FLII and MLL1 Cooperatively Regulate Aryl Hydrocarbon Receptor-Mediated Transcription in ARPE-19 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. RNA Interference
2.3. Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
2.4. Protein-Binding Assay
2.5. Chromatin Immunoprecipitation (ChIP) Assay
2.6. Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE)-qPCR
2.7. Statistical Analysis
3. Results
3.1. FLII and MLL1 Are Involved in AHR-Mediated Transcription in ARPE-19 Cells
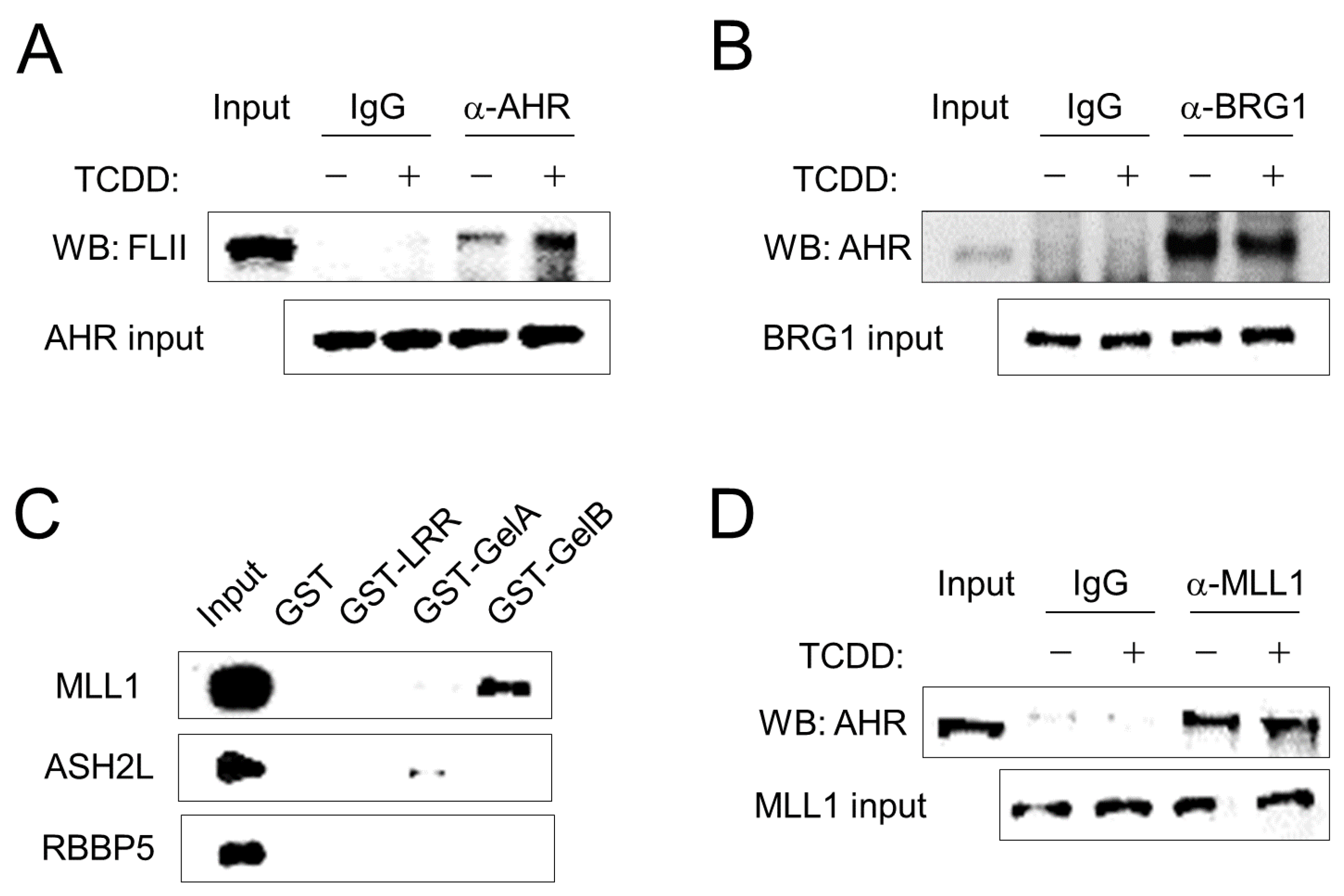
3.2. FLII Interacts with AHR and MLL1 in ARPE-19 Cells
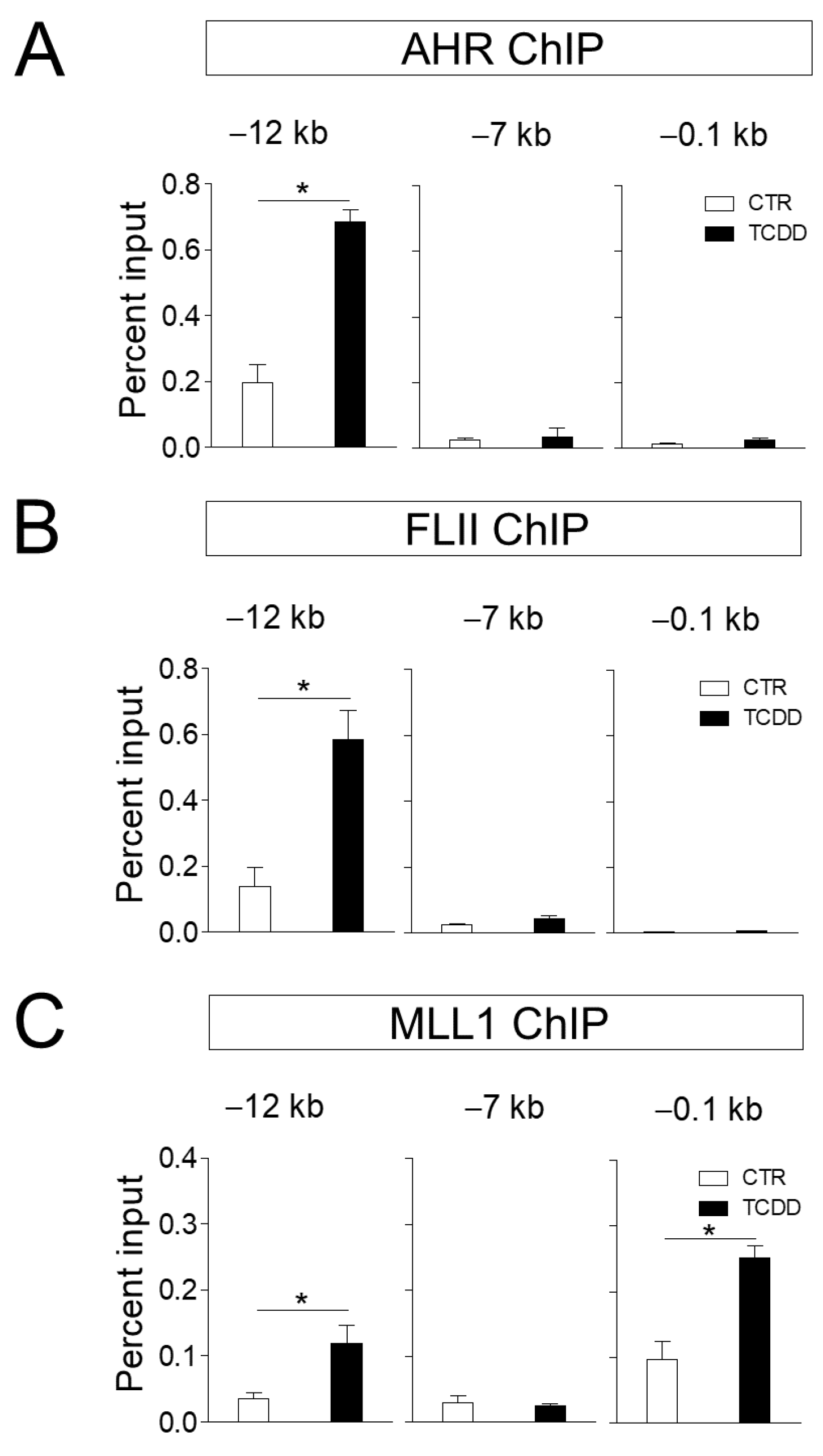
3.3. FLII and MLL1 Are Recruited to the CYP1A1 in ARPE-19 Cells
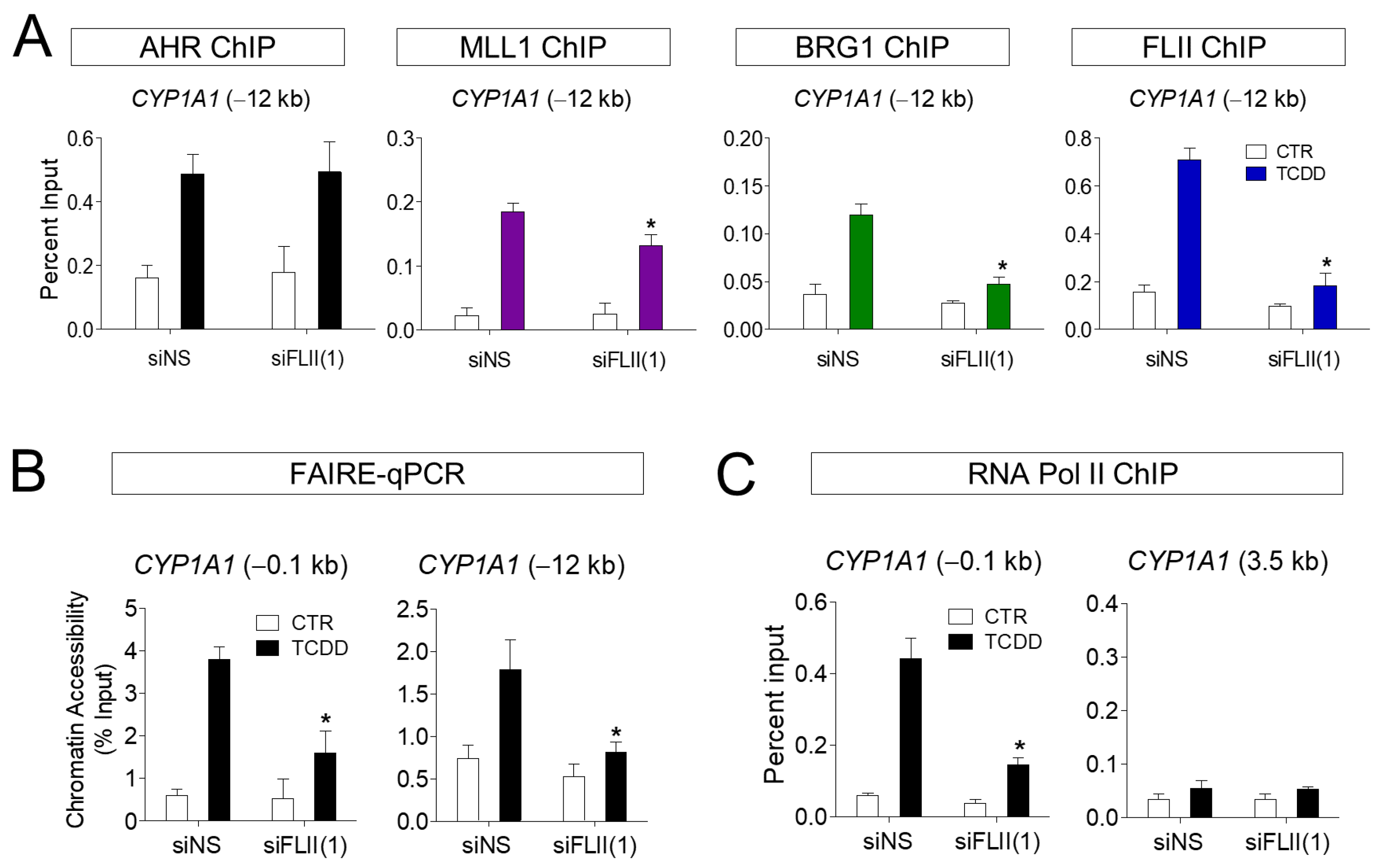
3.4. FLII Is Required for the Chromatin Structure at the CYP1A1 Locus in ARPE-19 Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Klein, R.; Lee, K.E.; Gangnon, R.E.; Klein, B.E. Incidence of visual impairment over a 20-year period: The Beaver Dam Eye Study. Ophthalmology 2013, 120, 1210–1219. [Google Scholar] [CrossRef] [Green Version]
- Sarks, S.; Cherepanoff, S.; Killingsworth, M.; Sarks, J. Relationship of Basal laminar deposit and membranous debris to the clinical presentation of early age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2007, 48, 968–977. [Google Scholar] [CrossRef]
- Curcio, C.A.; Millican, C.L. Basal linear deposit and large drusen are specific for early age-related maculopathy. Arch. Ophthalmol. 1999, 117, 329–339. [Google Scholar] [CrossRef]
- Anderson, D.H.; Radeke, M.J.; Gallo, N.B.; Chapin, E.A.; Johnson, P.T.; Curletti, C.R.; Hancox, L.S.; Hu, J.; Ebright, J.N.; Malek, G.; et al. The pivotal role of the complement system in aging and age-related macular degeneration: Hypothesis re-visited. Prog. Retin. Eye Res. 2010, 29, 95–112. [Google Scholar] [CrossRef] [Green Version]
- Crabb, J.W.; Miyagi, M.; Gu, X.; Shadrach, K.; West, K.A.; Sakaguchi, H.; Kamei, M.; Hasan, A.; Yan, L.; Rayborn, M.E.; et al. Drusen proteome analysis: An approach to the etiology of age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2002, 99, 14682–14687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Clark, M.E.; Crossman, D.K.; Kojima, K.; Messinger, J.D.; Mobley, J.A.; Curcio, C.A. Abundant lipid and protein components of drusen. PLoS ONE 2010, 5, e10329. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.H. Polycyclic aromatic hydrocarbons in the diet. Mutat. Res. 1999, 443, 139–147. [Google Scholar] [CrossRef]
- Kim, S.Y.; Yang, H.J.; Chang, Y.S.; Kim, J.W.; Brooks, M.; Chew, E.Y.; Wong, W.T.; Fariss, R.N.; Rachel, R.A.; Cogliati, T.; et al. Deletion of aryl hydrocarbon receptor AHR in mice leads to subretinal accumulation of microglia and RPE atrophy. Investig. Ophthalmol. Vis. Sci. 2014, 55, 6031–6040. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Herrmann, R.; Bednar, A.; Saloupis, P.; Dwyer, M.A.; Yang, P.; Qi, X.; Thomas, R.S.; Jaffe, G.J.; Boulton, M.E.; et al. Aryl hydrocarbon receptor deficiency causes dysregulated cellular matrix metabolism and age-related macular degeneration-like pathology. Proc. Natl. Acad. Sci. USA 2013, 110, E4069–E4078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Li, S.; Huang, L.; Yang, Y.; Zhang, L.; Yang, M.; Liu, W.; Ramasamy, K.; Jiang, Z.; Sundaresan, P.; et al. A splicing mutation in aryl hydrocarbon receptor associated with retinitis pigmentosa. Hum. Mol. Genet. 2018, 27, 2587. [Google Scholar] [CrossRef]
- Tsai, C.H.; Lee, Y.; Li, C.H.; Cheng, Y.W.; Kang, J.J. Down-regulation of aryl hydrocarbon receptor intensifies carcinogen-induced retinal lesion via SOCS3-STAT3 signaling. Cell Biol. Toxicol. 2020, 36, 223–242. [Google Scholar] [CrossRef]
- Esfandiary, H.; Chakravarthy, U.; Patterson, C.; Young, I.; Hughes, A.E. Association study of detoxification genes in age related macular degeneration. Br. J. Ophthalmol. 2005, 89, 470–474. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, M.A.; Davis, S.S.; Rosko, A.; Nguyen, S.M.; Mitchell, K.P.; Mateen, S.; Neves, J.; Garcia, T.Y.; Mooney, S.; Perdew, G.H.; et al. A novel AhR ligand, 2AI, protects the retina from environmental stress. Sci. Rep. 2016, 6, 29025. [Google Scholar] [CrossRef]
- Zapadka, T.E.; Lindstrom, S.I.; Batoki, J.C.; Lee, C.A.; Taylor, B.E.; Howell, S.J.; Taylor, P.R. Aryl Hydrocarbon Receptor Agonist VAF347 Impedes Retinal Pathogenesis in Diabetic Mice. Int. J. Mol. Sci. 2021, 22, 4335. [Google Scholar] [CrossRef]
- Wang, S.; Hankinson, O. Functional involvement of the Brahma/SWI2-related gene 1 protein in cytochrome P4501A1 transcription mediated by the aryl hydrocarbon receptor complex. J. Biol. Chem. 2002, 277, 11821–11827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, H.L.; Jeong, K.W. Regulation of aryl hydrocarbon receptor-mediated transcription in human retinal pigmented epithelial cells. Biochem. Biophys. Res. Commun. 2016, 472, 366–372. [Google Scholar] [CrossRef]
- Jeong, K.W.; Lee, Y.H.; Stallcup, M.R. Recruitment of the SWI/SNF chromatin remodeling complex to steroid hormone-regulated promoters by nuclear receptor coactivator flightless-I. J. Biol. Chem. 2009, 284, 29298–29309. [Google Scholar] [CrossRef] [Green Version]
- Chung, Y.S.; Jin, H.L.; Jeong, K.W. Cell-specific expression of ENACalpha gene by FOXA1 in the glucocorticoid receptor pathway. Int. J. Immunopathol. Pharmacol. 2020, 34, 2058738420946192. [Google Scholar] [CrossRef]
- Yang, L.; Jin, M.; Park, S.J.; Seo, S.Y.; Jeong, K.W. SETD1A Promotes Proliferation of Castration-Resistant Prostate Cancer Cells via FOXM1 Transcription. Cancers 2020, 12, 1736. [Google Scholar] [CrossRef]
- Yang, L.; Jin, M.; Jung, N.; Jeong, K.W. MLL2 regulates glucocorticoid receptor-mediated transcription of ENACalpha in human retinal pigment epithelial cells. Biochem. Biophys. Res. Commun. 2020, 525, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.L.; Choi, Y.; Jeong, K.W. Crosstalk between Aryl Hydrocarbon Receptor and Glucocorticoid Receptor in Human Retinal Pigment Epithelial Cells. Int. J. Endocrinol. 2017, 2017, 5679517. [Google Scholar] [CrossRef]
- Endler, A.; Chen, L.; Shibasaki, F. Coactivator recruitment of AhR/ARNT1. Int. J. Mol. Sci. 2014, 15, 11100–11110. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Jin, M.; Jeong, K.W. Histone H3K4 Methyltransferases as Targets for Drug-Resistant Cancers. Biology 2021, 10, 581. [Google Scholar] [CrossRef]
- Choudhary, M.; Kazmin, D.; Hu, P.; Thomas, R.S.; McDonnell, D.P.; Malek, G. Aryl hydrocarbon receptor knock-out exacerbates choroidal neovascularization via multiple pathogenic pathways. J. Pathol. 2015, 235, 101–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, R.T.; Wang, F.; Hsu, E.L.; Hankinson, O. Roles of coactivator proteins in dioxin induction of CYP1A1 and CYP1B1 in human breast cancer cells. Toxicol. Sci. Off. J. Soc. Toxicol. 2009, 107, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, M.S.; Jeong, K.W. Role of Flightless-I (Drosophila) homolog in the transcription activation of type I collagen gene mediated by transforming growth factor beta. Biochem. Biophys. Res. Commun. 2014, 454, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Nayak, A.; Reck, A.; Morsczeck, C.; Muller, S. Flightless-I governs cell fate by recruiting the SUMO isopeptidase SENP3 to distinct HOX genes. Epigenetics Chromatin 2017, 10, 15. [Google Scholar] [CrossRef] [Green Version]

| Name | Assay Type | Forward | Reverse |
|---|---|---|---|
| 18S | RT-qPCR | GAGGATGAGGTGGAACGTGT | TCTTCAGTCGCTCCAGGTCT |
| CYP1A1 | RT-qPCR | TGAACCCCAGGGTACAGAGA | GGCCTCCATATAGGGCAGAT |
| CYP1B1 | RT-qPCR | AACGTACCGGCCACTATCAC | CCACGACCTGATCCAATTCT |
| AHRR | RT-qPCR | AGACTCCAGGACCCACAAAG | CATCCTCACTGTGCTTTCCC |
| CYP1A1 (−12 kb) | ChIP | AGTGGCTCACGCCAGTAATC | CGTGTTAGCCAGGATGGTCT |
| CYP1A1 (−7 kb) | ChIP | GTAGAGACGGGGTTTCACCA | GTGGCTCACGCCTATAATCC |
| CYP1A1 (−0.1 kb) | ChIP | CGTACAAGCCCGCCTATAAA | CTGGGATCACAAGGATCAGG |
| CYP1A1 (3.5 kb) | ChIP | CATGTCGGCCACGGAGTTTCTTC | ACAGTGCCAGGTGCGGGTTCTTTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, K.W. FLII and MLL1 Cooperatively Regulate Aryl Hydrocarbon Receptor-Mediated Transcription in ARPE-19 Cells. Curr. Issues Mol. Biol. 2021, 43, 1623-1631. https://doi.org/10.3390/cimb43030115
Jeong KW. FLII and MLL1 Cooperatively Regulate Aryl Hydrocarbon Receptor-Mediated Transcription in ARPE-19 Cells. Current Issues in Molecular Biology. 2021; 43(3):1623-1631. https://doi.org/10.3390/cimb43030115
Chicago/Turabian StyleJeong, Kwang Won. 2021. "FLII and MLL1 Cooperatively Regulate Aryl Hydrocarbon Receptor-Mediated Transcription in ARPE-19 Cells" Current Issues in Molecular Biology 43, no. 3: 1623-1631. https://doi.org/10.3390/cimb43030115






