CgMFS1, a Major Facilitator Superfamily Transporter, Is Required for Sugar Transport, Oxidative Stress Resistance, and Pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
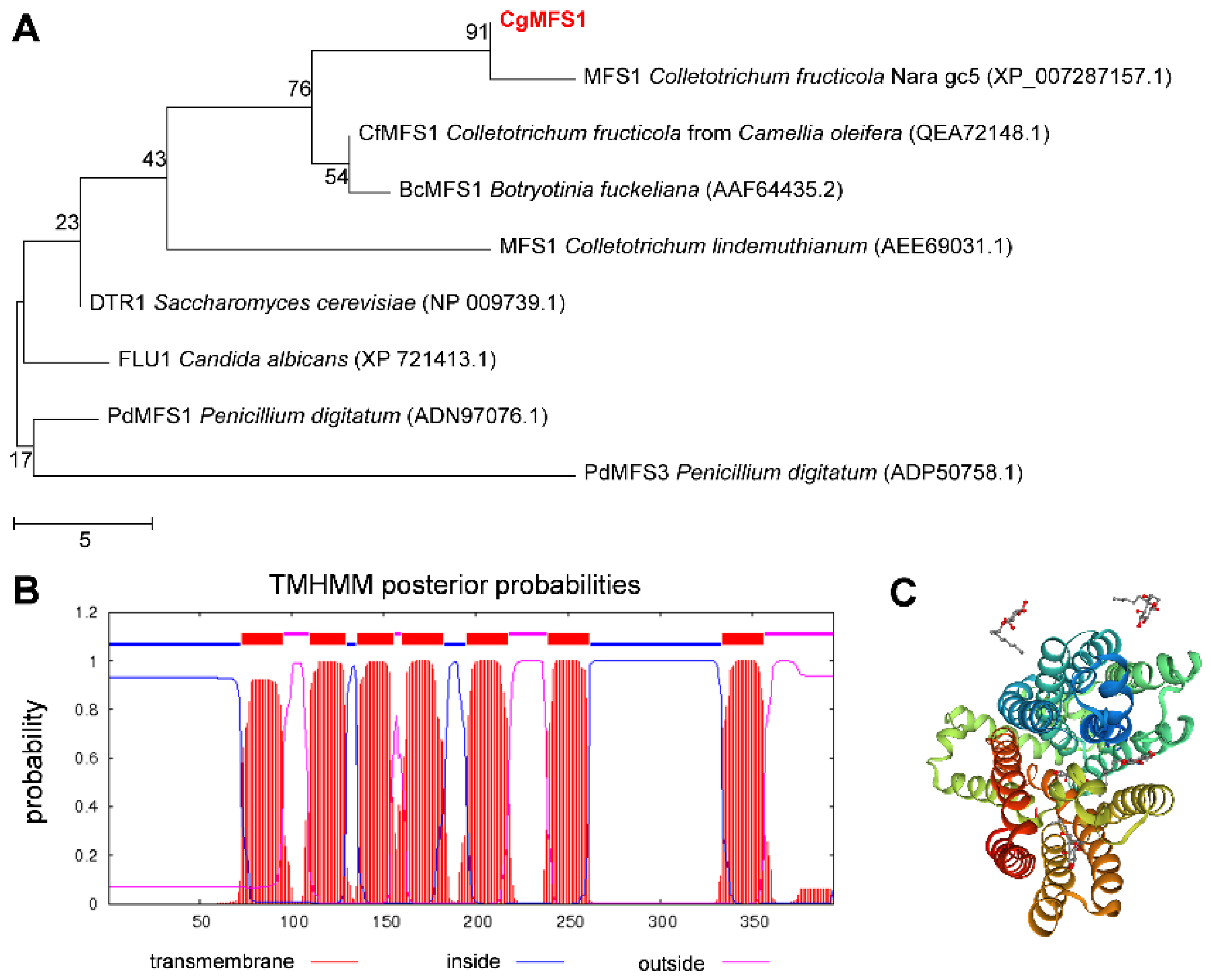
2.2. Bioinformatics Analysis
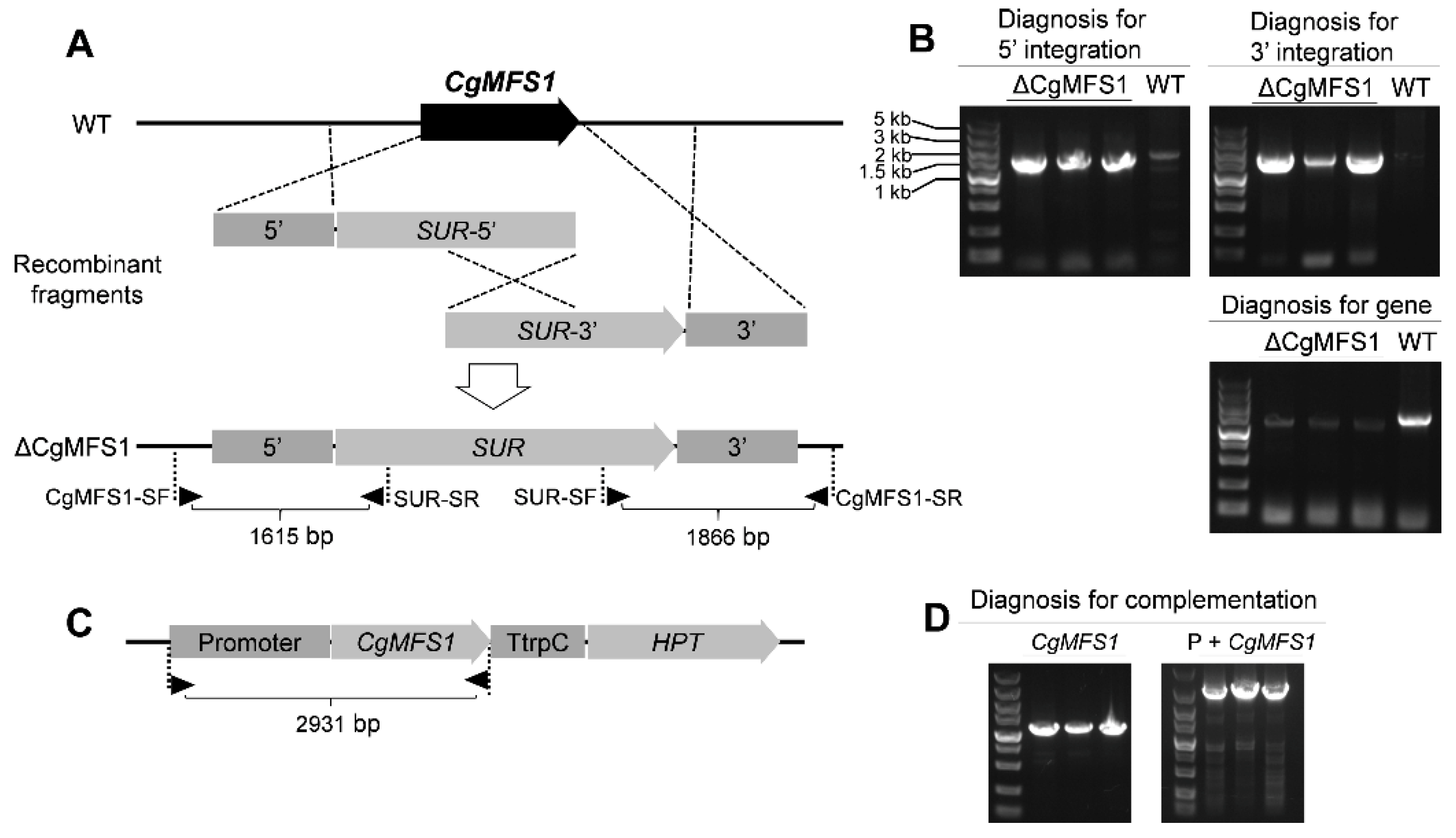
2.3. Construction of Knock-Out and Complementation Mutants
2.4. Colony Growth Assay
2.5. Stresses and Sensitivity Tests
2.6. Quantitative RT-PCR Analysis
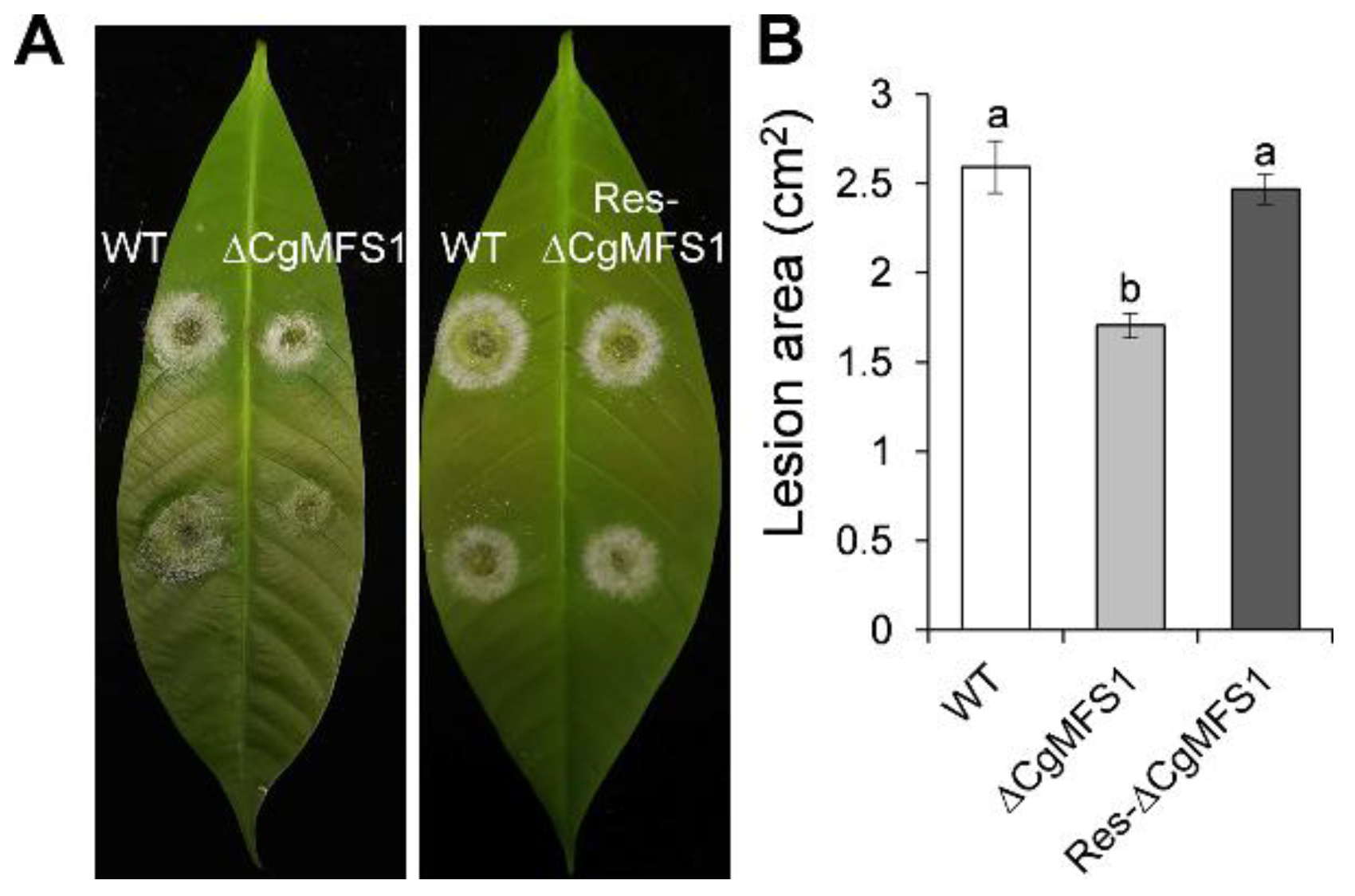
2.7. Pathogenicity Assay
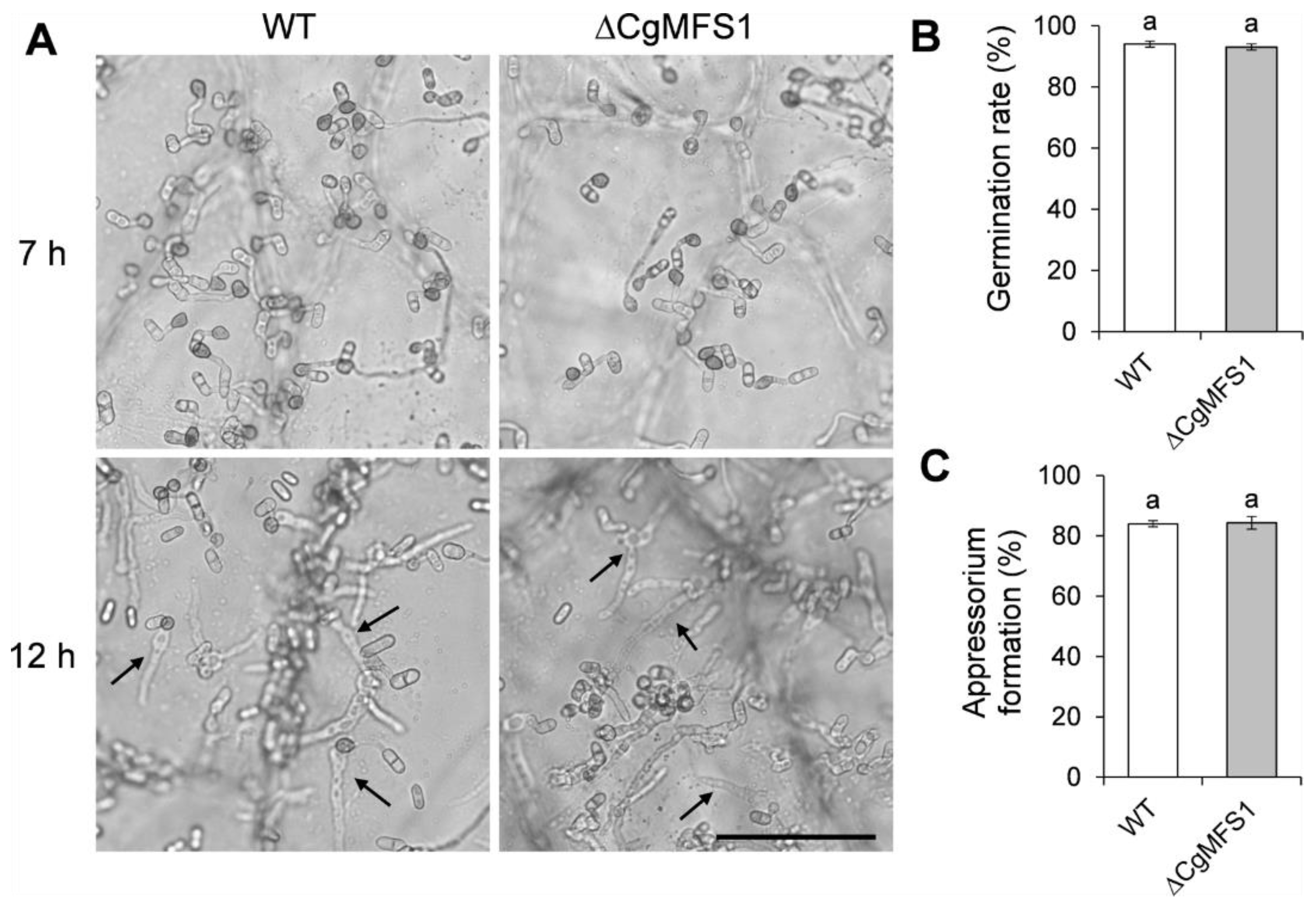
2.8. Appressorium Formation Assay
2.9. Statistical Analysis
3. Results
3.1. Identification of CgMFS1
3.2. Generation of Knock-Out and Complementation Mutants
3.3. CgMFS1 Is Required for Carbon Utilization
3.4. CgMFS1 Is Required for Tolerance to Oxidative and Fungicide Stresses
3.5. CgMFS1 Plays a Role in Fungal Pathogenicity
3.6. CgMFS1 Is Not Required for Conidial Germination and Appressorium Formation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connell, R.J.; Thon, M.R.; Hacquard, S.; Amyotte, S.G.; Kleemann, J.; Torres, M.F.; Damm, U.; Buiate, E.A.; Epstein, L.; Alkan, N. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 2012, 44, 1060–1065. [Google Scholar] [CrossRef] [PubMed]
- Prusky, D. Pathogen quiescence in postharvest diseases. Annu. Rev. Phytopathol. 1996, 34, 413–434. [Google Scholar] [CrossRef] [Green Version]
- Gan, P.; Ikeda, K.; Irieda, H.; Narusaka, M.; O’Connell, R.J.; Narusaka, Y.; Takano, Y.; Kubo, Y.; Shirasu, K. Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol. 2013, 197, 1236–1249. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Shlykov, M.A.; Castillo, R.; Sun, E.I.; Saier, M.H. The major facilitator superfamily (MFS) revisited. FEBS J. 2012, 279, 2022–2035. [Google Scholar] [CrossRef]
- Marger, M.D.; Saier, M.H., Jr. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem. Sci. 1993, 18, 13–20. [Google Scholar] [CrossRef]
- Yan, N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 2013, 38, 151–159. [Google Scholar] [CrossRef]
- Wang, S.C.; Davejan, P.; Hendargo, K.J.; Javadi-Razaz, I.; Chou, A.; Yee, D.C.; Ghazi, F.; Lam, K.; Conn, A.M.; Madrigal, A.; et al. Expansion of the Major Facilitator Superfamily (MFS) to include novel transporters as well as transmembrane-acting enzymes. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183277. [Google Scholar] [CrossRef]
- Calabrese, D.; Bille, J.; Sanglard, D. A novel multidrug efflux transporter gene of the major facilitator superfamily from Candida albicans (FLU1) conferring resistance to fluconazole. Microbiology 2000, 146, 2743–2754. [Google Scholar] [CrossRef] [Green Version]
- Cannon, R.D.; Lamping, E.; Holmes, A.R.; Niimi, K.; Baret, P.V.; Keniya, M.V.; Tanabe, K.; Niimi, M.; Goffeau, A.; Monk, B.C. Efflux-mediated antifungal drug resistance. Clin. Microbiol. Rev. 2009, 22, 291–321. [Google Scholar] [CrossRef] [Green Version]
- Felder, T.; Bogengruber, E.; Tenreiro, S.; Ellinger, A.; Sá-Correia, I.; Briza, P. Dtrlp, a multidrug resistance transporter of the major facilitator superfamily, plays an essential role in spore wall maturation in Saccharomyces cerevisiae. Eukaryot. Cell 2002, 1, 799–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.Y.; Sun, X.P.; Lin, L.Y.; Zhang, T.Y.; Ma, Z.H.; Li, H.Y. PdMfs1, a major facilitator superfamily transporter from Penicilliun digitatum, is partially involved in the imazalil-resistance and pathogenicity. Afr. J. Microbiol. Res. 2012, 6, 95–105. [Google Scholar]
- Wu, Z.; Wang, S.; Yuan, Y.; Zhang, T.; Liu, J.; Liu, D. A novel major facilitator superfamily transporter in Penicillium digitatum (PdMFS2) is required for prochloraz resistance, conidiation and full virulence. Biotechnol. Lett. 2016, 38, 1349–1357. [Google Scholar] [CrossRef]
- De Ramón-Carbonell, M.; López-Pérez, M.; González-Candelas, L.; Sánchez-Torres, P. PdMFS1 transporter contributes to Penicilliun digitatum fungicide resistance and fungal virulence during citrus fruit infection. J. Fungi 2019, 5, 100. [Google Scholar] [CrossRef] [Green Version]
- Pereira, M.F.; de Araújo Dos Santos, C.M.; de Araújo, E.F.; de Queiroz, M.V.; Bazzolli, D.M. Beginning to understand the role of sugar carriers in Colletotrichum lindemuthianum: The function of the gene mfs1. J. Microbiol. 2013, 51, 70–81. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, G.; Liu, J. A major facilitator superfamily transporter in Colletotrichum fructicola (CfMfs1) is required for sugar transport, appressorial turgor pressure, conidiation and pathogenicity. Forest Pathol. 2019, 49, e12558. [Google Scholar] [CrossRef]
- Hayashi, K.; Schoonbeek, H.J.; De Waard, M.A. Bcmfs1, a novel major facilitator superfamily transporter from Botrytis cinerea, provides tolerance towards the natural toxic compounds camptothecin and cercosporin and towards fungicides. Appl. Environ. Microbiol. 2002, 68, 4996–5004. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.H.; Tsai, H.C.; Yu, P.L.; Chung, K.R. A Major Facilitator superfamily transporter-mediated resistance to oxidative stress and fungicides requires Yap1, Skn7, and MAP kinases in the citrus fungal pathogen Alternaria alternata. PLoS ONE 2017, 12, e0169103. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; An, B.; Hou, X.; Guo, Y.; Luo, H.; He, C. Dicer-like proteins regulate the growth, conidiation, and pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis. Front. Microbiol. 2018, 8, 2621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Yuan, Y.; Huang, J.; Zhang, S.; Luo, S.; Wang, N.; Pu, D.; Zhao, N.; Tang, Q.; Hirata, K.; et al. Structural basis for blocking sugar uptake into the malaria parasite Plasmodium falciparum. Cell 2020, 183, 258–268.e12. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Xu, C.; Sun, P.; Wu, J.; Yan, C.; Hu, M.; Yan, N. Crystal structure of the human glucose transporter GLUT1. Nature 2014, 510, 121–125. [Google Scholar] [CrossRef]
- Wisedchaisri, G.; Park, M.S.; Iadanza, M.G.; Zheng, H.; Gonen, T. Proton-coupled sugar transport in the prototypical major facilitator superfamily protein XylE. Nat. Commun. 2014, 5, 4521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998, 62, 1–34. [Google Scholar] [CrossRef] [Green Version]
- Molina, L.; Kahmann, R. An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 2007, 19, 2293–2309. [Google Scholar] [CrossRef] [Green Version]
- Samalova, M.; Meyer, A.J.; Gurr, S.J.; Fricker, M.D. Robust anti-oxidant defences in the rice blast fungus Magnaporthe oryzae confer tolerance to the host oxidative burst. New Phytol. 2014, 201, 556–573. [Google Scholar] [CrossRef] [Green Version]
- Lushchak, V.I. Adaptive response to oxidative stress: Bacteria, fungi, plants and animals. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011, 153, 175–190. [Google Scholar] [CrossRef]
- Nakaune, R.; Hamamoto, H.; Imada, J.; Akutsu, K.; Hibi, T. A novel ABC transporter gene, PMR5, is involved in multidrug resistance in the phytopathogenic fungus Penicillium digitatum. Mol. Genet. Genomics. 2002, 267, 179–185. [Google Scholar] [CrossRef]
- Wojtaszek, P. Oxidative burst: An early plant response to pathogen infection. Biochem. J. 1997, 322, 681–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, N.; Wang, Q.; He, C.; An, B. CgMFS1, a Major Facilitator Superfamily Transporter, Is Required for Sugar Transport, Oxidative Stress Resistance, and Pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis. Curr. Issues Mol. Biol. 2021, 43, 1548-1557. https://doi.org/10.3390/cimb43030109
Liu N, Wang Q, He C, An B. CgMFS1, a Major Facilitator Superfamily Transporter, Is Required for Sugar Transport, Oxidative Stress Resistance, and Pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis. Current Issues in Molecular Biology. 2021; 43(3):1548-1557. https://doi.org/10.3390/cimb43030109
Chicago/Turabian StyleLiu, Na, Qiannan Wang, Chaozu He, and Bang An. 2021. "CgMFS1, a Major Facilitator Superfamily Transporter, Is Required for Sugar Transport, Oxidative Stress Resistance, and Pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis" Current Issues in Molecular Biology 43, no. 3: 1548-1557. https://doi.org/10.3390/cimb43030109
APA StyleLiu, N., Wang, Q., He, C., & An, B. (2021). CgMFS1, a Major Facilitator Superfamily Transporter, Is Required for Sugar Transport, Oxidative Stress Resistance, and Pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis. Current Issues in Molecular Biology, 43(3), 1548-1557. https://doi.org/10.3390/cimb43030109





