugarcane under low nitrogen stress. Environ. Exp. Bot. 2019, 163, 97–111. [Google Scholar] [CrossRef]
- Que, Y.; Su, Y.; Guo, J.; Wu, Q.; Xu, L. A global view of transcriptome dynamics during Sporisorium scitamineum challenge in sugarcane by RNA-seq. PLoS ONE 2014, 9, e106476. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Hou, M.; Liu, Y.; Pei, Y.; Ye, M.; Zhou, Y.; Huang, C.; Zhao, Y.; Ma, H. Genome-wide identification, characterization and expression analysis of the non-specific lipid transfer proteins in potato. BMC Genom. 2019, 20, 375. [Google Scholar] [CrossRef]
- Brooks, M.J.; Rajasimha, H.K.; Roger, J.E.; Swaroop, A. Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl−/− retinal transcriptomes. Mol. Vis. 2011, 17, 3034–3054. [Google Scholar]
- Galili, T.; O’Callaghan, A.; Sidi, J.; Sievert, C. Heatmaply: An R package for creating interactive cluster heatmaps for online publishing. Bioinformatics 2018, 34, 1600–1602. [Google Scholar] [CrossRef]
- Wang, Z.; Ren, H.; Xu, F.; Lu, G.; Cheng, W.; Que, Y.; Xu, L. Genome-wide characterization of lectin receptor kinases in Saccharum spontaneum L. and their responses to Stagonospora tainanensis infection. Plants 2021, 10, 322. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta CT) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, C.; Zhang, M.; Yang, C.; Bao, Y.; Wang, D.; Chen, Y.; Chen, Q. Genome-wide analysis and expression profiling of the Phospholipase D gene family in Solanum tuberosum. Biology 2021, 10, 741. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.; Xu, L.; Wang, Z.; Peng, Q.; Yang, Y.; Chen, Y.; Que, Y. Comparative proteomics reveals that central metabolism changes are associated with resistance against Sporisorium scitamineum in sugarcane. BMC Genom. 2016, 17, 800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karve, R.; Liu, W.; Willet, S.G.; Torii, K.U.; Shpak, E.D. The presence of multiple introns is essential for ERECTA expression in Arabidopsis. RNA 2011, 17, 1907–1921. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Washida, H.; Onodera, Y.; Harada, K.; Takaiwa, F. Quantitative nature of the Prolamin-box, ACGT and AACA motifs in a rice glutelin gene promoter: Minimal cis-element requirements for endosperm-specific gene expression. Plant J. 2000, 23, 415–421. [Google Scholar] [CrossRef]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchishinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Ora, H.; Hardtke, C.S. CLAVATA 1-type receptors in plant development. J. Exp. Bot. 2016, 16, 4827–4833. [Google Scholar]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Canirrd, A.; Jones, J.D.G.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Owens, D.K.; Alerding, A.B.; Crosby, K.C.; Bandara, A.B.; Westwood, J.H.; Winkel, B.S.J. Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol. 2008, 147, 1046–1061. [Google Scholar] [CrossRef] [Green Version]
- Gómezgómez, L.; Boller, T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Hu, H.; Xiong, L.; Yang, Y. Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection. Planta 2005, 222, 107–117. [Google Scholar] [CrossRef]
- Gou, X.; Yin, H.; He, K.; Du, J.; Yi, J.; Xu, S.; Lin, H.; Clouse, S.; Li, J. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 2012, 8, e1002452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrecht, C.; Russinova, E.; Kemmerling, B.; Kwaaitaal, M.; de Vries, S.C. Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiol. 2008, 148, 611–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).